Abstract
Mobile genetic elements are major contributing factors to the generation of genetic diversity in prokaryotic organisms. For example, insertion sequence (IS) elements have been shown to specifically contribute to niche adaptation by promoting a variety of genetic rearrangements. The complete genome sequence of the cheese culture Lactobacillus helveticus DPC 4571 was determined and revealed significant conservation compared to three nondairy gut lactobacilli. Despite originating from significantly different environments, 65 to 75% of the genes were conserved between the commensal and dairy lactobacilli, which allowed key niche-specific gene sets to be described. However, the primary distinguishing feature was 213 IS elements in the DPC 4571 genome, 10 times more than for the other lactobacilli. Moreover, genome alignments revealed an unprecedented level of genome stability between these four Lactobacillus species, considering the number of IS elements in the L. helveticus genome. Comparative analysis also indicated that the IS elements were not the primary agents of niche adaptation for the L. helveticus genome. A clear bias toward the loss of genes reported to be important for gut colonization was observed for the cheese culture, but there was no clear evidence of IS-associated gene deletion and decay for the majority of genes lost. Furthermore, an extraordinary level of sequence diversity exists between copies of certain IS elements in the DPC 4571 genome, indicating they may represent an ancient component of the L. helveticus genome. These data suggest a special unobtrusive relationship between the DPC 4571 genome and its mobile DNA complement.
Lactobacillus species are employed in the production of a wide range of fermented milk, meat, and plant products and are also routinely isolated from the vagina and gastrointestinal (GI) tract. This broad range of environmental niches is reflected in the diversity of species belonging to the genus Lactobacillus. The genomes of a number of lactobacilli have been determined (2, 9, 13, 24, 27, 35, 46), and comparative analysis has provided considerable insight into the evolution of this diverse group of microbes. Reconstruction of the evolution of the group highlighted extensive loss of ancestral genes since divergence from a common Lactobacillales ancestor and subsequent lineage-specific gene loss (27). Many of the changes seem to be related to the transition to a nutritionally rich environment. The general trend toward metabolic simplification is reflected in changes in sugar metabolism genes, the loss of genes for the biosynthesis of cofactors and amino acids, and increased numbers of transporters and peptidases. However, direct comparison of species from different niches has been less informative due to the level of interspecies diversity at the genome level (4, 8). We report here the genome sequence of Lactobacillus helveticus DPC 4571, a Swiss cheese isolate that has been thoroughly investigated as a starter and adjunct culture in cheese manufacture and that demonstrates a number of highly desirable traits, including rapid autolysis, reduced bitterness, and increased flavor notes (19, 20, 23). The genome of this commercially valuable cheese culture is of additional interest due to its close relationship with a group of Lactobacillus strains that have gained prominence as health-promoting probiotic organisms. Probiotics are defined as living microorganisms that, upon ingestion in certain numbers, exert health benefits beyond inherent nutrition and have potential applications for conditions such as GI infections and certain bowel disorders (30), although they are successfully marketed to healthy consumers with more general health-promoting claims. It should be noted that although L. helveticus is not considered a probiotic culture, certain strains have been shown to exert beneficial effects through the proteolytic generation of antihypertensive peptides from milk during fermentation (45).
To date, seven insertion sequence (IS) elements have been described for L. helveticus in the IS element database (http://www-is.biotoul.fr/is.html), and these elements vary in copy number (2 to 21 copies/genome) depending on the strain (36). IS elements are short (1- to 2-kb), phenotypically cryptic segments of DNA with a simple genetic organization and are capable of inserting at multiple sites in a target molecule (26). Insertions of IS elements can cause gene inactivation and/or can have strong polar effects. Besides these local effects, IS elements are recognized by the recombination machinery of the cell and have been shown to participate in chromosomal rearrangements (26). Microbial-evolution experiments demonstrated that IS elements can contribute substantially to the generation of genetic diversity and help promote adaptation of microbial populations (37). Moreover, IS-mediated rearrangements have been postulated to be critical in the evolution of pathogenesis in Bordetella (31), Burkholderia (29), Francisella (33), Mycobacterium (41), Shigella (49), and Yersinia (10) species that carry high numbers of IS elements. Sequencing of the L. helveticus DPC 4571 genome determined a 2.08-Mb genome saturated with IS elements and confirmed the close genetic relationship between this dairy culture and lactobacilli that inhabit the GI tract. Interestingly, the overall gene order between the strains is highly conserved despite the extensive L. helveticus IS insertions. The relatedness between the strains also permitted the first detailed delineation of the gene differences between a dairy-evolved bacterium and a gut-colonizing probiotic bacterium.
MATERIALS AND METHODS
Genome sequencing of L. helveticus DPC 4571.
The complete genome sequence of DPC 4571 was determined by random shotgun sequencing of a small (∼1-kb) and a larger (2- to 2.5-kb) insert library to 7.7-fold redundancy by MWG-Biotech AG (Ebersberg, Germany). Directed sequencing of the larger inserts was performed where required. The sequence traces were assembled using the Staden sequence analysis package (40). The universal vectorette system (Sigma-Aldrich Corporation, St. Louis, MO) and combinatorial PCR were used for gap closure. The assembly was verified by pulsed-field gel electrophoresis analysis of the chromosome by single and double digests using ApaI, AscI, I-CeuI, NotI, SgrAI, SmaI, and SrfI restriction enzymes (New England Biolabs, Beverly, MA).
Bioinformatic analysis.
The complete genome sequence was processed with the automatic annotation software GAMOLA (1). The gene model was determined with GLIMMER, and sequence similarity analyses were performed with a gapped BLASTP algorithm (3) by using the nonredundant database provided by NCBI (ftp://ftp.ncbi.nih.gov/BLAST/db). The automated annotation was manually verified using Artemis (http://www.sanger.ac.uk) as a sequence-viewing and editing tool. Start positions were reviewed and altered based on protein sequence alignments and ribosomal binding sites. Functional analysis was aided by the ERGO bioinformatics suite (Integrated Genomics, Inc., Chicago, IL). The phylogenetic supertree was constructed using 47 ribosomal proteins from 19 completely sequenced species that were individually aligned using ClustalW, and protein trees were built using the PHYLIP package (17). The best supertree was found using the most similar supertree (dfit) and maximum quartet fit (qfit) analysis methods from the Clann package (14). Genome comparisons were performed using the Artemis Comparison Tool (ACT), also available from the Sanger Institute. The differences in the DPC 4571 and Lactobacillus acidophilus NCFM gene sets were compiled visually using ACT and from GAMOLA BLASTP outputs. The strain-specific gene sets were verified by FASTA searches of the DPC 4571 and NCFM sequence data using the Kodon software package (Applied Maths, Inc.). Nucleotide repeats were also identified and visualized using the Kodon software package. tRNAs were identified using TRNASCAN-SE with stringent parameter sets (25). Genome atlas views were generated with GenomeViz (18).
Nucleotide sequence accession number.
The L. helveticus DPC 4571 genome sequence has been deposited in GenBank under accession number CP000517.
RESULTS
General features.
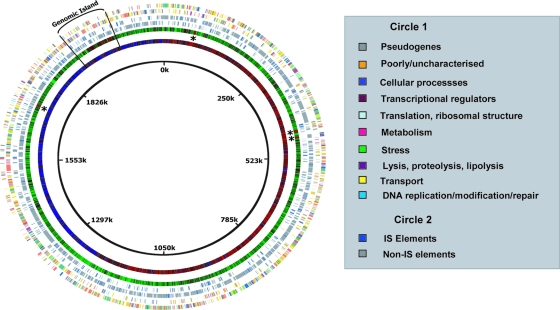
The L. helveticus DPC 4571 whole-genome-sequencing project produced a circular chromosomal sequence of 2,080,931 nucleotides with an average GC content of 37.73% (Fig. 1). The size and restriction enzyme profiles of the final sequence assembly were consistent with the pulsed-field gel electrophoresis patterns generated with a range of restriction enzymes (see Materials and Methods). This assembly verification was particularly important given the large number of repeat elements identified by in silico sequence analysis (see below). The genome included a total of 2,065 GLIMMER-predicted ORFs, but manual annotation resulted in 19% (388 ORFs) being reclassified as representing a large complement of 217 pseudogenes. Interestingly, the same percentage of ORFs were classified as pseudogenes in the genome of another species of lactic acid bacteria (LAB), Streptococcus thermophilus (6). The associated loss-of-function events suggested the mechanism of adaptation by S. thermophilus to the dairy niche. However, a significant proportion (36%) of pseudogenes in DPC 4571 had similarity to transposase enzyme genes, indicating they belonged to IS elements. Transposase inactivation is a common feature of IS elements and is recognized as a mechanism of transposition regulation (26). The 141 non-transposase-encoding pseudogenes of DPC 4571 were compiled (see Table S1 in the supplemental material). In common with S. thermophilus, two of the most highly decayed functional groups in the L. helveticus genome were predicted transport protein and energy metabolism genes, 19 and 11 pseudogenes, respectively. However, a significant number of pseudogenes encoded putative regulators (15 pseudogenes) and amino acid metabolism (9 pseudogenes), and a number of nucleotide metabolism genes (6 pseudogenes) also appeared to be inactivated (see Table S1 in the supplemental material).
FIG. 1.
Genome atlas of L. helveticus DPC 4571. The circles represent the following (from outer to inner). Circle 1 (two strands, positive and negative) shows predicted ORFs assigned and colored according to 10 functional groupings based on BLASTP (3) similarity searches. Circle 2 (two strands, positive and negative) shows IS elements (blue) distinguished from all other features (gray). Circle 3 shows percent GC content (red represents high GC content, and green represents low GC content). Circle 4 shows GC skew. The positions of the putative genomic island and rRNA operon (asterisks) are also indicated. The atlas was created using GenomeViz software (18).
Based on in silico analyses of biosynthetic capabilities, DPC 4571 can synthesize only 4 amino acids (aspartate, asparagine, cysteine, and serine), either de novo or as derivatives. In common with other LAB, DPC 4571 also appears to be incapable of synthesizing most of the vitamins and cofactors necessary for its growth. Overall, the in silico analyses predicted a dependency on external supplies of amino acids and cofactors for the dairy culture similar to that described for closely related GI tract isolates, L. acidophilus NCFM and Lactobacillus johnsonii NC533 (additional information on biosynthetic capabilities can be found in the supplemental material). Two of the most important attributes of L. helveticus DPC 4571 as a cheese culture are its high peptidolytic activity and ability to lyse rapidly in the cheese matrix. The genetic bases for these traits were investigated, and nine genes with potential lytic activity were identified (see Table S2 in the supplemental material). A similar complement of genes is present in closely related lactobacilli, indicating that the rapid lysis of the L. helveticus strain is not simply due to numbers and types of genes with lytic potential. Regarding proteolysis, the DPC 4571 genome was found to include 24 genes with significant homology (>34% identity at the amino acid level) to known peptidase genes (see Table S3 in the supplemental material). Many of the genes were described in L. helveticus previously, but the genome sequence included novel genes with homology to the PepE, PepQ, PepT, and PepD enzyme genes. Of particular interest for dairy industry cultures are the homologues of the proline-specific PepQ, due to the relatively high number of proline residues in casein, and the PepE endopeptidase homologue, because certain endopeptidases have roles in reducing bitter defects during cheese ripening (11, 39). Two additional genes, designated pcp (Lhv202) and pepM (Lhv655), were identified that potentially encode novel L. helveticus aminopeptidase enzymes. The DPC 4571 pepM gene encodes the cobalt-binding signature sequence of methionine aminopeptidases (PROSITE accession number PS00680) and could account for the recently reported uncharacterized methionine-specific activity of L. helveticus (12). The pcp gene product has significant homology to characterized pyrrolidone carboxyl peptidase (60% at the amino acid level) of Lactococcus lactis (15). The pcp gene product may account for the previously described low level of activity against Pyr-AMC (22) and the presence of pyroglutamic acid in cheeses ripened with L. helveticus (28).
The genome contains no complete prophage but does include a single group II intron of the Bh.Int group (44), although the recombinase associated with these group II introns (Lhv1086) appears to be inactivated by a frameshift. Analysis of the GC content (Fig. 1) identified six regions that deviated from the average GC content, with rRNA operons accounting for four of the GC content spikes. The other two spikes were associated with a gene cluster predicted to synthesize exopolysaccharide (Lhv1794 to Lhv1807) and a 100-kb region that displayed the characteristics of a genomic island (see below). In addition, a series of repeats composing a spacer-interspersed direct-repeat locus in L. acidophilus NCFM is conserved in DPC4 4571 but is flanked by the processing machinery of both spacer-interspersed direct-repeat and clustered regularly interspaced short palindromic repeat loci (Lhv1616 to Lhv1621). These features of bacterial genomes provide acquired resistance to bacteriophage (3a).
IS elements.
The annotation of the DPC 4571 genome sequence identified 223 genes and pseudogenes with similarity to a variety of characterized and predicted transposase enzymes. Examination of these sequences revealed that 213 of the gene loci could be classified as complete or partial IS elements (Table 1). The previously reported L. helveticus IS elements, IS1201 (43), ISL2 (51), ISLhe1 and ISLhe15 (7) were among the most numerous elements in the genome, with 32, 18, 19, and 23 partial or complete copies identified, respectively. There were also copies of IS elements previously reported in Lactobacillus delbrueckii (ISL5 and ISL7), L. johnsonii (ISLjo1 and ISLjo5), and Leuconostoc mesenteroides (IS1165) and the IS element that forms part of transposon Tn3692 of Lactobacillus crispatus (see Table S4 in the supplemental material). The majority of the remaining ORFs with transposase enzyme gene similarity were grouped and classified as new IS elements based on the following criteria. First, the ORF(s) encoded an enzyme with significant similarity (>40% identity at the amino acid level) to known elements or transposase enzymes; secondly, inverted repeats flanked the transposase enzyme; and thirdly, the proposed IS elements was present in more than one copy in the genome sequence. These criteria allowed the majority of repeat regions that did not constitute previously reported IS elements to be classified as members of 11 new types of IS elements. Most of the new elements appear to have close relatives in other lactobacilli (see Table S4 in the supplemental material). There were an additional 10 genes with transposase gene similarity that did not meet the three criteria. These genes were present only in single copies on the genome and had low similarity (less than 35% at the amino acid level) to characterized transposase enzyme genes. However, six did have repeat sequences flanking the predicted transposase and may represent new IS elements. The majority of DPC 4571 IS elements were located in noncoding sequences between predicted ORFs and appeared to be randomly distributed (Fig. 1). There were no obvious hot spots for insertion, although it was common for more than one element to be inserted at a single locus, and in certain cases, IS elements were integrated within the boundaries of other elements (see Fig. S1 in the supplemental material). The proximity of two identical IS elements at a number of loci also suggested the possibility that DPC 4571 may encode composite transposon elements (see Fig. S1 in the supplemental material) that have been described in L. crispatus (42) but had not been reported in L. helveticus previously.
TABLE 1.
IS elements in the L. helveticus DPC 4571 genome
| Name | Size (bp) | No. of ORFs | Inverted repeats (bp)a | Direct repeats (bp) | Family | No. of complete copies (pseudogenes)b | No. of partial copies | Transposase divergence (%)c | Total no. of copies |
|---|---|---|---|---|---|---|---|---|---|
| IS1201 | 1,387 | 1 | 20/24 | 8 | IS256 | 30 (8) | 2 | 9 | 32 |
| ISL2 | 858 | 1 | 17 | 2 | IS5 | 17 (17) | 1 | 1 | 18 |
| ISL5 | 1,590 | 1 | 48/64 | 0 | IS4 | 2 | 2 | ||
| ISL7 | 1,239 | 1 | 22/28 | NDd | IS30 | 1 | 1 | ||
| IS1165 | 1,553 | 1 | 20/39 | 0 | ISL3 | 1 | 1 | 2 | |
| ISLjo1-like | 1,195 | 1 | 24/32 | 0 | IS30 | 9 (7) | 46 | 9 | |
| ISLjo5 | 2,101 | 2 | 0 | 0 | IS200-IS605 | 2 | 2 | ||
| IS of Tn3692 | 1,482 | 1 | 24/29 | 4 | IS3 | 2 (2) | 1 | 3 | |
| ISLhe1 | 982 | 1 | 27/28 | 8 | IS982 | 18 (9) | 1 | 27 | 19 |
| ISLhe2 | 1,462 | 1 | 10/10e | 7 | ISL3 | 27 (6) | 2 | 15 | 29 |
| ISLhe4 | 1,392 | 1 | 15/20 | 0 | IS110 | 5 (1) | 5 | ||
| ISLhe6 | 1,305 | 1 | 24/29 | 4 | IS3 | 4 (1) | 4 | ||
| ISLhe61 | 1,465 | 1 | 21/24 | 4 | IS3 | 5 (4) | 5 | ||
| ISLhe12 | 1,533 | 1 | 7/10 | 0 | IS607 | 6 | 1 | 6 | |
| ISLhe9 | 2,607 | 2 | 13/17e | 0 | IS605 | 3 | 3 | ||
| ISLhe15 | 1,620 | 1 | 17/21 | 12 | ND | 21 (5) | 2 | 15 | 23 |
| ISLhe30 | 1,041 | 1 | 20/25 | 0 | IS30 | 2 (2) | 1 | 3 | |
| ISLhe65 | 1,426 | 1 | 9/11e | 0 | IS605 | 27 (15) | 1 | 10 | 28 |
| ISLhe66 | 2,280 | 13/4 | 27/27 | 5-10 | IS66 | 2 | 2 | ||
| ISLhe60 | 2,302 | 2 | 13/16 | 0 | IS605 | 5 (1) | 1 | 6 | |
| ISLhe63 | 1,898 | 1 | 21/22 | 0 | ND | 10 (1) | 1 | 3 | 11 |
| Total | 197 (79) | 16 | 213 |
Number of base pairs conserved between right and left end repeats/length of repeat.
Copies with interrupted transposase genes.
Percent divergence between nonpseudogene transposase proteins as determined by ClustalV alignments.
ND, not defined.
Repeats within 15 to 20 bp of the terminus.
It is interesting that the L. helveticus DPC 4571 genome includes significantly more IS elements than have been reported for the other six lactobacillus genomes published to date (see Table S5 in the supplemental material). The phenomenon of large differences in numbers of IS elements between closely related bacterial species has been confirmed by genome sequencing for a variety of microbes. Among pathogenic species, examples include Escherichia coli and its pathovar Shigella (49), Yersinia pestis and Yersinia pseudotuberculosis (10), and Bordetella bronchiseptica and Bordetella pertussis (31). In many of these cases, the IS elements have been associated with genome rearrangements that are proposed to increase the virulence of the IS-loaded microorganism for its specific host. Overall, L. helveticus DPC 4571 is second only to Shigella dysenteriae strain 197 in the frequency of IS elements occurring in the genome (as measured per megabase of genome). However, DPC 4571 includes a greater diversity of IS elements (at least 21 and possibly 27 different types of IS elements) among the eubacterial genomes sequenced to date. Only the sequenced archaeon Sulfolobus solfataricus P2 shows the same level of diversity in types of IS elements on a single genome (38).
Comparative analysis.
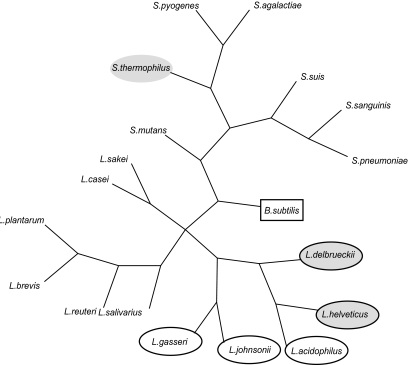
L. helveticus belongs to the largest of eight proposed phylogenetic Lactobacillus subgroups, the L. acidophilus-L. delbrueckii group (16). This subgroup consists largely of GI tract-associated species, and the 16S rRNA sequence of L. helveticus shares 98.4% identity with L. acidophilus, which suggested that the DPC 4571 cheese culture is particularly closely related to the probiotic commensal L. acidophilus strain NCFM. We confirmed the positioning of L. helveticus within the group by constructing a phylogenetic supertree with 47 ribosomal proteins (Fig. 2), a robust method of phylogenetic analysis (14). The relationship between L. helveticus and L. acidophilus is reflected in separate branching from the sequenced genomes of dairy L. delbrueckii and probiotic Lactobacillus gasseri/L. johnsonii species. More importantly, this relatedness is reflected in the fact that 75% of predicted DPC 4571 ORFs have orthologues in the L. acidophilus NCFM genome (orthology was defined as a BLASTP E value of <10−20). Considering the level of conservation between the DPC 4571 and NCFM genomes and the dramatically different environments that the strains inhabit, it was of interest to define the gene sets that differentiated the two closely related organisms (summarized in Table 2). Each genome included approximately 500 predicted genes that were not conserved in the other genome. The majority of these differentiating genes have an assigned function from in silico analyses (the complete lists of genes are compiled in Table S6 and Table S7 in the supplemental material). More than half the genes with predicted functions that were acquired by DPC 4571 or lost from NCFM were, unsurprisingly, for transposase enzymes. However, four restriction/modification (R/M) systems (Lhv27-Lhv28, Lhv260, Lhv1152 to -1158, and Lhv1978-Lhv1979) and two independent putative restriction endonucleases (Lhv1031 and Lhv1478-Lhv1479) are DPC 4571 specific, as well as a large section involved in fatty acid metabolism (Lhv1922 to Lhv1933). The R/M systems may account for the poor transformation efficiencies we observed for DPC 4571 compared to other lactobacilli (data not shown). The presence of additional fatty acid biosynthesis genes in dairy LAB cultures that are absent from L. acidophilus and L. gasseri has been noted previously (27, 46). In addition, a number of DPC 4571-specific amino acid metabolism genes were also identified (see Table S6 in the supplemental material). These DPC 4571-specific genes tend to be clustered within two large 100-kb sections and a number of 15- to 30-kb sections. This clustering is indicative of lateral gene transfer events, and GC content distribution analysis of the genome highlighted a difference at one of the 100-kb regions (Fig. 1). The region is characterized by a GC content of 42% (5% higher than the rest of the genome), and it is flanked by IS elements and unique 12-bp direct-repeat (TCATCTACTTTC) sequences. In addition, the region is not conserved in L. acidophilus or L. johnsonii. These data are consistent with recently acquired chromosomal regions or genomic islands (5) that have been described in Lactobacillus plantarum (24) but not in lactobacilli of the acidophilus complex. The putative genomic island (GEI) includes the lipid biosynthesis genes described above, in addition to predicted restriction endonuclease and amino acid metabolism genes (cysteine synthase and serine acetyltransferase).
FIG. 2.
Phylogenetic supertree of sequenced lactobacilli and streptococci. Members of the L. acidophilus-L. delbrueckii group are circled, dairy cultures are shaded, and the Bacillus subtilis outgroup is boxed. The supertree was calculated from 32 individual ribosomal protein trees. All branches are supported at >75% bootstrap values.
TABLE 2.
General features of L. helveticus DPC 4571 and L. acidophilus NCFM genomes
| Property | Value
|
|
|---|---|---|
| DPC 4571 | NCFM | |
| Size (nt) | 2,080,931 | 1,993,564 |
| Total no. of ORFs | 2,065 | 1,864 |
| No. of pseudogenes | 217 | 0 |
| No. of rRNAs/tRNAs | 4/61 | 4/61 |
| No. of IS elements | 213 | 17 |
| No. of PTS systems | 9 | 20 |
| No. of cell wall proteins | 9 | 22 |
| No. of mucus binding proteins | 0 | 7 |
| No. of glucosidases | 3 | 13 |
| No. of R/M systems | 5 | 2 |
| No. of peptidases | 24 | 23 |
| No. of lysis genes | 9 | 9 |
Since diverging from its GI tract relative, DPC 4571 has lost a range of genes that were highlighted by Altermann et al. (2) and Pridmore et al. (35) as encoding features likely to contribute to the ability of probiotic lactobacilli to colonize and interact with the intestinal mucosa and microbiota (Table 2). Fewer putative cell wall-anchoring proteins have been detected in the genome of L. helveticus DPC 4571 (see Table S8 in the supplemental material) than in that of L. acidophilus NCFM. Nine proteins with the motif LPXTG were found (22 have been described for L. acidophilus), and none of them has a predicted gram-positive anchoring domain. Regarding ORFs with the LPQTXE motif, only two of the four in NCFM were detected (with the motif LPQTGE). Both have low homology to much larger surface proteins, and one appears to be inactivated due to a frameshift. A further six surface proteins from L. acidophilus NCFM are lacking in L. helveticus DPC 4571 (see Table S7 in the supplemental material), including the LPXTG motif-containing proteins Lba1633, Lba1634, and SlpA (Lba169), which has been described as being involved in adherence (6a), although it was noted that another protein involved in adhesion, fibronectin binding protein FbpA (Lba1148), is highly conserved. The predicted mucus binding proteins (Lba1020, Lba1377, Lba1392, Lba1460, Lba1609, Lba1652, and Lba1709), some of which encode anchoring motifs, are all missing from DPC 4571 (see Table S7 in the supplemental material). In fact, no complete mucus binding proteins were detected in the L. helveticus genome. Another feature of the probiotic L. acidophilus NCFM strain is the ability to utilize a wide range of sugars as energy sources. NCFM has 20 phosphoenol pyruvate-dependent phosphotransferase (PEP-PTS) systems by which the sugars are transported into the cytoplasm and phosphorylated. DPC 4571 demonstrates the classic limited L. helveticus fermentation profile (see Table S9 in the supplemental material), which is reflected in the fact that only nine individual PEP-PTS systems were identified in the genome. Moreover, the transporters for the complex dietary carbohydrates raffinose and fructooligosaccharides and most of the predicted glucosidase enzymes (10 of 13 genes) in the NCFM genome are missing or inactivated (see Table S7 in the supplemental material). Other deletions that were of interest were the three NCFM potential autonomous units. Neither NCFM nor DPC 4571 has complete prophage sequences (47), but NCFM does encode three potential autonomous units, designated pauLAI, -II. and -III (2). The core seven ORFs of pauLAII and pauLAIII and adjacent R/M system genes are all missing from DPC 4571 (see Table S7 in the supplemental material). The core seven ORFs for pauLAI and surrounding ORFs, including the prophage maintenance killer system and antidote genes, are also missing from DPC 4571, although one type III R/M system is encoded at the DPC 4571 locus, corresponding to pauLAI. Finally, the contributions of IS element-mediated events to gene interruption and deletion were investigated by examining the disruption of local gene synteny between the NCFM and DPC 4571 genomes by using ACT. Ten interrupted genes and deletions at 31 loci were predicted (see Tables S6 and S7 and text in the supplemental material). However, the IS-associated differences account for only a fraction of the differences observed between the DPC 4571 and NCFM genomes despite their abundance in the L. helveticus strain.
Genome stability.
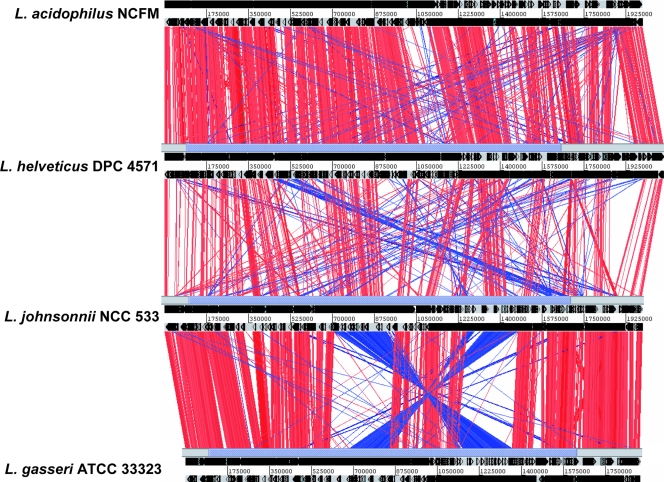
IS elements are recognized by the recombination machinery of the cell and have been shown to participate in chromosomal rearrangements (26). Linear genomic comparisons of the IS-loaded genomes of Bordetella (31), Burkholderia (29), Francisella (33), Mycobacterium (41), Shigella (49) and Yersinia (10) species revealed multiple IS-associated rearrangements (for examples, see Fig. S2 in the supplemental material). In many of these cases, the IS elements have been associated with genome rearrangements that are proposed to increase the virulence of the IS-loaded microorganisms for their specific hosts. In contrast, whole-genome alignments of DPC 4571 and other L. acidophilus-L. delbrueckii group species confirmed that DPC 4571 maintained the whole-genome order previously reported for the L. acidophilus group (4) and revealed no evidence of chromosomal rearrangements between L. acidophilus NCFM and DPC 4571 despite the abundant IS elements in the DPC 4571 genome (Fig. 3). This extraordinary level of whole-genome conservation between genomes with such different IS element contents suggested that the IS elements may have been recently acquired. However, the level of sequence divergence observed for the IS element copies in the genome indicates that at least some elements are not recently acquired (see below). Alternatively, a missing or malfunctioning recombination enzyme(s) encoded by the DPC 4571 bacterium may have limited intragenomic rearrangements without affecting chromosomal segregation and repair of damaged DNA. The L. helveticus genome encodes a complete set of enzymes with significant similarity to the recognized bacterial recombination pathways in gram-positive bacteria (50). The DPC 4571 genome encodes predicted AddAB (Lhv1273-Lhv1274) and RecFOR (Lhv4, Lhv1300, and Lhv402, respectively) pathway proteins, in addition to orthologues of RecG (Lhv1394), RecJ (Lhv1329), RecQ (Lhv1098), RecN (Lhv1410), RecU (Lhv1266), RecX (Lhv678), and RuvAB (Lhv429-Lhv430). Moreover, a large chromosomal rearrangement around the replication terminus in the L. johnsonii and L. gasseri genomes (Fig. 3) indicates that the capacity of these species for intragenomic rearrangements is not limited. Comparison of the DPC4571, L. johnsonii, and L. gasseri genomes revealed no obvious differences in the predicted complements of recombination enzymes. Interestingly, comparison of the L. helveticus and L. acidophilus genomes did reveal one striking difference. The product encoded by Lhv1134 has no homologue in NCFM and is a protein with similarity to MutS2 proteins that have recently been shown to specifically limit homologous recombination in Helicobacter (21, 34). However, it should be noted that Lhv1134 is conserved in L. johnsonii and L. gasseri. Irrespective of the mechanism promoting the stability of the IS-loaded DPC 4571 genome, the conservation of the whole-genome order between the L. helveticus and L. acidophilus strains suggests that one of the three acknowledged promoters of genetic variation in microbial populations, intragenomic recombination, did not facilitate or promote the evolution of these two microbial species from a common ancestor.
FIG. 3.
Alignment of L. helveticus DPC 4571, L. acidophilus NCFM, L. johnsonii NCC533, and L. delbrueckii ATCC 11842 genomes using ACT. The lines between the genomes represent DNA-DNA similarities (BLASTN matches) between the two sequences.
Wagner (48) studied the main representative of each major family of IS elements in 376 completely sequenced bacterial chromosomes and plasmids and found that 68% of ISs within individual genomes are completely identical. Periodic extinctions of particular elements in a bacterial lineage was proposed to explain the low DNA divergence. In contrast to the findings of Wagner (48), a significant level of transposase sequence divergence was observed for the majority of high-copy-number DPC 4571 IS elements (see Fig. S3 in the supplemental material). This divergence suggests that these elements represent more ancient components of the L. helveticus genome. However, the absence of information on sequence substitution rates and the potential for lateral transfer of IS elements make it difficult to establish how long individual IS elements have existed in this isolated genome. It has also been suggested that IS expansion occurred in Yersinia and Bordetella species during an evolutionary bottleneck in which the effective population size was reduced and that a lower level of intraspecies competition allowed clones with increased numbers of IS elements to survive (31, 32). The protected environment within the dairy plant could represent such an evolutionary bottleneck. This could certainly explain the expansion of ISLhe8 and ISL2 elements, since the copies of these elements in the genome are nearly identical (Table 1). However, based on our current understanding of IS element biology, the level of sequence divergence indicates that most of the DPC 4571 elements have been present in the genome significantly longer than the estimated 8,000 years that bacterial cultures have been employed in fermented-food production.
DISCUSSION
L. helveticus and L. acidophilus are easily distinguished as species from a classical microbiological prospective. L. helveticus is a specialist dairy culture with limited sugar fermentation capacity, whereas L. acidophilus strains are isolated from mucosal surfaces and demonstrate a broader fermentation profile. However, the 16S sequences of these bacteria, which are valued for two distinct capabilities, health promotion through GI tract colonization and cheese production, differ by just 1.6%. These strains represent the most closely related dairy and probiotic LAB cultures sequenced to date. Analysis of the L. helveticus DPC 4571 genome provided important insights into the evolution of dairy cultures and related probiotic bacteria. The DPC 4571 strain was selected because it makes good cheese, which was attributed to the characteristics of rapid lysis and high proteolytic activity. The NCFM and DPC 4571 genomes include basically the same set of peptidase and lysis genes, although there have been significant and possibly important changes in the peptidase protein sequences. In contrast, a definite bias toward the loss of functions associated with probiotic functionality in NCFM was observed. Half the PTS systems, cell wall-anchoring proteins, and all the mucus binding proteins predicted in NCFM were deleted or classed as pseudogenes in DPC 4571. The L. helveticus genome sequence confirms that the previously reported selective loss of membrane protein and sugar metabolism genes from the S. thermophilus dairy culture genome (6) also holds for Lactobacillus. A consistent pattern of specialization for the milk niche has now been documented for both species. From the perspective of probiotic-culture research, the selective loss of functionally related groups of genes from the DPC 4571 genome points to the importance of those genes in the probiotic genome and the presence of selective pressure to maintain them in the gut environment.
From the perspective of microbial genome architecture, the comparative genomics of the probiotic and dairy strains revealed an even more astonishing feature of conservation. The adaptation capacities of bacterial species are related to the genetic diversity from which the fittest variants are selected. The three mechanisms to generate this variability are recognized as point mutations, horizontal gene transfer, and intragenomic rearrangements. We report here that it is possible for two related genomes as distinct as those of NCFM and DPC 4571 to evolve without generating intragenomic rearrangements. Interestingly, adaptation to the milk niche was reported to promote stabilization of genome structure for the dairy S. thermophilus culture (6), revealing a further commonality between the dairy lactobacilli and streptococci. Moreover, the IS-loaded DPC 4571 genome demonstrates exceptional stability, since IS elements are generally viewed as facilitators of increased genomic rearrangement, conferring an advantage in variant generation. Overall, the DPC 4571 IS elements appear to be particularly unobtrusive considering their prominence in the genome.
Supplementary Material
Acknowledgments
We thank Berthold Fartmann for the sequence determination at MWG-Biotech and Marcus Claesson, Sinead Leahy, and Eric Altermann for assistance with bioinformatics analysis.
This work was funded in part by the Department of Agriculture and Food, Ireland, under the Food Institutional Research Measure, project reference numbers 01/R&D/TD/191 and 04/R&D/TD/311.
Footnotes
Published ahead of print on 9 November 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altermann, E., and T. R. Klaenhammer. 2003. GAMOLA: a new local solution for sequence annotation and analyzing draft and finished prokaryotic genomes. Omics 7161-169. [DOI] [PubMed] [Google Scholar]
- 2.Altermann, E., W. M. Russell, M. A. Azcarate-Peril, R. Barrangou, B. L. Buck, O. McAuliffe, N. Souther, A. Dobson, T. Duong, M. Callanan, S. Lick, A. Hamrick, R. Cano, and T. R. Klaenhammer. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. USA 1023906-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Barrangou, R., C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero, and P. Horvath. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 3151709-1712. [DOI] [PubMed] [Google Scholar]
- 4.Berger, B., R. D. Pridmore, C. Barretto, F. Delmas-Julien, K. Schreiber, F. Arigoni, and H. Brussow. 2007. Similarity and differences in the Lactobacillus acidophilus group identified by polyphasic analysis and comparative genomics. J. Bacteriol. 1891311-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binnewies, T. T., Y. Motro, P. F. Hallin, O. Lund, D. Dunn, T. La, D. J. Hampson, M. Bellgard, T. M. Wassenaar, and D. W. Ussery. 2006. Ten years of bacterial genome sequencing: comparative-genomics-based discoveries. Funct. Integr. Genomics 6165-185. [DOI] [PubMed] [Google Scholar]
- 6.Bolotin, A., B. Quinquis, P. Renault, A. Sorokin, S. D. Ehrlich, S. Kulakauskas, A. Lapidus, E. Goltsman, M. Mazur, G. D. Pusch, M. Fonstein, R. Overbeek, N. Kyprides, B. Purnelle, D. Prozzi, K. Ngui, D. Masuy, F. Hancy, S. Burteau, M. Boutry, J. Delcour, A. Goffeau, and P. Hols. 2004. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 221554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Buck, B. L., E. Alterman, T. Svinegerod, and T. R. Klaenhammer. 2005. Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 718344-8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callanan, M. J., T. P. Beresford, and R. P. Ross. 2005. Genetic diversity in the lactose operons of Lactobacillus helveticus strains and its relationship to the role of these strains as commercial starter cultures. Appl. Environ. Microbiol. 711655-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canchaya, C., M. J. Claesson, G. F. Fitzgerald, D. van Sinderen, and P. W. O'Toole. 2006. Diversity of the genus Lactobacillus revealed by comparative genomics of five species. Microbiology 1523185-3196. [DOI] [PubMed] [Google Scholar]
- 9.Chaillou, S., M. C. Champomier-Verges, M. Cornet, A. M. Crutz-Le Coq, A. M. Dudez, V. Martin, S. Beaufils, E. Darbon-Rongere, R. Bossy, V. Loux, and M. Zagorec. 2005. The complete genome sequence of the meat-borne lactic acid bacterium Lactobacillus sakei 23K. Nat. Biotechnol. 231527-1533. [DOI] [PubMed] [Google Scholar]
- 10.Chain, P. S., E. Carniel, F. W. Larimer, J. Lamerdin, P. O. Stoutland, W. M. Regala, A. M. Georgescu, L. M. Vergez, M. L. Land, V. L. Motin, R. R. Brubaker, J. Fowler, J. Hinnebusch, M. Marceau, C. Medigue, M. Simonet, V. Chenal-Francisque, B. Souza, D. Dacheux, J. M. Elliott, A. Derbise, L. J. Hauser, and E. Garcia. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 10113826-13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen, J. E., J. R. Broadbent, and J. L. Steele. 2003. Hydrolysis of casein-derived peptides αS1-casein(f1-9) and beta-casein(f193-209) by Lactobacillus helveticus peptidase deletion mutants indicates the presence of a previously undetected endopeptidase. Appl. Environ. Microbiol. 691283-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen, J. E., and J. L. Steele. 2003. Impaired growth rates in milk of Lactobacillus helveticus peptidase mutants can be overcome by use of amino acid supplements. J. Bacteriol. 1853297-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Claesson, M. J., Y. Li, S. Leahy, C. Canchaya, J. P. van Pijkeren, A. M. Cerdeno-Tarraga, J. Parkhill, S. Flynn, G. C. O'Sullivan, J. K. Collins, D. Higgins, F. Shanahan, G. F. Fitzgerald, D. van Sinderen, and P. W. O'Toole. 2006. Multireplicon genome architecture of Lactobacillus salivarius. Proc. Natl. Acad. Sci. USA 1036718-6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Creevey, C. J., and J. O. McInerney. 2005. Clann: investigating phylogenetic information through supertree analyses. Bioinformatics 21390-392. [DOI] [PubMed] [Google Scholar]
- 15.Curley, P., and D. van Sinderen. 2000. Identification and characterisation of a gene encoding aminoacylase activity from Lactococcus lactis MG1363. FEMS Microbiol. Lett. 183177-182. [DOI] [PubMed] [Google Scholar]
- 16.Dellaglio, F., and G. E. Felis. 2005. Taxonomy of Lactobacilli and Bifidobacteria., p. 48-73. In G. W. Tannock (ed.), Probiotics and prebiotics: scientific aspects. Caister Academic Press, Wymondham, Norfolk, United Kingdom.
- 17.Felsenstein, J. 1997. An alternating least squares approach to inferring phylogenies from pairwise distances. Syst. Biol. 46101-111. [DOI] [PubMed] [Google Scholar]
- 18.Ghai, R., T. Hain, and T. Chakraborty. 2004. GenomeViz: visualizing microbial genomes. BMC Bioinform. 5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hannon, J. A., M. G. Wilkinson, C. M. Delahunty, J. M. Wallace, P. A. Morrissey, and T. P. Beresford. 2003. Use of autolytic starter systems to accelerate the ripening of Cheddar cheese. Int. Dairy J. 13313-323. [Google Scholar]
- 20.Hickey, D. K., K. N. Kilcawley, T. P. Beresford, E. M. Sheehan, and M. G. Wilkinson. 2006. Starter strain related effects on the biochemical and sensory properties of Cheddar cheese. J. Dairy Res. 749-17. [DOI] [PubMed] [Google Scholar]
- 21.Kang, J., S. Huang, and M. J. Blaser. 2005. Structural and functional divergence of MutS2 from bacterial MutS1 and eukaryotic MSH4-MSH5 homologs. J. Bacteriol. 1873528-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kenny, O., R. J. FitzGerald, G. O'Cuinn, T. Beresford, and K. Jordan. 2003. Growth phase and growth medium effects on the peptidase activities of Lactobacillus helveticus. Int. Dairy J. 13509-516. [Google Scholar]
- 23.Kiernan, R. C., T. P. Beresford, G. O'Cuinn, and K. N. Jordan. 2000. Autolysis of lactobacilli during Cheddar cheese ripening. Irish J. Agric. Food Res. 3995-106. [Google Scholar]
- 24.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 1001990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. M. Goodstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J. H. Lee, I. Diaz-Muniz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, J. Broadbent, R. Hutkins, D. O'Sullivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 10315611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mucchetti, G., F. Locci, P. Massara, R. Vitale, and E. Neviani. 2002. Production of pyroglutamic acid by thermophilic lactic acid bacteria in hard-cooked mini-cheeses. J. Dairy Sci. 852489-2496. [DOI] [PubMed] [Google Scholar]
- 29.Nierman, W. C., D. DeShazer, H. S. Kim, H. Tettelin, K. E. Nelson, T. Feldblyum, R. L. Ulrich, C. M. Ronning, L. M. Brinkac, S. C. Daugherty, T. D. Davidsen, R. T. Deboy, G. Dimitrov, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, H. Khouri, J. F. Kolonay, R. Madupu, Y. Mohammoud, W. C. Nelson, D. Radune, C. M. Romero, S. Sarria, J. Selengut, C. Shamblin, S. A. Sullivan, O. White, Y. Yu, N. Zafar, L. Zhou, and C. M. Fraser. 2004. Structural flexibility in the Burkholderia mallei genome. Proc. Natl. Acad. Sci. USA 10114246-14251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Sullivan, G. C., P. Kelly, S. O'Halloran, C. Collins, J. K. Collins, C. Dunne, and F. Shanahan. 2005. Probiotics: an emerging therapy. Curr. Pharm. Des. 113-10. [DOI] [PubMed] [Google Scholar]
- 31.Parkhill, J., M. Sebaihia, A. Preston, L. D. Murphy, N. Thomson, D. E. Harris, M. T. Holden, C. M. Churcher, S. D. Bentley, K. L. Mungall, A. M. Cerdeno-Tarraga, L. Temple, K. James, B. Harris, M. A. Quail, M. Achtman, R. Atkin, S. Baker, D. Basham, N. Bason, I. Cherevach, T. Chillingworth, M. Collins, A. Cronin, P. Davis, J. Doggett, T. Feltwell, A. Goble, N. Hamlin, H. Hauser, S. Holroyd, K. Jagels, S. Leather, S. Moule, H. Norberczak, S. O'Neil, D. Ormond, C. Price, E. Rabbinowitsch, S. Rutter, M. Sanders, D. Saunders, K. Seeger, S. Sharp, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, L. Unwin, S. Whitehead, B. G. Barrell, and D. J. Maskell. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 3532-40. [DOI] [PubMed] [Google Scholar]
- 32.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413523-527. [DOI] [PubMed] [Google Scholar]
- 33.Petrosino, J. F., Q. Xiang, S. E. Karpathy, H. Jiang, S. Yerrapragada, Y. Liu, J. Gioia, L. Hemphill, A. Gonzalez, T. M. Raghavan, A. Uzman, G. E. Fox, S. Highlander, M. Reichard, R. J. Morton, K. D. Clinkenbeard, and G. M. Weinstock. 2006. Chromosome rearrangement and diversification of Francisella tularensis revealed by the type B (OSU18) genome sequence. J. Bacteriol. 1886977-6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinto, A. V., A. Mathieu, S. Marsin, X. Veaute, L. Ielpi, A. Labigne, and J. P. Radicella. 2005. Suppression of homologous and homeologous recombination by the bacterial MutS2 protein. Mol. Cell 17113-120. [DOI] [PubMed] [Google Scholar]
- 35.Pridmore, R. D., B. Berger, F. Desiere, D. Vilanova, C. Barretto, A. C. Pittet, M. C. Zwahlen, M. Rouvet, E. Altermann, R. Barrangou, B. Mollet, A. Mercenier, T. Klaenhammer, F. Arigoni, and M. A. Schell. 2004. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. USA 1012512-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ricci, G., and M. G. Fortina. 2006. Characterization of Lactobacillus helveticus strains isolated from cheeses by distribution studies of insertion sequences. Int. J. Food Microbiol. 112112-119. [DOI] [PubMed] [Google Scholar]
- 37.Schneider, D., and R. E. Lenski. 2004. Dynamics of insertion sequence elements during experimental evolution of bacteria. Res. Microbiol. 155319-327. [DOI] [PubMed] [Google Scholar]
- 38.She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, G. Allard, M. J. Awayez, C. C. Chan-Weiher, I. G. Clausen, B. A. Curtis, A. De Moors, G. Erauso, C. Fletcher, P. M. Gordon, I. Heikamp-de Jong, A. C. Jeffries, C. J. Kozera, N. Medina, X. Peng, H. P. Thi-Ngoc, P. Redder, M. E. Schenk, C. Theriault, N. Tolstrup, R. L. Charlebois, W. F. Doolittle, M. Duguet, T. Gaasterland, R. A. Garrett, M. A. Ragan, C. W. Sensen, and J. Van der Oost. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 987835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sridhar, V. R., J. E. Hughes, D. L. Welker, J. R. Broadbent, and J. L. Steele. 2005. Identification of endopeptidase genes from the genomic sequence of Lactobacillus helveticus CNRZ32 and the role of these genes in hydrolysis of model bitter peptides. Appl. Environ. Microbiol. 713025-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Staden, R. 1996. The Staden sequence analysis package. Mol. Biotechnol. 5233-241. [DOI] [PubMed] [Google Scholar]
- 41.Stinear, T. P., T. Seemann, S. Pidot, W. Frigui, G. Reysset, T. Garnier, G. Meurice, D. Simon, C. Bouchier, L. Ma, M. Tichit, J. L. Porter, J. Ryan, P. D. Johnson, J. K. Davies, G. A. Jenkin, P. L. Small, L. M. Jones, F. Tekaia, F. Laval, M. Daffe, J. Parkhill, and S. T. Cole. 2007. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 17192-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stroman, P., C. C. Muller, and K. I. Sorensen. 2003. Heat shock treatment increases the frequency of loss of an erythromycin resistance-encoding transposable element from the chromosome of Lactobacillus crispatus CHCC3692. Appl. Environ. Microbiol. 697173-7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tailliez, P., S. D. Ehrlich, and M. C. Chopin. 1994. Characterization of IS1201, an insertion sequence isolated from Lactobacillus helveticus. Gene 14575-79. [DOI] [PubMed] [Google Scholar]
- 44.Takami, H., C. G. Han, Y. Takaki, and E. Ohtsubo. 2001. Identification and distribution of new insertion sequences in the genome of alkaliphilic Bacillus halodurans C-125. J. Bacteriol. 1834345-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takano, T. 2002. Anti-hypertensive activity of fermented dairy products containing biogenic peptides. Antonie van Leeuwenhoek 82333-340. [PubMed] [Google Scholar]
- 46.van de Guchte, M., S. Penaud, C. Grimaldi, V. Barbe, K. Bryson, P. Nicolas, C. Robert, S. Oztas, S. Mangenot, A. Couloux, V. Loux, R. Dervyn, R. Bossy, A. Bolotin, J. M. Batto, T. Walunas, J. F. Gibrat, P. Bessieres, J. Weissenbach, S. D. Ehrlich, and E. Maguin. 2006. The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc. Natl. Acad. Sci. USA 1039274-9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ventura, M., C. Canchaya, V. Bernini, E. Altermann, R. Barrangou, S. McGrath, M. J. Claesson, Y. Li, S. Leahy, C. D. Walker, R. Zink, E. Neviani, J. Steele, J. Broadbent, T. R. Klaenhammer, G. F. Fitzgerald, P. W. O'Toole, and D. van Sinderen. 2006. Comparative genomics and transcriptional analysis of prophages identified in the genomes of Lactobacillus gasseri, Lactobacillus salivarius, and Lactobacillus casei. Appl. Environ. Microbiol. 723130-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wagner, A. 2006. Periodic extinctions of transposable elements in bacterial lineages: evidence from intragenomic variation in multiple genomes. Mol. Biol. Evol. 23723-733. [DOI] [PubMed] [Google Scholar]
- 49.Yang, F., J. Yang, X. Zhang, L. Chen, Y. Jiang, Y. Yan, X. Tang, J. Wang, Z. Xiong, J. Dong, Y. Xue, Y. Zhu, X. Xu, L. Sun, S. Chen, H. Nie, J. Peng, J. Xu, Y. Wang, Z. Yuan, Y. Wen, Z. Yao, Y. Shen, B. Qiang, Y. Hou, J. Yu, and Q. Jin. 2005. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 336445-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zuniga-Castillo, J., D. Romero, and J. M. Martinez-Salazar. 2004. The recombination genes addAB are not restricted to gram-positive bacteria: genetic analysis of the recombination initiation enzymes RecF and AddAB in Rhizobium etli. J. Bacteriol. 1867905-7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zwahlen, M. C., and B. Mollet. 1994. ISL2, a new mobile genetic element in Lactobacillus helveticus. Mol. Gen. Genet. 245334-338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.