Abstract
Proteins of two-component systems (TCS) have essential functions in the sensing of external and self-generated signals in bacteria and in the generation of appropriate output responses. Accordingly, in Myxococcus xanthus, TCS are important for normal motility and fruiting body formation and sporulation. Here we analyzed the M. xanthus genome for the presence and genetic organization of genes encoding TCS. Two hundred seventy-two TCS genes were identified, 251 of which are not part of che gene clusters. We report that the TCS genes are unusually organized, with 55% being orphan and 16% in complex gene clusters whereas only 29% display the standard paired gene organization. Hybrid histidine protein kinases and histidine protein kinases predicted to be localized to the cytoplasm are overrepresented among proteins encoded by orphan genes or in complex gene clusters. Similarly, response regulators without output domains are overrepresented among proteins encoded by orphan genes or in complex gene clusters. The most frequently occurring output domains in response regulators are involved in DNA binding and cyclic-di-GMP metabolism. Our analyses suggest that TCS encoded by orphan genes and complex gene clusters are functionally distinct from TCS encoded by paired genes and that the connectivity of the pathways made up of TCS encoded by orphan genes and complex gene clusters is different from that of pathways involving TCS encoded by paired genes. Experimentally, we observed that orphan TCS genes are overrepresented among genes that display altered transcription during fruiting body formation. The systematic analysis of the 25 orphan genes encoding histidine protein kinases that are transcriptionally up-regulated during development showed that 2 such genes are likely essential for viability and identified 7 histidine protein kinases, including 4 not previously characterized that have important function in fruiting body formation or spore germination.
A fundamental property of cells is their ability to sense and respond to external stimuli and self-generated signals. In the case of bacteria, this ability maximizes their chances of survival. Signal transduction proteins have essential functions in stimulus sensing, information processing, and the generation of output responses. Despite the multitude of cues that bacteria need to monitor, the signal transduction schemes involved center on a few types (11, 65): ligand-regulated one-component systems, which consist of single protein molecules containing both a sensing domain and an output domain; cyclic-di-GMP synthetases, phosphodiesterases, adenylate and guanylate cyclases, which act by modifying the level of secondary messenger molecules; methyl-accepting chemotaxis proteins that modulate the activity of chemosensory systems; and systems in which information transfer depends on covalent modification by phosphorylation/dephosphorylation by means of either Ser/Thr/Tyr protein kinases or histidine protein kinases (HPKs). HPKs are the more common type of protein kinase in bacteria and function as part of two-component systems (TCS) (11). Despite the fact that one-component systems are the numerically dominant sensing systems in bacteria (65), TCS are essential for bacteria in order to sense and respond to changes in the environment (58).
A typical TCS consists of an HPK and a cognate response regulator (RR) which are encoded in the same operon and architecturally organized in a simple linear 1:1 phosphotransfer signal transduction pathway (58). HPKs are multidomain proteins and generally contain a nonconserved sensor domain, which is responsible for detecting a particular stimulus, and a highly conserved kinase domain, which can be further subdivided into the HisKA domain, which is involved in dimerization and contains the conserved phosphorylatable histidine residue, and the HATPase_c domain. Typical RRs are either single-domain proteins consisting only of the conserved receiver domain, which contains the conserved aspartate residue that accepts the phosphoryl group from the histidine residue in the cognate HPK, or multidomain proteins consisting of a receiver domain and a variable output domain. HPKs autophosphorylate in a stimulus-dependent manner on the conserved histidine residue in the HisKA domain using ATP as a phosphodonor. Subsequently, the phosphoryl group is transferred to the conserved aspartate residue in the receiver domain of the cognate RR, resulting in activation of the RR and the generation of an appropriate output response. The phosphorelay systems constitute a structurally more complex type of TCS, with phosphotransfer occurring in three sequential steps (3). Generally, the architecture of these systems is similar to that of typical TCS and involves a linear phosphotransfer scheme. Specifically, the phosphoryl group is first transferred from the conserved histidine in the HPK to the conserved aspartate in a receiver domain, next to a conserved histidine in a phosphotransferase domain (Hpt), and finally to the conserved aspartate in a second RR. The domain organization of proteins in phosphorelays is highly modular. Thus, all four domains involved in phosphotransfer may reside in separate proteins (5), the kinase and first receiver may be present in the same protein (42), or the kinase, the first receiver, and the Hpt domain may be present in the same protein (64). It has been argued that the specific advantage of phosphorelays over typical TCS is that they allow the integration of several sensory inputs in one signal transduction pathway (9). In TCS, as well as in phosphorelays, the output response is determined by the phosphorylation status of the final RR and typically involves alterations in gene expression, in protein-protein interactions, or in enzyme activity. In addition to the linear 1:1 TCS and 1:1:1:1 phosphorelays, TCS and phosphorelays may also be organized as branched pathways. In the one-to-many pathways (54), one HPK has several cognate RRs, as exemplified by CheA (58) and ArcB (41) in Escherichia coli. This pathway connectivity would be suited for generating several responses to one input signal. In the many-to-one pathway (54), one RR has several cognate HPKs, as exemplified by Spo0F in Bacillus subtilis (5) and DosR in Mycobacterium tuberculosis (31). This pathway connectivity would be suited for integrating several input signals to generate one output response.
Here we have focused on an analysis of TCS in the gram-negative deltaproteobacterium Myxococcus xanthus. M. xanthus has a highly complex life cycle that involves social behavior (23). In the presence of nutrients, cells grow and divide, and if present on a solid surface, they form coordinately spreading colonies. In response to starvation, a developmental program is initiated that culminates in the formation of multicellular, spore-filled fruiting bodies. This developmental program involves two temporally and spatially coordinated morphogenetic events, aggregation of cells into fruiting bodies and sporulation of cells that have accumulated inside fruiting bodies. Cells that remain outside the fruiting bodies do not sporulate (43). Fruiting body formation is initiated by the RelA-dependent accumulation of the intracellular signaling molecule(s) guanosine penta- and tetraphosphate [(p)ppGpp] (16, 53). In addition, two intercellular signals, the A- and C-signals, are important for fruiting body formation. The A-signal consists of a mixture of amino acids and peptides (32) and is part of a system that monitors the density of starving cells (32, 33). The C-signal is a 17-kDa protein (27, 38) that induces and coordinates aggregation and sporulation (26, 30, 37). Fruiting body formation is accompanied by changes in gene expression, and several of the genes required for fruiting body formation are transcriptionally regulated in response to starvation (19, 29). The formation of spreading colonies, as well as fruiting body formation, depends on the functionality of the A- and S-gliding motility systems (18).
Forward- and reverse-genetics approaches have identified 35 TCS genes that are important for fruiting body formation (see Tables S3, S4, and S5 in the supplemental material for a list of these genes). These effects on fruiting body formation, in several cases, e.g., the RRs DigR (44) and AglZ (67), are likely to be indirect and caused by an effect on gliding motility. However, most other TCS mutants display only developmental defects, suggesting that the corresponding proteins are directly involved in development. More than half of the TCS genes required for fruiting body formation are encoded by orphan TCS genes or by TCS genes in complex gene clusters (See Materials and Methods for a definition of TCS genes), and the architecture of the corresponding signal transduction pathways remains poorly understood.
To begin to address the function of the remaining TCS genes in M. xanthus, we used bioinformatics in combination with functional analyses. We report that 71% of the TCS genes are organized in unusual manners as orphan genes or in complex gene clusters whereas the remaining 29% display the standard paired gene organization. Our bioinformatics analyses suggest that TCS proteins encoded by orphan genes and complex gene clusters are functionally distinct from TCS proteins encoded by paired genes and that the connectivity of the pathways made up of TCS proteins encoded by orphan genes and complex gene clusters is different from that of pathways involving TCS proteins encoded by paired genes. Experimentally, we found that orphan TCS genes are overrepresented among TCS genes that display altered transcription during fruiting body formation. The systematic analysis of 25 orphan HPKs that are transcriptionally up-regulated during development led to the identification of 2 HPKs that are likely essential for viability and 4 new HPKs that have important function in fruiting body formation or spore germination.
MATERIALS AND METHODS
Identification and phylogenetic analysis of TCS proteins.
TCS proteins can generally be divided into three groups: HPKs, which contain either a HisKA, HisKA_2, HisKA_3 (hereafter these three domains will be collectively referred to as the HisKA domain), or Hpt domain and the conserved HATPase_c domain; HPK-like proteins, which contain the HisKA domain and lack the HATPase_c domain or vice versa; and RRs, which contain the conserved receiver domain. HPK, HPK-like proteins, and receiver domain-containing proteins were identified using the TIGR Comprehensive Microbial Resource database (http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi) and HMMER (HMMER 2.3; http://selab.janelia.org/) searches on the M. xanthus proteome using the pfam HisKA, HisKA_2, HisKA_3, Hpt, and HATPase_c matrices for domains in HPKs and the Response_reg matrix for receiver domains. The E-value cutoffs were adopted from the SMART web service domain identification tool (51) and were 0.02 for the HisKA, Hpt, and Response_reg domains and 1e-5 for the HATPase_c domain. Sequences from proteins containing one or more of the homologous matrices of HisKA, Hpt, HATPase_c, and Response_reg were extracted based on boundaries identified using the HMMER results, SMART analysis results (36), and manual curation using the domain boundary definitions described by Grebe and Stock (15). To generate phylogenetic trees, the M. xanthus TCS domains were compared to the domain sets used by Grebe and Stock (15) by aligning all HPK domains and, separately, all receiver domains using the CLUSTALW 1.83 alignment software (gap open penalty = 50; gap extension penalty = 0.5; Gonnet protein matrix) (http://www.ebi.ac.uk/clustalw) (62). The resulting alignments were improved by manual curation and were then used to generate phylogenetic trees using the neighbor-joining method (49) (1,000 bootstrapping replications implemented).
To identify output domains in RR, HMMER searches were performed on all M. xanthus proteins bearing receiver domains using all matrices provided in the pfam database (http://pfam.sanger.ac.uk/) (10). Results from the searches were inspected, and based on the annotations provided for each pfam matrix, all hits from known output domains were noted. For those proteins for which the above search failed to identify an output domain, possible cryptic, conserved domains were searched for as follows: peptide sequences leading and trailing the receiver domains from these proteins were collected and subjected to a BLAST search of the NCBI nonredundant protein database. Results were manually analyzed, but no new domains were identified.
Transmembrane helices were predicted using the TMHMM 2.0c software package with the provided default model and options (http://www.cbs.dtu.dk/services/TMHMM) (56). If a protein was predicted to contain only a single transmembrane helix, the location of the transmembrane helix was inspected. If the helix was located within the first 20 amino acid residues and would thus overlap with the signal peptide, the HPK was categorized as cytoplasmic.
Classification of TCS proteins in M. xanthus based on genetic organization.
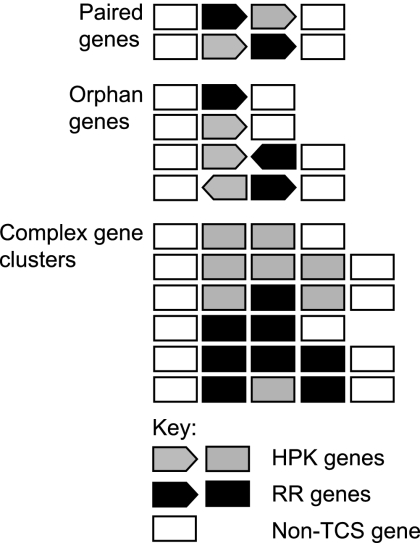
Based on the genetic organization of TCS genes, a set of criteria was developed to classify these genes into three groups (Fig. 1), as follows.
FIG. 1.
Classification scheme for two-component system genes. Schematic diagram of classification schemes for two-component system genes. The definition of paired and orphan TCS genes includes information about transcription direction as indicated by the arrow symbols. Complex TCS gene clusters include clusters containing two or more RR genes, clusters containing two or more HPK or HPK-like genes, and clusters with three or more TCS genes irrespective of transcription direction, as indicated by the box symbols. For complex gene clusters, only the most common gene organizations are shown (see Tables S3, S4, and S5 in the supplemental material for all gene organizations found in these clusters).
Complex gene clusters.
Complex gene clusters were defined as gene clusters containing two or more RR genes, independent of their transcriptional direction, gene clusters containing two or more HPK or HPK-like genes, independent of their transcriptional direction, and gene clusters containing three or more TCS genes, independent of their transcriptional direction.
Paired genes.
Paired genes were defined as two adjacent genes encoding an HPK or an HPK-like protein and an RR and transcribed in the same direction.
Orphan genes.
All other gene organizations were considered orphan genes. Three che gene clusters are flanked by three orphan genes (MXAN6953, MXAN6955, and MXAN6966) encoding HPKs and one gene (MXAN2687) encoding an HPK-like protein that are not CheA-like, and these genes are classified as orphan.
Cell growth and development for DNA microarray analysis.
M. xanthus DK1622 wild-type cells were grown in liquid 1% CTT medium (18) at 32°C to a density of 5 × 108 cells/ml, harvested, and resuspended in prewarmed (32°C) MC7 (10 mM morpholinepropanesulfonic acid [pH 7.0], 1 mM CaCl2) to a calculated density of 5 × 109 cells/ml. The cell suspension was diluted 1:8 with MC7 and 35 ml of the resulting suspension transferred to a sterile 145-mm culture dish (Greiner Bio-One) and incubated at 32°C. After 0, 2, 4, 6, 9, 12, 15, 18, or 24 h of development, cells were harvested, and immediately frozen in liquid nitrogen and stored at −80°C. For the preparation of total RNA from exponentially growing cells, cells were grown as described above, harvested at a density of 5 × 108 cells/ml, and immediately frozen in liquid nitrogen and stored at −80°C. Total RNA from these cells served as “reference RNA.” Three biological replicate time course experiments were performed.
Isolation of total RNA and DNase I treatment.
Total RNA was isolated from cell pellets using the hot-phenol method (44). One hundred micrograms total RNA was treated with 20 U RNase-free DNase I (Fermentas) for 60 min at 37°C. RNA was purified using the RNeasy minikit (Qiagen). The absence of DNA was verified by PCR. RNA quality was checked using agarose gel electrophoresis (50).
cDNA synthesis, fluorescent labeling, and hybridization.
cDNA synthesis was carried out using 25 μg of DNA-free total RNA for each experimental sample (RNA from developing cells) and 25 μg of the DNA-free reference sample as described previously (20, 44) with the following modifications. cDNA synthesis was carried out in the presence of 0.5 mM (each) dATP, dCTP, and dGTP, 0.1 mM dTTP, and 0.4 mM aminoallyl-dUTP in a total volume of 30 μl. Subsequently, RNA was hydrolyzed by addition of 10 μl of 1 M NaOH and 10 μl of 0.5 M EDTA and incubated at 65°C for 15 min, followed by addition of 10 μl of 1 M HCl. cDNA was purified using a Zymo kit (Zymo Research), vacuum dried, and resuspended in 13 μl of fresh 100 mM sodium bicarbonate, pH 9. cDNA of the reference probe was labeled with Cy5 and cDNA of the developmental probe with Cy3 as described previously (20). The M. xanthus DNA microarrays cover 88% of all protein-coding genes in the M. xanthus genome and were printed at the Stanford DNA microarray printing facility and postprocessed as described previously (8, 44). Briefly, each open reading frame (ORF) is represented on the microarray as a 275- to 325-bp PCR fragment. Hybridizations were carried out as described previously (44).
Data acquisition and analysis.
Microarrays were scanned simultaneously at two wavelengths (for Cy3, 532 nm; for Cy5, 635 nm) using a GenePix 4000B microarray scanner (Axon Instruments, Inc.). Image analysis and processing was performed using the GenePix Pro 6.0 software package (Axon Instruments, Inc.). The ratio-normalized data set (mean ratio of medians = 1) containing median signal intensity and median signal background from each channel was further analyzed using Aquity 4.0 software (Axon Instruments, Inc.) and the Significance Analysis of Microarrays (SAM) software (version 2.0), which assigns a score to each feature on a microarray on the basis of changes in gene expression relative to the standard deviation of repeated measurements (63). A filtered subset of all features printed on the array was selected based on the following criteria: (i) found by the Genepix Pro 6.0 spot-finding algorithm (“Flags” ≥ 0) and (ii) signal-to-noise ratio of either the Cy3 (532 nm) or Cy5 (635 nm) channel was greater than 2. For statistical significance analysis of the selected data points, SAM was used to calculate a t-like statistic (d) based on the estimated variance of the data. The cutoff value of the SAM analysis was chosen as the value where the median false discovery rate became 0%. For the selected features, ratios were averaged and subjected to a 1.5-fold cutoff criterion (at least in one time point during development) and were further selected for the ones where data points were present for all developmental time points and the expression value at 0 h was between +1.5 and −1.5 (not regulated).
Quantitative real-time PCR.
Verification of gene expression data obtained by DNA microarray analysis was carried out using quantitative real-time PCR (qRT-PCR) as described for M. xanthus (44). Briefly, cDNA was synthesized with the cDNA Archive kit (ABI) from 1.0 μg of DNA-free total RNA from six different developmental time points (0, 6, 12, 18, and 24 h) from one biological experiment from the DNA microarray analyses. Primers for qRT-PCR (see Table S1 in the supplemental material) were designed with Primer Express 2.0 (ABI) to give fragments with sizes of 60 to 150 bp. The qRT-PCRs were carried out in triplicate in a total volume of 25 μl containing 12.5 μl SYBR green PCR master mix (ABI), 1 μl of each primer (10 μM), 0.1 μl cDNA, and 11.9 μl H2O. qRT-PCRs were performed on an AB 7300 real-time PCR detection system using standard conditions. Expression ratios were calculated as the absolute expression level in developing cells over the absolute expression level in vegetative cells. The efficiency of each primer pair was determined using four different concentrations of DK1622 chromosomal DNA (10 ng/μl, 1.0 ng/μl, 0.1 ng/μl, and 0.01 ng/μl) as a template in qRT-PCRs.
Construction of mutants in genes coding for HPKs.
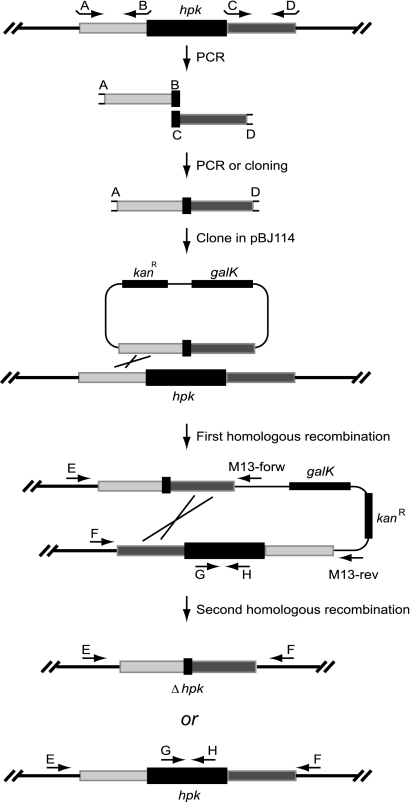
In-frame deletion mutants were constructed by two-step homologous recombination (Fig. 2). Briefly, approximately 1,100-bp PCR products containing the in-frame deletions were cloned in the plasmid pBJ114 (22), which contains the galK gene for counterselection. Primers used for the constructions are listed in Table S2 in the supplemental material. Four primers (A, B, C, and D) were designed to amplify the 1,100-bp fragment carrying an hpk in-frame deletion by PCR with M. xanthus chromosomal DNA as a template. Briefly, primers A and B were used to amplify the upstream flanking region of the hpk gene. Primer A contained a restriction site for cloning in pBJ114, and primer B contained either a restriction site for cloning or a region complementary to the downstream flanking PCR fragment. Primers C and D were used to amplify the downstream flanking fragment of the hpk gene. Primer C contained either a restriction site for cloning or a region complementary to the upstream flanking PCR fragment, and primer D contained a restriction site for cloning in pBJ114. The fragments AB and CD were used to generate the full-length in-frame deletion fragment either by direct cloning or in a second PCR with primers A and D and the two flanking PCR fragments as templates. The hpk in-frame fragments were cloned in the plasmid pBJ114 and transformed into E. coli Top 10 (Invitrogen) (F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araΔ139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG) and checked by sequencing. Correct plasmids were introduced into the M. xanthus wild-type strain DK1622 by electroporation (25). The insertion of plasmids after the first homologous recombination was confirmed by PCRs with three primer pair combinations (primers are listed in Table S2 in the supplemental material): primers E (binds upstream of primer A) and F (binds downstream of primer D), primers E and M13-forward (hybridizes to pBJ114), and primers F and M13-reverse (hybridizes to pBJ114). For each in-frame construct, at least one clone with the insertion of the plasmid in the upstream flanking region of the hpk gene and one clone with the insertion in the downstream flanking region of the hpk gene were chosen for the second homologous recombination. To isolate clones containing the in-frame deletion, cells were plated on CTT plates with 1% or 2% galactose (Sigma) for counterselection. Galactose-resistant and kanamycin-sensitive colonies were screened out and checked by two PCRs with the primers E and F and the primers G and H, which bind to the deleted part of the hpk gene, to verify the in-frame deletion. For MXAN3036 and MXAN4988, no in-frame deletions were obtained with the 1,100-bp in-frame deletion constructs using the primers labeled “short” in Table S2 in the supplemental material, i.e., all galactose-resistant clones contained in the intact genes. Therefore, for both genes in-frame deletion constructs on 1,400-bp fragments were generated using the primers labeled “long” in Table S2 in the supplemental material. Also, with these constructs, no in-frame deletions were isolated. In the case of MXAN0060, primers A and B were used to generate an internal fragment of MXAN0060, which was cloned into pBGS18 (57) to generate pXS011, which was used to generate an insertion mutation in MXAN0060. M. xanthus strains are listed in Table 1.
FIG. 2.
Outline of strategy for generating in-frame deletions in genes encoding histidine protein kinases. For each gene to be deleted, a plasmid was constructed by PCR and cloning with approximately 550-bp regions of homology upstream (light gray) or downstream (dark gray) flanking the in-frame construct. In a two-step procedure, deletion strains were isolated by first selecting for kanamycin resistance, followed by galactose counterselection using the galK gene carried on the plasmid and screening for loss of kanamycin resistance. Cells harboring galK die in the presence of galactose. Candidate clones for carrying the in-frame deletion were identified as Galr and KanS. Clones carrying the in-frame deletion were identified by PCR using the primer pairs EF and GH. For simplicity, the first homologous recombination is shown only for the recombination occurring in the upstream flanking region.
TABLE 1.
M. xanthus strains used in this work
| Strain | Genotype | Reference |
|---|---|---|
| DK1622 | Wild type | 24 |
| MS1512 | ΔMXAN1014 (= Δsdek) | 46 |
| PH1017 | ΔMXAN6855 (= ΔespC) | B. Lee and P. Higgsa |
| SA2107 | MXAN0060::pXS011 | This study |
| SA2112 | ΔMXAN3290 | This study |
| SA2115 | ΔMXAN6315 | This study |
| SA2117 | ΔMXAN0340 | This study |
| SA2118 | ΔMXAN0736 | This study |
| SA2119 | ΔMXAN7123 | This study |
| SA2120 | ΔMXAN0928 | This study |
| SA2121 | ΔMXAN6994 | This study |
| SA2124 | ΔMXAN2763 | This study |
| SA2126 | ΔMXAN5483 | This study |
| SA2130 | ΔMXAN3098 | This study |
| SA2131 | ΔMXAN0571 | This study |
| SA2133 | ΔMXAN7398 | This study |
| SA2134 | ΔMXAN4465 | This study |
| SA2136 | ΔMXAN0176 | This study |
| SA2137 | ΔMXAN6941 | This study |
| SA2138 | ΔMXAN0712 | This study |
| SA2139 | ΔMXAN0931 (= ΔespA) | This study |
| SA2142 | ΔMXAN0706 | This study |
| SA2143 | ΔMXAN7206 (= ΔmokA) | This study |
| SA2144 | ΔMXAN6996 (= ΔasgD) | This study |
Max Planck Institute for Terrestrial Microbiology.
Development of M. xanthus, spore assays, and motility tests.
To test for developmental defects, M. xanthus cells were grown as described above and developed as described previously in submerged culture, on clone fruiting (CF) agar or on TPM agar (55). Spore numbers were determined as the number of spores formed after 72 h and 120 h of starvation by harvesting 5 × 108 cells from CF agar. Cells were placed for 2 h at 50°C and briefly sonicated to disperse fruiting bodies. Spores were counted in a hemacytometer (depth, 0.1 mm; Marienfeld). Three biological experiments were performed for each strain to determine the sporulation efficiency. To determine the number of germinating spores, spore solutions were plated on 1.0% CTT agar plates. The number of fruiting bodies formed was determined after 120 h of starvation by manually counting fruiting bodies formed in three 20-μl cell aliquots on CF agar in three biological experiments. The surface area covered by individual fruiting bodies was calculated by measuring the total area of fruiting bodies formed in three 20-μl cell aliquots on CF agar after 120 h of starvation in three biological experiments using the area measurement tool in Metamorph (Molecular Devices), followed by division by the total number of fruiting bodies formed.
To test for motility defects, cells were grown as described above and plated on 0.5% CTT medium supplemented with 0.5% or 1.5% agar as described previously (52).
Microarray data accession number.
The microarray data discussed in this publication have been deposited in the NCBIs Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through Gene Expression Omnibus Series accession number GSE9477.
RESULTS
Identification of TCS genes in M. xanthus.
We analyzed the M. xanthus genome and identified 272 genes that encode proteins of TCS (Materials and Methods). Among these genes, 21 are localized in 8 loci that encode proteins which are part of Che-like systems, i.e., these clusters encode homologues of Che proteins and the HPK has a domain structure similar to that found in CheA (See Table S3 and S4 in the supplemental material for a list of these genes). In this report, we specifically focus on the 251 TCS genes encoding proteins that are not part of Che-like systems. These 251 genes include 118 HPK, 119 RR, and 14 HPK-like genes (Table 2). HPK genes are predicted to encode bona fide kinases which contain the conserved HATPase_c domain and a HisKA domain with the phosphorylatable His residue. RR genes are predicted to encode bona fide RRs which either consist of a single receiver domain or are multidomain proteins which in addition to the receiver domain also contain an output domain. HPKs that contained one or more receiver domains were classified as hybrid HPK. HPK-like genes encode proteins that either contain a HisKA domain with the phosphorylatable His residue and lack the HATPase_c domain or vice versa. The HATPase_c domain is also found in other ATPases, such as DNA gyrase B and the DNA repair protein MutL (11). To avoid annotating such proteins as HPK-like proteins, proteins were classified as HPK-like proteins only if they were encoded by genes located next to a TCS gene (e.g., MXAN0461 and MXAN4203) or if they contained one or more receiver domains (e.g., MXAN0230 and MXAN4432) (See Table S5 in the supplemental material for the domain structures of these proteins).
TABLE 2.
Summary of two-component system genes and proteins in M. xanthus
| Category | Total no. of genes | No. of complex gene clusters | No. of orphan genes | No. of paired genes |
|---|---|---|---|---|
| HPK genes/proteins | 118 (31 hybrid) | 18 (8 hybrid) | 68 (22 hybrid) | 32 (1 hybrid) |
| RR genes/proteins | 119 | 18 | 64 | 37 |
| HPK-like genes/proteins | 14 | 3 | 6 | 5 |
| Total TCS genes/proteins | 251 | 39 | 138 | 74 |
Given the large number of TCS genes in M. xanthus, we next asked whether the kinase and receiver domains belong to the established kinase and receiver families as defined by Grebe and Stock (15). To analyze the relationship between the M. xanthus TCS proteins, we aligned all kinase domains (encompassing the HisKA and HATPase_c domains) and receiver domains in RR, including receiver domains in hybrid HPKs and in HPK-like proteins, with those used by Grebe and Stock (15), and phylogenetic trees were generated. Analysis of the alignments and trees showed that all the kinase and receiver domains in the M. xanthus TCS proteins belong to the established Grebe and Stock families (See Table S3 and Table S4 in the supplemental material for family assignment). Thus, despite the large number of TCS proteins, there is no evidence that M. xanthus has evolved new kinase or receiver domains. Rather, M. xanthus appears to use domains also found in other species.
Genetic organization of two-component genes in M. xanthus.
During the analysis of M. xanthus TCS genes, we noticed that many of these genes are not organized as pairs as is typically reported. To analyze the genetic organization of TCS genes, we developed a set of criteria according to which TCS genes were divided into three categories: complex gene clusters, orphan genes, and paired genes (Fig. 1) (see Materials and Methods). The genetic organization of TCS genes in M. xanthus diverges significantly from the standard paired organization (Table 2; Fig. 3, 4, and 5): 55% (138 genes out of 251 total) of TCS genes are orphans, 16% (39 out of 251) of TCS genes are located in complex gene clusters, and only 29% (74 out of 251) are found as paired genes.
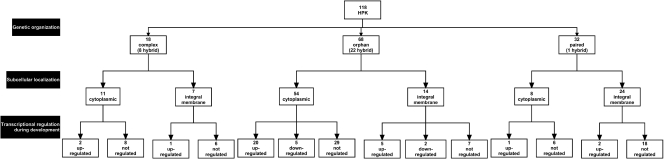
FIG. 3.
Organization diagram of histidine protein kinase genes and proteins in the M. xanthus genome. HPK genes were divided into three categories based on genetic organization. The number of hybrid HPKs is indicated in parentheses for each category. In the second layer, HPKs are divided according to their likely subcellular localization based on the presence (integral membrane) or absence (cytoplasmic) of transmembrane helices. In the third layer, HPK genes are divided according to transcriptional regulation during development. Note that in the third layer, not all HPK genes are included because they are not all present on the M. xanthus DNA microarray or they were not tested by qRT-PCR.
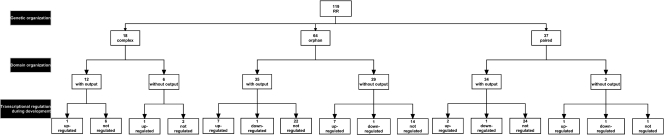
FIG. 4.
Organization diagram of response regulator genes and proteins in the M. xanthus genome. RR genes were divided into three categories based on genetic organization. In the second layer, RRs are divided according to the presence or absence of output domains. In the third layer, RRs are divided according to transcriptional regulation during development. Note that in the third layer, not all RR genes are included because they are not all present on the M. xanthus DNA microarray or they were not tested by qRT-PCR.
FIG. 5.
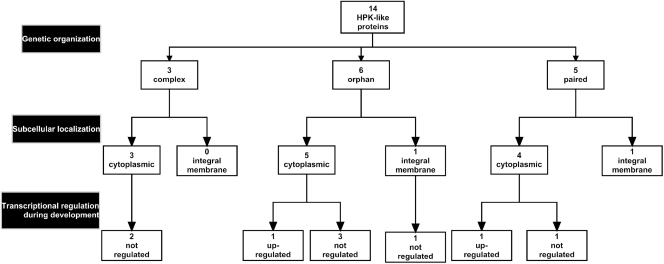
Organization diagram of histidine protein kinase-like genes and proteins in the M. xanthus genome. Genes encoding HPK-like proteins were divided into three categories based on genetic organization. In the second layer, HPK-like proteins are divided according to their likely subcellular localization based on the presence (integral membrane) or absence (cytoplasmic) of transmembrane helices. Note that in the third layer, not all HPK-like genes are included because they are not all present on the M. xanthus DNA microarray or they were not tested by qRT-PCR.
Histidine protein kinases.
As a first step in understanding how M. xanthus TCS proteins are connected, we divided the 118 HPKs into HPKs and hybrid HPKs, which in addition to the HisKA and HATPase_c domains also contain one or more receiver domains. We identified 31 hybrid kinases (26% of the total number of HPKs), which contain between 1 and 3 receiver domains (Table 2; Fig. 3) (see Table S3 in the supplemental material for detailed domain organization). The distribution of the 31 hybrid HPKs is highly biased. Eight of the hybrid HPKs are encoded by complex gene clusters (corresponding to 44% of all HPKs encoded by these genes), 22 of the hybrid HPKs are encoded by orphan genes (corresponding to 32% of all HPKs encoded by these genes), and only 1 hybrid HPK is encoded by a paired gene (corresponding to 3% of all HPKs encoded by these genes). Among the 31 hybrid HPKs, only 1, MXAN2317, is predicted to contain an Hpt domain.
HPKs are generally described as being integral membrane proteins (58). To determine whether HPKs in M. xanthus conform to this general description, we analyzed HPKs for the presence or absence of transmembrane helices. Among the 118 HPKs, 45 (38% of the total number of HPKs) are likely to be integral membrane proteins and 73 are likely to be cytoplasmic (Fig. 3) (see Table S3 in the supplemental material for detailed results). Also in this analysis, we found a biased distribution of the two types of HPKs. Thus, 11 of the predicted cytoplasmic HPKs are encoded by complex gene clusters (corresponding to 61% of all HPKs encoded by these genes), 54 of the predicted cytoplasmic HPKs are encoded by orphan genes (corresponding to 79% of all HPKs encoded by these genes), and only 8 of the predicted cytoplasmic HPKs are encoded by paired genes (corresponding to 25% of all HPKs encoded by these genes).
RRs and output domains.
As a second step in understanding the connectivity of TCS in M. xanthus, we divided the RRs into single-domain RRs, which consist only of the receiver domain, and multidomain RRs, which in addition to the receiver domain contain an output domain. The 119 RRs can be divided into 38 without (32% of the total number of RRs) and 81 with output domains (Table 3; Fig. 4) (see Table S4 in the supplemental material for the detailed domain organization). All che gene clusters contain either a CheAY hybrid kinase or CheY homologs (see Tables S3 and S4 in the supplemental material for the detailed description of proteins encoded by che gene clusters). Thus, the remaining single-domain RRs are not likely to be CheY paralogs.
TABLE 3.
Summary of output domains in RRs encoded in the M. xanthus genome
| Output domain category | Total no. of RRs with domains | No. of RRs encoded by:
|
||
|---|---|---|---|---|
| Complex gene clusters | Orphan genes | Paired genes | ||
| None | 38 | 6 | 29 | 3 |
| DNA binding | 50 | 5 | 13 | 32 |
| GGDEF | 10 | 5 | 5 | 0 |
| PilZ | 2 | 0 | 2 | 0 |
| Other domainsa | 5 | 0 | 5 | 0 |
| DUF | 14 | 2 | 10 | 2 |
This group consists of various output domains present only once.
The distribution of RRs without output domains is highly biased. The complex gene clusters include 18 RR genes encoding 12 RR with and 6 RR without output domains (corresponding to 33%). The output domains comprise five DNA binding domains, five GGDEF domains, which are involved in cyclic-di-GMP synthesis and regulation (21, 47), and two domains of unknown function (DUF). The 64 orphan RR genes include 35 encoding RR with and 29 without output domains (corresponding to 45%). The four largest categories of output domains for the orphan RR comprise 13 DNA binding domains, five GGDEF domains, two PilZ domains, which bind cyclic-di-GMP (2, 48), and 10 DUF. Finally, for the 37 RR encoded by paired genes, 34 contain an output domain and only 3 are without an output domain (corresponding to 9%). The 34 output domains comprise 32 DNA binding domains and 2 DUF. The distribution of the output domains suggests that the two main outputs from TCS in M. xanthus are gene regulation (50 domains) and cyclic-di-GMP regulation (12 domains). RRs with DUF are overrepresented among RRs encoded by complex gene clusters and orphan genes.
Transcriptional regulation of TCS genes during development.
Thirty-five TCS genes have been identified as important for fruiting body formation and sporulation (see Table S3, Table S4, and Table S5 in the supplemental material for the specific genes). Several of these genes are developmentally regulated at the transcriptional level during development. Thus, we reasoned that one approach to identify TCS proteins that may have a function in fruiting body formation would be to analyze the expression profiles of TCS genes during development.
For these experiments, we used an M. xanthus DNA microarray covering 88% of the 7,380 ORFs on the M. xanthus genome (see Materials and Methods). The detailed analysis of the experiments will be described elsewhere (N. Hamann, S. Wegener-Feldbrügge, L. Søgaard-Andersen, and R. Hedderich, data not shown). Total RNA was isolated from mid-exponentially growing wild-type cells (DK1622) and from different time points during development (0, 2, 4, 6, 9, 12, 15, 18, and 24 h), and cDNA was prepared, labeled with Cy3 (samples from development) and Cy5 (reference), and competitively hybridized to the microarray. As a reference, the RNA isolated from mid-exponentially growing wild-type cells was used. A total of three biological experiments were performed. Thus, data analysis (Materials and Methods) was carried out on three experimental values for each gene. The ratio-normalized data set was analyzed using Acuity 4.0 software (Axon Instruments) and SAM software, version 2.0, which assigns a score to each feature on a microarray on the basis of changes in gene expression relative to the standard deviation of repeated measurements (63). Genes called to be significantly regulated during development were selected by a delta value of the SAM analysis where the false discovery rate became 0% in combination with a 1.5-fold cutoff and data points for all time points.
Among the 200 TCS genes present on the array, 50 displayed altered expression during development (Fig. 3, 4, and 5; see Table S3, Table S4, and Table S5 in the supplemental material for details on the expression of individual genes). Developmentally regulated genes exhibited expression ratios in the range of 10.9-fold up-regulation to 6.6-fold down-regulation. The changes in gene expression during development were asymmetric: 46 genes were up-regulated, and 4 genes were down-regulated. To validate the significance of the expression data obtained from the DNA microarrays, qRT-PCR was applied to 11 genes (10 genes up-regulated during development and 1 gene not showing regulation). The transcriptional differences determined in the microarray experiments were confirmed by the qRT-PCR analysis (see Table S3, Table S4, and Table S5 in the supplemental material for details of qRT-PCR data).
To generate a complete data set on the expression profile for an entire category of TCS genes, we tested by qRT-PCR the expression during development of the 13 orphan HPK genes for which no expression data were available from the DNA microarray experiments. Among these genes, we found that six were up-regulated and seven were down-regulated during development (see Table S3 in the supplemental material for details of the qRT-PCR data).
In total, we identified 63 TCS genes out of 213 TCS genes tested either in microarray analyses or by qRT-PCRC that were transcriptionally regulated during development (Fig. 3, Fig. 4 and Fig. 5) (see Tables S3, S4, and S5 in the supplemental material for the expression of individual genes). Fifty-two genes were up-regulated and 11 genes down-regulated. Thirty-six percent and 13% of the tested orphan and complex genes, respectively, were transcriptionally regulated during development, whereas only 12% of the tested paired genes were transcriptionally regulated during development.
Genetic analysis of transcriptionally up-regulated, orphan histidine protein kinase genes.
The gene expression profiling experiments indicate that orphan TCS may have important functions in fruiting body formation. To test this hypothesis and to potentially identify novel TCS genes required for development, we focused on the 25 orphan HPK genes that are transcriptionally up-regulated during development (Fig. 3) (Table 4 contains a list of all 25 genes, including expression data). These 25 genes include espA (7), sdeK (13), espC (34), asgD (6), and mokA (28), which have previously been suggested to be important for development. To analyze the importance of the remaining 20 genes for fruiting body formation, we sought to generate in-frame deletions in 19 of these genes using a two-step recombination procedure (Fig. 2); for MXAN0060, an insertion mutant was constructed. In-frame deletions were preferred over insertion mutants to avoid polar effects on downstream genes. In addition, we generated or obtained in-frame deletions of the five previously identified orphan HPKs important for development in order to systematically compare developmental defects. For each of the genes MXAN3036 and MXAN4988, more than 200 galactose-resistant, kanamycin-sensitive clones were tested (see Materials and Methods). For both genes, all colonies tested contained the intact HPK gene. These observations suggest that MXAN3036 and MXAN4988 are essential genes for viability. For the remaining 22 genes, we obtained stable in-frame deletion mutants. For MXAN0060, we obtained a stable insertion mutant.
TABLE 4.
Orphan histidine protein kinase genes in M. xanthus transcriptionally up-regulated during development
| TIGR_MXAN (gene symbol) | Max. expression ratio (microarrays)/induction time (h)a,b | Max. expression ratio (qRT-PCR)/induction time (h)a,b | No. of TM helicesc | Domain organizationd |
|---|---|---|---|---|
| MXAN0060 | NOA | 19.3×-up/0-6 | 2 | HisKA-HATPase_c |
| MXAN0176 | NOA | 14.8×-up/6-12 | 0 | HisKA-HATPase_c |
| MXAN0340 | 2.7×-up/4-6 | ND | 0 | HisKA-HATPase_c |
| MXAN0571 | 1.9×-up/2-4 | ND | 0 | HisKA-HATPase_c |
| MXAN0706 | NOA | 3.4×-up/0-6 | 0 | HisKA-HATPase_c |
| MXAN0712 | 1.6×-up/12-15 | ND | 0 | HisKA-HATPase_c-RR-RR |
| MXAN0736 | 2.2×-up/0-2 | 128×-up/0-6 | 2 | HisKA-HATPase_c |
| MXAN0928 | 10.9×-up/2-4 | 29.7×-up/0-6 | 0 | HisKA-HATPase_c |
| MXAN0931 (= espA) | 4.4×-up/0-2 | ND | 0 | HisKA-HATPase_c-RR |
| MXAN1014 (= sdeK) | 2.6×-up/0-2 | ND | 0 | HisKA-HATPase_c |
| MXAN2763 | 2.4×-up/0-2 | ND | 0 | HisKA-HATPase_c-RR |
| MXAN3036 | 2.1×-up/4-6 | 29.8×-up/0-6 | 3 | HisKA-HATPase_c |
| MXAN3098 | 1.9×-up/0-2 | 4.1×-up/0-6 | 0 | HisKA-HATPase_c |
| MXAN3290 | 2.8×-up/0-2 | 4.4×-up/0-6 | 0 | HisKA-HATPase_c-RR |
| MXAN4465 | 2.1×-up/4-6 | ND | 0 | HisKA-HATPase_c-RR |
| MXAN4988 | 1.6×-up/0-2 | 2.8×-up/0-6 | 8 | HisKA-HATPase_c |
| MXAN5483 | 2.2×-up/4-6 | ND | 0 | HisKA-HATPase_c |
| MXAN6315 | 4.2×-up/0-2 | 36.5×-up/0-6 | 0 | HisKA-HATPase_c-RR |
| MXAN6855 (espC) | 1.7×-up/12-15 | ND | 8 | HisKA-HATPase_c-RR |
| MXAN6941 | NOA | 7.5×-up/6-12 | 0 | HisKA-HATPase_c |
| MXAN6994 | 4.4×-up/2-4 | 76.4×-up/0-6 | 0 | HisKA-HATPase_c |
| MXAN6996 (asgD) | 1.8×-up/12-15 | ND | 0 | RR-HisKA-HATPase_c |
| MXAN7123 | 4.0×-up/2-4 | 143×-up/0-6 | 0 | HisKA-HATPase_c |
| MXAN7206 (mokA) | NOA | 9.2×-up/0-6 | 0 | HisKA-HATPase_c-RR |
| MXAN7398 | NOA | 3.8×-up/0-6 | 0 | HisKA-HATPase_c |
Expression ratios were calculated as the expression in developing cells over the expression in vegetative cells. Maximum (max.) expression ratios indicate the maximum expression ratio for a particular gene during all time points tested using DNA microarrays (0, 2, 4, 6, 9, 12, 15, 18, and 24 h) and qRT-PCR (0, 6, 12, 18, and 24 h). Induction time indicates the time interval in which the expression ratio began to change relative to that observed in vegetative cells. “-up” indicates up-regulation.
NOA, gene not represented or not detected on DNA microarray; ND, not determined.
TM, transmembrane.
All domains present in a protein are indicated except for domains in the sensor domain.
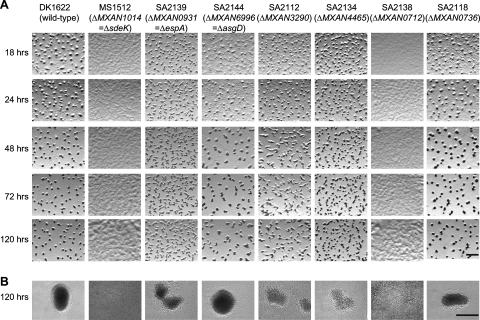
We next examined the phenotypes of the wild-type strain (DK1622) and the 23 HPK mutants. To test for motility defects, cell spreading was examined on 0.5% CTT medium containing 0.5% agar, which favors motility by means of the S-motility system, or 1.5% agar, which favors motility by means of the A-motility system (52). None of the 23 mutants displayed motility defects on these two types of surfaces (data not shown). To test the mutants for developmental defects, cells were exposed to starvation under three different conditions, i.e., CF starvation agar, TPM starvation agar, and in submerged culture and the sporulation frequencies determined after 72 h and 120 h of starvation on CF agar. Moreover, levels of germinating spores were determined after 72 h and 120 h of starvation on CF agar. Individual mutants displayed similar phenotypes under all three developmental conditions (data from development on CF agar and in submerged culture are shown in Fig. 6 and Table 5). DK1622 wild-type cells had formed fruiting bodies at 24 h, and at 48 h the fruiting bodies had darkened (Fig. 6; Table 5). Sixteen mutants displayed normal development, sporulation, and spore germination (data not shown). As previously reported (13, 46), MS1512 carrying ΔMXAN1014 (= Δsdek) was unable to aggregate to form fruiting bodies and was strongly reduced in sporulation. Also as previously reported, a mutant carrying ΔMXAN0931 (= ΔespA) (SA2139) displayed early aggregation with the formation of many small and irregularly shaped fruiting bodies; also, SA2139 displayed early sporulation with many spores localized outside the fruiting bodies (Fig. 6B). SA2144 carrying ΔMXAN6996 (= ΔasgD) displayed delayed aggregation but normal levels of sporulation. This is in disagreement with a previous report, in which an insertion in asgD was reported to cause aggregation as well as sporulation defects (6). We attribute these differences to differences in strain backgrounds and mutations being analyzed in the previous report and the data presented here. SA2112 carrying ΔMXAN3290 displayed delayed aggregation and formation of abnormally shaped fruiting bodies. SA2112 sporulated at wild-type levels, however, many of the spores were localized outside fruiting bodies. SA2134 carrying ΔMXAN4465 displayed normal timing of aggregation and sporulation. However, this mutant formed 1.7-fold ± 0.4-fold more fruiting bodies than the wild type and individual fruiting bodies were smaller than those formed by the wild type, covering an area of only 60% ± 7% of that of a wild-type fruiting body. Moreover, the fruiting bodies formed by SA2134 in submerged culture were less condensed than those formed by the wild type (Fig. 6B). SA2138 carrying ΔMXAN0712 was unable to aggregate to form fruiting bodies and was strongly reduced in sporulation. Finally, SA2118 carrying ΔMXAN0736 displayed normal aggregation and fruiting body formation and sporulated at wild-type levels. However, these spores germinated at a level threefold lower than that observed with the wild type. It should be noted that the two mutants carrying in-frame deletions of espC (PH1017) and mokA (SA2143) developed in a manner indistinguishable from that of the DK1622 wild type under all conditions tested and sporulated at wild-type levels and with the formation of germination-proficient spores (data not shown). Inactivation of these two genes has previously been reported to cause developmental defects (28, 34). We attribute the difference between published results and our results to differences in strain backgrounds used and to different mutations being analyzed.
FIG. 6.
Developmental phenotypes of mutants containing in-frame deletions of orphan histidine protein kinase genes. (A) Developmental phenotypes on CF agar. The indicated strains were starved on CF agar for the indicated periods of time. All strains analyzed are derivatives of DK1622; below strain numbers, the in-frame deletion present in a particular strain is indicated. Scale bar, 1.0 mm. (B) Developmental phenotypes in submerged culture. The same strains as in panel A were exposed to starvation in submerged culture for 120 h. Scale bar, 100 μm.
TABLE 5.
Sporulation frequencies of M. xanthus wild-type and HPK mutants
| Strain | Genotype | Sporulation frequency (%) at:a
|
|
|---|---|---|---|
| 72 h | 120 h | ||
| DK1622 | Wild type | 66 ± 4 | 100 ± 6b |
| MS1512 | ΔMXAN1014 (= Δsdek) | <0.001 | <0.001 |
| SA2139 | ΔMXAN0931 (= ΔespA) | 94 ± 9 | 151 ± 15 |
| SA2144 | ΔMXAN6996 (= ΔasgD) | 56 ± 8 | 96 ± 2 |
| SA2112 | ΔMXAN3290 | 47 ± 9 | 80 ± 16 |
| SA2118 | ΔMXAN0736 | 49 ± 9 | 71 ± 8c |
| SA2134 | ΔMXAN4465 | 56 ± 17 | 78 ± 19 |
| SA2138 | ΔMXAN0712 | <0.001 | <0.001 |
Sporulation frequencies are presented relative to the sporulation level in the wild-type strain DK1622 after 120 h of starvation. Values are means and standard deviations from three experiments.
The absolute sporulation level of DK1622 was 14.7% ± 1.0% at 120 h.
Spores of SA2118 germinated at a threefold-lower frequency than wild-type spores.
DISCUSSION
Here we report the identification of 251 TCS genes, which are not part of che gene clusters, in the M. xanthus genome. These 251 TCS genes make up 3.4% of the 7,380 ORFs (14) in the M. xanthus genome. To our knowledge, this number of TCS genes is the highest reported so far for any organism. However, when corrected for genome size, the number of TCS genes in M. xanthus falls within that reported for other bacteria and thus complies with the general rule that the number of TCS proteins per genome increases with the square of the genome size (11, 12). The number of TCS genes in M. xanthus is also in line with the general notion that growth of bacterial genome size is accompanied by accumulation of paralogous protein families.
The M. xanthus TCS genes could be divided into three classes based on their genetic organization. Fifty-five percent and 16% of all the TCS genes are organized as orphan genes or in complex gene clusters, respectively, and only 29% are found as paired genes. We compared this genetic organization of TCS genes to that in other bacteria and found that of the 59 TCS genes in E. coli, 20% are orphan, 8% are in complex gene clusters, and 72% are paired; of the 63 TCS genes in B. subtilis, 14% are orphan, 0% in complex gene clusters, and 86% paired; of the 86 TCS genes in Caulobacter crescentus, 63% are orphan, 7% in complex gene clusters, and 30% paired; of the 126 TCS genes in Pseudomonas aeruginosa, 35% are orphan, 8% in complex gene clusters, and 57% paired; and of the 186 TCS genes in Streptomyces coelicolor, 25% are orphan, 9% in complex gene clusters, and 66% paired (S. Huntley and L. Søgaard-Andersen, unpublished data). This comparison shows that the genetic organization of TCS genes shows large interspecies variations. Moreover, it is evident that the three categories of TCS genes are not peculiar to the M. xanthus genome.
The genome size of M. xanthus is 9.14 Mb, and it has been suggested that lineage-specific gene family expansions (LSE) were major contributors to the genomic expansion (14). Our preliminary analyses suggest that a large fraction of TCS genes in M. xanthus may have arisen by LSE (Huntley and Søgaard-Andersen, unpublished). Alm et al. (1) reported that deltaproteobacteria have a propensity for LSE of TCS genes. In agreement with this, we find that the complements of TCS genes in the three Myxococcales species Sorangium cellulosum (genome sequence provided by Rolf Müller, University of Saarland, Saarbrücken, Germany), Stigmatella aurantiaca (genome at http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi), and Anaeromyxobacter dehalogenans (genome at http://genome.jgi-psf.org/finished_microbes/anade/anade.home.html) also contain a large fraction of TCS genes that likely arose by LSE (Huntley and Søgaard-Andersen, unpublished). Interestingly, however, the complements of TCS genes that have expanded by LSE in the four Myxococcales species appear to be different, suggesting that for each species the particular genes amplified provide that species with some selective benefits. A detailed description of LSE of TCS genes in Myxococcales will be presented elsewhere (Huntley and Søgaard-Andersen, unpublished).
We identified three groups of structurally remarkable TCS proteins in M. xanthus. One group consists of 14 HPK-like proteins, which contain only a HisKA domain or a HATPase_c domain (see Table S5 in the supplemental material). Four lines of evidence suggest that these genes are not pseudogenes but code for functional proteins. First, three of these genes (MXAN0461 [= redE] [17], MXAN2670 [= asgA] [45], and MXAN5123 [= mrpA] [59, 60]) are required for development. Second, at least nine of the genes were found to be expressed in global transcriptional profiling experiments. Third, two of the genes are transcriptionally up-regulated during development. Finally, the HPK-like protein YojN, which functions in a phosphorelay with RcsB and RcsC (61) in Escherichia coli, provides evidence that HPK-like proteins may be functional. A second group of proteins with interesting structural features consists of proteins that have organizations of signal transduction domains, which have not been reported previously and which raise interesting questions in terms of how they function in phosphotransfer reactions. For instance, the orphan HPKs MXAN2606 and MXAN2317 are predicted to have the domain structures HisKA-HATPase_c-RR-HisKA-HATPase_c and HisKA-HATPase_c-RR-RR-Hpt, respectively, and the RR MXAN7362, which is encoded by a complex gene cluster, is predicted to have the domain structure RR-Hpt-RR-RR-GGDEF. The third group of TCS proteins with interesting structural features consists of 14 RR with output DUF. These RRs are overrepresented among RRs encoded by complex gene clusters and orphan genes, i.e., 10 and 2 of these domains are found in RRs encoded by orphan and complex genes, respectively. Interestingly, four of these proteins are orphan RRs involved in regulating gliding motility (MXAN2991 [= aglZ] [40, 67], MXAN4149 [= frzS] [39, 66], MXAN4461 [= romR] [35], and MXAN6627 [= sgnC] [68]).
The analysis of M. xanthus TCS proteins revealed structural features that have functional implications. First, 73 out of the 118 HPKs are predicted to be cytoplasmic, suggesting that the many HPKs in M. xanthus may be involved not in monitoring external stimuli or intercellular signals but rather in monitoring cytoplasmic stimuli. Alternatively, they could indirectly be involved in monitoring external stimuli or intercellular signals by interacting with membrane proteins. Second, the analysis of output domains in RRs suggests that the output responses from TCS systems in M. xanthus center on three types, regulation of gene expression, regulation of cyclic-di-GMP metabolism, and unknown functions.
Interestingly, we found strongly biased distributions of different types of TCS proteins encoded by paired genes and orphan genes and in complex gene clusters. These biased distributions have several functional implications, as discussed below. For paired TCS genes, the main implication is that a large fraction of the corresponding proteins are part of simple 1:1 TCS with an integral membrane HPK and a cognate RR that is involved in regulation of gene expression. The predicted membrane localization of the paired HPKs suggests that they are primarily involved in monitoring external stimuli. Moreover, the underrepresentation of these genes among transcriptionally regulated genes during development indicates that they may be functionally active in vegetative cells. Clearly, the latter implication does not preclude a function during fruiting body formation of these proteins. Consistently, 15 paired TCS genes have been identified which are important for development (see Tables S3, S4, and S5 in the supplemental material for the identities of these genes).
For TCS proteins encoded by orphan genes or genes in complex gene clusters, the biased distribution of protein characteristics and expression profiles suggests that the corresponding HPKs are primarily involved in monitoring cytoplasmic stimuli (due to the overrepresentation of HPKs predicted to be cytoplasmic) and that the main output responses from the corresponding pathways are regulation of gene expression, regulation of cyclic-di-GMP metabolism, and unknown functions (as indicated by the overrepresentation of RRs with DUF output domains). Moreover, the overrepresentation of these genes among those that are transcriptionally regulated during development suggests that many of these genes encode TCS proteins with a function only during development. Consistently, 16 orphan TCS genes (including the 4 identified in this report) and 6 TCS genes encoded in complex gene clusters have been shown to be important for development or spore germination (see Tables S3, S4, and S5 in the supplemental material for the identities of these genes). It should be noted that transcriptional regulation during development does not preclude a function in vegetative cells.
A question that remains to be addressed focuses on the connectivity of the TCS proteins in M. xanthus. As mentioned for the paired TCS genes, the almost complete absence of hybrid HPKs and single-domain RRs in the corresponding proteins suggests that the paired genes encode proteins that make up simple, linear 1:1 pathways. For TCS proteins encoded by orphan genes and in complex gene clusters, the connectivity has been analyzed experimentally only for the RedCDEF proteins (= MXAN0459 to MXAN0462), and the data suggest that these four proteins may constitute a complex phosphorelay (17). Since the connectivity of TCS proteins cannot be predicted based on sequence conservation alone (54), this question, therefore, remains open for most of the TCS proteins encoded by orphan genes and in complex gene clusters. The close to 1:1 numerical ratio of HPKs and RRs encoded by these genes could lead to the notion that they could be organized in 1:1 pathways. However, two observations argue against this notion. First, hybrid HPKs are overrepresented among these proteins. Second, many of the RRs encoded by these genes are single-domain RRs. The overrepresentation of hybrid HPKs and RRs without output domains among the proteins encoded by complex gene clusters and orphan genes strongly suggests that the signal transduction pathways encoded by these genes are structured as phosphorelays and/or are branched. Phosphorelays would likely depend on the presence of Hpt domain-containing proteins. In addition to CheA kinases, we identified only two proteins containing Hpt domains, the hybrid orphan HPK MXAN2317 and the RR MXAN7362, which is encoded in a complex gene cluster. It has been argues that Hpt domains are difficult to identify due to the low level of sequence conservation (4); thus, M. xanthus may indeed encode more proteins containing Hpt domains. Clearly, experimental analyses are needed to address the question of the connectivity of the M. xanthus TCS proteins.
We directly tested genetically the hypothesis that orphan developmentally up-regulated genes could be important for development by focusing on the 25 orphan HPK genes that are up-regulated at the transcriptional level during development. Among these genes, we found two (MXAN3036 and MXAN4988) that are likely to be essential for viability and seven that are important for development without having vegetative defects. These seven genes include MXAN0931 (= espA) (7), MXAN1014 (= sdeK) (13, 46), and MXAN6996 (= asgD) (6), which have previously been shown to be important for development. In addition, we identified MXAN0712, MXAN0736, MXAN3290, and MXAN4465 as important for development or spore germination. Finally, inactivation of MXAN6855 (= espC) (34) and MXAN7206 (= mokA) (28), which have previously been reported to be important for development, did not display developmental defects under our conditions. How these seven proteins function in development remains to be determined. Clearly, the lack of developmental defects in the remaining 16 mutants could be caused by functional redundancy among HPKs. Nevertheless, our data have two implications: first, the transcriptional up-regulation of a TCS gene does not necessarily mean that this gene has an essential function during development (at least not under the three conditions tested here). This notion is supported by the observation that several other genes that are transcriptionally up-regulated during development also do not have essential functions during development (29). Second, even though TCS proteins clearly have important functions in development, the large number of TCS genes in M. xanthus may not have evolved solely to regulate fruiting body formation.
Supplementary Material
Acknowledgments
We thank B. Lee, P. Higgs, and M. Singer for strains and R. Müller, University of Saarland, for access to the Sorangium cellulosum genome sequence prior to publication.
The International Max Planck Research School for Environmental, Cellular and Molecular Microbiology and the Max Planck Society supported this work.
Footnotes
Published ahead of print on 9 November 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alm, E., K. Huang, and A. Arkin. 2006. The evolution of two-component systems in bacteria reveals different strategies for niche adaptation. PLoS Comput. Biol. 2e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amikam, D., and M. Y. Galperin. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 13-6. [DOI] [PubMed] [Google Scholar]
- 3.Appleby, J. L., J. S. Parkinson, and R. B. Bourret. 1996. Signal transduction via the multi-step phosphorelay: not necessarily a road less traveled. Cell 86845-848. [DOI] [PubMed] [Google Scholar]
- 4.Biondi, E. G., S. J. Reisinger, J. M. Skerker, M. Arif, B. S. Perchuk, K. R. Ryan, and M. T. Laub. 2007. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature 444899-904. [DOI] [PubMed] [Google Scholar]
- 5.Burbulys, D., K. A. Trach, and J. A. Hoch. 1991. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 64545-552. [DOI] [PubMed] [Google Scholar]
- 6.Cho, K., and D. R. Zusman. 1999. AsgD, a new two-component regulator required for A-signalling and nutrient sensing during early development of Myxococcus xanthus. Mol. Microbiol. 34268-281. [DOI] [PubMed] [Google Scholar]
- 7.Cho, K., and D. R. Zusman. 1999. Sporulation timing in Myxococcus xanthus is controlled by the espAB locus. Mol. Microbiol. 34714-725. [DOI] [PubMed] [Google Scholar]
- 8.Diodati, M. E., F. Ossa, N. B. Caberoy, I. R. Jose, W. Hiraiwa, M. M. Igo, M. Singer, and A. G. Garza. 2006. Nla18, a key regulatory protein required for normal growth and development of Myxococcus xanthus. J. Bacteriol. 1881733-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabret, C., V. A. Feher, and J. A. Hoch. 1999. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J. Bacteriol. 1811975-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finn, R., J. Mistry, B. Schuster-Böckler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, S. R. Eddy, E. L. L. Sonnhammer, and A. Bateman. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34D247-D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galperin, M. 2005. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galperin, M. Y. 2006. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J. Bacteriol. 1884169-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garza, A. G., J. S. Pollack, B. Z. Harris, A. Lee, I. M. Keseler, E. F. Licking, and M. Singer. 1998. SdeK is required for early fruiting body development in Myxococcus xanthus. J. Bacteriol. 1804628-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldman, B. S., W. C. Nierman, D. Kaiser, S. C. Slater, A. S. Durkin, J. A. Eisen, C. M. Ronning, W. B. Barbazuk, M. Blanchard, C. Field, C. Halling, G. Hinkle, O. Iartchuk, H. S. Kim, C. Mackenzie, R. Madupu, N. Miller, A. Shvartsbeyn, S. A. Sullivan, M. Vaudin, R. Wiegand, and H. B. Kaplan. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. USA 10315200-15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grebe, T. W., and J. B. Stock. 1999. The histidine protein kinase superfamily. Adv. Microb. Physiol. 41139-227. [DOI] [PubMed] [Google Scholar]
- 16.Harris, B. Z., D. Kaiser, and M. Singer. 1998. The guanosine nucleotide (p)ppGpp initiates development and A-factor production in Myxococcus xanthus. Genes Dev. 121022-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higgs, P. I., K. Cho, D. E. Whitworth, L. S. Evans, and D. R. Zusman. 2005. Four unusual two-component signal transduction homologs, RedC to RedF, are necessary for timely development in Myxococcus xanthus. J. Bacteriol. 1878191-8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): two gene systems control movement. Mol. Gen. Genet. 171177-191. [Google Scholar]
- 19.Inouye, M., S. Inouye, and D. R. Zusman. 1979. Gene expression during development of Myxococcus xanthus: pattern of protein synthesis. Dev. Biol. 68579-591. [DOI] [PubMed] [Google Scholar]
- 20.Jakobsen, J. S., L. Jelsbak, L. Jelsbak, R. D. Welch, C. Cummings, B. Goldman, E. Stark, S. Slater, and D. Kaiser. 2004. σ54 enhancer binding proteins and Myxococcus xanthus fruiting body development. J. Bacteriol. 1864361-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenal, U., and J. Malone. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40385-407. [DOI] [PubMed] [Google Scholar]
- 22.Julien, B., A. D. Kaiser, and A. Garza. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 979098-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaiser, D. 2004. Signaling in myxobacteria. Annu. Rev. Microbiol. 5875-98. [DOI] [PubMed] [Google Scholar]
- 24.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 765952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kashefi, K., and P. L. Hartzell. 1995. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF defect. Mol. Microbiol. 15483-494. [DOI] [PubMed] [Google Scholar]
- 26.Kim, S. K., and D. Kaiser. 1991. C-factor has distinct aggregration and sporulation thresholds during Myxococcus development. J. Bacteriol. 1731722-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, S. K., and D. Kaiser. 1990. C-factor: a cell-cell signaling protein required for fruiting body morphogenesis of M. xanthus. Cell 6119-26. [DOI] [PubMed] [Google Scholar]
- 28.Kimura, Y., H. Nakano, H. Terasaka, and K. Takegawa. 2001. Myxococcus xanthus mokA encodes a histidine kinase-response regulator hybrid sensor required for development and osmotic tolerance. J. Bacteriol. 1831140-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kroos, L., A. Kuspa, and D. Kaiser. 1986. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev. Biol. 117252-266. [DOI] [PubMed] [Google Scholar]
- 30.Kruse, T., S. Lobedanz, N. M. S. Berthelsen, and L. Søgaard-Andersen. 2001. C-signal: a cell surface-associated morphogen that induces and coordinates multicellular fruiting body morphogenesis and sporulation in M. xanthus. Mol. Microbiol. 40156-168. [DOI] [PubMed] [Google Scholar]
- 31.Kumar, A., J. C. Toledo, R. P. Patel, J. R. Lancaster, and A. J. C. Steyn. 2007. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc. Natl. Acad. Sci. USA 10411568-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuspa, A., L. Plamann, and D. Kaiser. 1992. Identification of heat-stable A-factor from Myxococcus xanthus. J. Bacteriol. 1743319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuspa, A., L. Plamann, and D. Kaiser. 1992. A-signalling and the cell density requirement for Myxococcus xanthus development. J. Bacteriol. 1747360-7369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, B., P. I. Higgs, D. R. Zusman, and K. Cho. 2005. EspC is involved in controlling the timing of development in Myxococcus xanthus. J. Bacteriol. 1875029-5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leonardy, S., G. Freymark, S. Hebener, E. Ellehauge, and L. Sogaard-Andersen. 2007. Coupling of protein localization & cell movements by a dynamically localized response regulator in Myxococcus xanthus. EMBO J. 264433-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Letunic, I., R. R. Copley, S. Schmidt, F. D. Ciccarelli, T. Doerks, J. Schultz, C. P. Ponting, and P. Bork. 2004. SMART 4.0: towards genomic data integration. 32D142-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, S., B.-U. Lee, and L. J. Shimkets. 1992. csgA expression entrains Myxococcus xanthus development. Genes Dev. 6401-410. [DOI] [PubMed] [Google Scholar]
- 38.Lobedanz, S., and L. Søgaard-Andersen. 2003. Identification of the C-signal, a contact dependent morphogen coordinating multiple developmental responses in Myxococcus xanthus. Genes Dev. 172151-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mignot, T., J. P. Merlie, and D. R. Zusman. 2005. Regulated pole-to-pole oscillations of a bacterial gliding motility protein. Science 310855-857. [DOI] [PubMed] [Google Scholar]
- 40.Mignot, T., J. W. Shaevitz, P. L. Hartzell, and D. R. Zusman. 2007. Evidence that focal adhesion complexes power bacterial gliding motility. Science 315853-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mika, F., and R. Hengge. 2005. A two-component phosphotransfer network involving ArcB, ArcA, and RssB coordinates synthesis and proteolysis of sigma(S) (RpoS) in E. coli. Genes Dev. 192770-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mok, K. C., N. S. Wingreen, and B. L. Bassler. 2003. Vibrio harveyi quorum sensing: a coincidence detector for two autoinducers controls gene expression. EMBO J. 22870-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Connor, K. A., and D. R. Zusman. 1991. Development in Myxococcus xanthus involves differentiation into two cell types, peripheral rods and spores. J. Bacteriol. 1733318-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Overgaard, M., S. Wegener-Feldbrügge, and L. Søgaard-Andersen. 2006. The orphan response regulator DigR is required for synthesis of extracellular matrix fibrils in Myxococcus xanthus. J. Bacteriol. 1884384-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plamann, L., Y. Li, B. Cantwell, and J. Mayor. 1995. The Myxococcus xanthus asgA gene encodes a novel signal transduction protein required for multicellular development. J. Bacteriol. 1772014-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pollack, J. S., and M. Singer. 2001. SdeK, a histidine kinase required for Myxococcus xanthus development. J. Bacteriol. 1833589-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Romling, U., and D. Amikam. 2006. Cyclic di-GMP as a second messenger. Curr. Opin. Microbiol. 9218-228. [DOI] [PubMed] [Google Scholar]
- 48.Ryjenkov, D. A., R. Simm, U. Romling, and M. Gomelsky. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 28130310-30314. [DOI] [PubMed] [Google Scholar]
- 49.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4406-425. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 51.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 955857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi, W., and D. R. Zusman. 1993. The two motility systems of Myxococcus xanthus show different selective advantages on various surfaces. Proc. Natl. Acad. Sci. USA 903378-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singer, M., and D. Kaiser. 1995. Ectopic production of guanosine penta- and teraphosphate can initiate early developmental gene expression in Myxococcus xanthus. Genes Dev. 91633-1644. [DOI] [PubMed] [Google Scholar]
- 54.Skerker, J. M., M. S. Prasol, S. P. Barrett, E. G. Biondi, and M. T. Laub. 2005. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 31770-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Søgaard-Andersen, L., F. J. Slack, H. Kimsey, and D. Kaiser. 1996. Intercellular C-signaling in Myxococcus xanthus involves a branched signal transduction pathway. Genes Dev. 10740-754. [DOI] [PubMed] [Google Scholar]
- 56.Sonnhammer, E. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6175-182. [PubMed] [Google Scholar]
- 57.Spratt, B. G., P. J. Hedge, S. te Heesen, A. Edelman, and J. K. Broome-Smith. 1986. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene 41337-342. [DOI] [PubMed] [Google Scholar]
- 58.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69183-215. [DOI] [PubMed] [Google Scholar]
- 59.Sun, H., and W. Shi. 2001. Analyses of mrp genes during Myxococcus xanthus development. J. Bacteriol. 1836733-6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun, H., and W. Shi. 2001. Genetic studies of mrp, a locus essential for cellular aggregation and sporulation of Myxococcus xanthus. J. Bacteriol. 1834786-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takeda, S., Y. Fujisawa, M. Matsubara, H. Aiba, and T. Mizuno. 2001. A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC to YojN to RcsB signalling pathway implicated in capsular synthesis and swarming behaviour. Mol. Microbiol. 40440-450. [DOI] [PubMed] [Google Scholar]
- 62.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 985116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uhl, M. A., and J. F. Miller. 1996. Central role of the BvgS receiver as a phosphorylated intermediate in a complex two-component regulatory phosphorelay. J. Biol. Chem. 27133176-33180. [DOI] [PubMed] [Google Scholar]
- 65.Ulrich, L. E., E. V. Koonin, and I. B. Zhulin. 2005. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol. 1352-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ward, M. J., H. Lew, and D. R. Zusman. 2000. Social motility in Myxococcus xanthus requires FrzS, a protein with an extensive coiled-coil domain. Mol. Microbiol. 371357-1371. [DOI] [PubMed] [Google Scholar]
- 67.Yang, R., S. Bartle, R. Otto, A. Stassinopoulos, M. Rogers, L. Plamann, and P. Hartzell. 2004. AglZ is a filament-forming coiled-coil protein required for adventurous gliding motility of Myxococcus xanthus. J. Bacteriol. 1866168-6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Youderian, P., and P. L. Hartzell. 2006. Transposon insertions of magellan-4 that impair social gliding motility in Myxococcus xanthus. Genetics 1721397-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.