Abstract
Autoinducer 2 (AI-2) is the only species-nonspecific autoinducer known in bacteria and is produced by both gram-negative and gram-positive organisms. Consequently, it is proposed to function as a universal quorum-sensing signal for interaction between bacterial species. AI-2 is produced as the by-product of a metabolic transformation carried out by the LuxS enzyme. To separate the metabolic function of the LuxS enzyme from the signaling role of AI-2, we carried out a global transcriptome analysis of a luxS null mutant culture of Streptococcus mutans UA159, an important cariogenic bacterium and a crucial component of the dental plaque biofilm community, in comparison to a luxS null mutant culture supplemented with chemically pure 4,5-dihydroxy-2,3-pentanedione, the precursor of AI-2. The data revealed fundamental changes in gene expression affecting 585 genes (30% of the genome) which could not be restored by the signal molecule AI-2 and are therefore not caused by quorum sensing but by lack of the transformation carried out by the LuxS enzyme in the activated methyl cycle. All functional classes of enzymes were affected, including genes known to be important for biofilm formation, bacteriocin synthesis, competence, and acid tolerance. At the same time, 59 genes were identified whose transcription clearly responded to the addition of AI-2. Some of them were related to protein synthesis, stress, and cell division. Three membrane transport proteins were upregulated which are not related to any of the known AI-2 transporters. Three transcription factors were identified whose transcription was stimulated repeatedly by AI-2 addition during growth. Finally, a global regulatory protein, the δ subunit of the RNA polymerase (rpoE), was induced 147-fold by AI-2, representing the largest differential gene expression observed. The data show that many phenotypes related to the luxS mutation cannot be ascribed to quorum sensing and have identified for the first time regulatory proteins potentially mediating AI-2-based signaling in gram-positive bacteria.
Streptococcus mutans is an important component of the dental plaque biofilm community and plays a key role in the development of caries, the potential result of a complex succession process beginning with the attachment of the bacteria to the tooth surface, followed by the development of a mixed-species biofilm (17). Tooth decay is triggered by a local reduction of pH following the fermentation of available carbohydrates. The main virulence factors of S. mutans (adhesion, acid tolerance, and acidogenicity) work coordinately to alter dental plaque ecology. The selection for a cariogenic flora increases the magnitude of the drop in pH and the rate of enamel demineralization (32).
Cell-cell communication is thought to play a large role in controlling the species composition and virulence properties of the dental plaque community (33, 34). In S. mutans, a signaling peptide (competence-stimulating peptide [CSP]) typical of quorum-sensing circuits in gram-positive bacteria regulates competence, bacteriocin production, and biofilm development in a density-dependent and species-specific way through a well-characterized signal transduction cascade (3, 12, 41, 73). In addition, S. mutans and several genera of oral gram-positive and gram-negative bacteria harbor the luxS gene (19, 33). It is responsible for the synthesis of autoinducer 2 (AI-2), the most broadly distributed interspecies signaling molecule known in bacteria (10, 74). AI-2 is able to elicit a specific response across species barriers (7, 80). It may therefore be particularly important for shaping multispecies biofilms such as those in dental plaque: recently it was reported that Streptococcus oralis and Actinomyces naeslundii (both of them carrying the luxS gene) produced very sparse biofilms when grown alone but developed dense, interdigitated biofilms when grown together. This good dual-species biofilm growth was abolished by knocking out luxS in S. oralis, but it could be restored by genetic complementation of the luxS mutant and by addition of synthetic AI-2 at very low concentrations (57).
In Streptococcus mutans, knockout of the luxS gene was shown to impair biofilm growth, as observed by scanning electron, light, and fluorescence microscopy (50, 78, 82). Moreover, a luxS-deficient strain exhibited reduced tolerance to acid killing, an important virulence trait, while it was more resistant against hydrogen peroxide and overexpressed the molecular chaperones GroEL and DnaK (78). The luxS gene was also shown to be involved in controlling the production of the lantibiotic mutacin I in S. mutans UA140 (49). However, at the moment there is no molecular mechanism described for sensing and responding to AI-2 in S. mutans or, to the best of our knowledge, in any gram-positive bacterium, with the exception of the ribose binding protein RbsB, a homologue of LuxP from Vibrio harveyi, which contributes to the internalization of AI-2 in the oral pathogen Actinobacillus actinomycetemcomitans (28).
The LuxS enzyme (S-ribosyl-L-homocysteinase) has a dual function. It carries out the second step of transforming the toxic intermediate S-adenosyl-L-homocysteine to homocysteine. This reaction is part of the central activated methyl cycle of the cell, which provides methyl groups to RNA, DNA, certain metabolites, and proteins (74, 79). As a by-product of the LuxS-catalyzed conversion, 4,5-dihydroxy-2,3-pentanedione (DPD) is produced, from which at least two compounds are formed spontaneously in solution, which are collectively called AI-2 (11, 51). In referring to this second function, the LuxS enzyme is also called AI-2 synthase. In genetic knockout of the luxS gene, both functions are impaired. Therefore, the phenotype of luxS mutants is pleiotropic and cannot a priori be ascribed to quorum-sensing-related signaling. Genetic complementation of a luxS-deficient strain also restores both functions of the LuxS enzyme. However, chemical complementation, i.e., addition of the synthetic pure autoinducer molecule to the culture medium of a null mutant for the signal synthase, in combination with whole-genome transcriptome analysis, allows identification of genes which are regulated by signaling and has been successfully performed to identify the quorum-sensing regulon in Pseudomonas aeruginosa (60) or Streptococcus pneumoniae (52).
Here we used this approach to identify AI-2-regulated genes in S. mutans. We report a genomewide transcriptome analysis of a luxS-deficient strain of S. mutans UA159 throughout growth both in the absence and in the presence of chemically synthesized AI-2 in comparison to the wild-type transcriptome. The data show that the expression of 30% of all genes of S. mutans was affected by the luxS mutation. Genes involved in biofilm formation, acid tolerance, bacteriocin synthesis, and oxidative stress tolerance were among them. The expression of these genes could not be restored by AI-2, and thus, they were not quorum sensing regulated. Their differential expression likely resulted from the disruption of the activated methyl cycle, in which the LuxS enzyme is an essential component. At the same time, we were able to identify 59 genes, among them transcriptional regulators and membrane transporters, whose transcription was clearly induced by AI-2. The data show that AI-2 elicits a distinct cellular response in S. mutans and indicate for the first time which proteins might be used for its uptake and for mediating its regulatory effect. However, the deep changes in the transcriptome of a luxS null mutant which are not quorum sensing regulated make it unsuitable for studying the role of AI-2 signaling and call for the development of novel experimental strategies to this end.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Microarray experiments were performed with the wild-type S. mutans strain UA159 (ATCC 700610) and a luxS null mutant which was constructed by allelic replacement of the luxS gene with an erythromycin resistance cassette using the PCR ligation mutagenesis strategy (39). Briefly, the location of luxS in the genome of S. mutans UA159 was identified using the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov). Approximately 1 kbp upstream and downstream of luxS were amplified using primers P1 and P2 (5′ flanking region) and P3 and P4 (3′ flanking region). Primer P2 contained a restriction site for AscI, and primer P3 had a restriction site for FseI (see Table S1 in the supplemental material for primer sequences). The amplified 5′and 3′ flanking regions were purified using the StrataPrep PCR purification kit (Stratagene) and singly digested with the respective restriction enzyme. The erythromycin resistance cassette was amplified from a plasmid pALN122 using primers pErm-19 and pErm-20, containing the restriction sites for AscI and FseI, respectively (see Table S1 in the supplemental material). The amplified resistance gene was doubly digested with AscI and FseI and then ligated to the amplified 5′ and 3′ flanking regions of luxS. For mutagenesis, wild-type S. mutans UA159 was cultured in Todd-Hewitt broth with yeast extract (THBY) (see below) at 37°C with 5% CO2 for 16 h. Cells were then diluted 1:10 with fresh THBY medium. CSP was added to a final concentration of 1 μg/ml. The DNA construct for allelic replacement was added to a final concentration of 1 μg/ml, and cells were incubated for 3 h at 37°C (5% CO2). Small aliquots were then spread on THBY agar plates containing erythromycin and incubated for 48 h as above. The lack of luxS was confirmed in mutant colonies by PCR and sequencing.
S. mutans strains were grown at 37°C without agitation under anaerobic (80% N2, 10% H2, 10% CO2) or aerobic conditions (with 5% CO2). Media used in this study were brain heart infusion (BHI) (Difco), containing 1% (wt/vol) sucrose when required; THBY (Difco); Luria-Bertani medium (LB) (Difco); and a biofilm medium (BM) slightly modified from that described in reference 43, supplemented with 0.5% (wt/vol) sucrose (BMS). BM contained 58 mM K2HPO4, 15 mM KH2PO4, 10 mM (NH4)2SO4, 35 mM NaCl, 0.2% (wt/vol) Casamino Acids, and 100 μM MnCl2·4H2O (pH 7.4) and was supplemented with filter-sterilized vitamins (0.04 mM nicotinic acid, 0.1 mM pyridoxine HCl, 0.01 mM pantothenic acid, 1 μM riboflavin, 0.3 μM thiamine HCl, and 0.05 μM d-biotin), amino acids (4 mM l-glutamic acid, 1 mM l-arginine HCl, 1.3 mM l-cysteine HCl, and 0.1 mM l-tryptophan), and 2 mM MgSO4·7H2O. All precultures of S. mutans were carried out in THBY medium without agitation at 37°C under anaerobic or aerobic conditions for anaerobic or aerobic cultures, respectively. For microarray experiments, S. mutans was grown at 37°C without agitation under anaerobic (80% N2, 10% H2, 10% CO2) conditions on BMS.
The AI-2 bioasssay was performed using Vibrio harveyi BB170 (Kmr; luxN::Tn5; AI-1 positive; AI-2 positive) (ATCC BAA-1117), V. harveyi BB152 (Kmr; luxLM::Tn5; AI-1 negative; AI-2 positive) (ATCC BAA-1119), and V. harveyi MM77 (Kmr; luxLM::Tn5, luxS::Tn5; AI-1 negative; AI-2 negative). All strains of V. harveyi were grown in autoinducer bioassay (AB) medium according to the method of Greenberg et al. (21) in 100-ml Erlenmeyer flasks with baffles at 30°C and agitation at 160 rpm. For solid media, 2% (wt/vol) agar (Difco) was added prior to autoclaving.
AI-2 bioassay.
Aliquots of 1.5-ml culture were withdrawn at various time points during cultivation, centrifuged (20,000 × g, 5 min, room temperature), and filter sterilized (Aerodisc 13-mm-syringe filter with 0.2-μm Supor Membrane; Pall Corporation, Ann Arbor, MI). The supernatants were immediately used for the AI-2 bioassay or stored at −20°C before use. The bioassay was carried out according to the procedure described by Greenberg et al. (21)and modified by Vilchez et al. (76). Twenty microliters of the test sample and 180 μl of the working dilution of V. harveyi BB170 were incubated in white microtiter plates (Nunc) at 30°C with agitation (650 rpm); luminescence was measured hourly for at least 6 h using a Victor Wallac3 multilabel counter (Perkin Elmer). Cell culture supernatants from V. harveyi BB152 and MM77 served as positive and negative controls, respectively. Chemically synthesized pure AI-2 (13) at a concentration of 0.4 μM (diluted in AB medium) was used as an additional positive control. All experiments were done in triplicate, and for every data point six measurements (technical replicas) were applied. The results, presented as n-fold induction values, were obtained by dividing the luminescence of the samples by the luminescence of sterile AB medium.
Whole-genome microarray of S. mutans UA159.
Each probe of the microarray consisted of an individual 50-mer oligonucleotide. The UA159 whole-genome microarray contained 1,952 oligonucleotides probing S. mutans open reading frames specifically at the genomic level. Five Arabidopsis thaliana sequences served as hybridization controls and for grid finding. Three replicates were immobilized randomly per oligonucleotide and array, and two arrays were placed on one slide. Design and synthesis of the oligonucleotides were performed by MWG Biotech (Ebersberg, Germany).
1957 individual 50-mer oligonucleotides were deposited on CodeLink activated slides (Amersham Biosciences) at a concentration of 25 μM in 1.5× sodium phosphate buffer in a contact-dependent manner using a MicroGrid TAS II spotter (BioRobotics, Freiburg, Germany). All 50-mers were amino modified at the 5′ end, enabling covalent linkage to reactive ester groups provided by the glass surface. Coupling of DNA was ensured by overnight incubation in a saturated sodium chloride chamber, and blocking of residual reactive groups was done as recommended by the manufacturer (Amersham Biosciences Europe, Freiburg, Germany). Until they were used, slides were maintained in a desiccated environment. To ensure complete spotting, SYBR green staining of three randomly selected microarrays of each printing batch was performed as previously described (8).
Sample preparation, hybridization, washing, staining, and scanning.
S. mutans UA159 wild type, a luxS null mutant, and a luxS null mutant complemented with synthetic AI-2 were cultured anaerobically in a defined synthetic medium (BM containing 0.5% sucrose) at 37°C in liquid culture (Ochs Ltd.) without shaking. The planktonic cells were harvested throughout growth at the following optical densities at 600 nm (OD600s): 0.05, 0.1, 0.2, 0.3, 0.5, 0.7, 0.9, 1.2, and 1.5. For the luxS mutant cultures, additional samples were obtained at OD600s of 2.0 and 2.5. Before sampling, culture flasks were agitated thoroughly. Samples were vortexed for approximately 5 s before the OD600 was determined. To obtain the same amounts of RNA from each growth stage, the volume of the cultures withdrawn for RNA extraction ranged from 200 ml (OD600 = 0.05) to 2.5 ml (OD600 > 1.2), resulting in approximately 600 mg of cell mass for each OD. The chemical complementation of the luxS mutation was carried out by two additions of DPD synthesized according to the method described by DeKeersmaecker (13), one at an OD600 of 0.05 to cover the early logarithmic phase of growth and the second one at an OD600 of 0.9 to cover the late logarithmic phase of growth. The DPD half-life in sterile BMS under anaerobic conditions was determined to be about 5 h (see Fig. S1 in the supplemental material). From a stock solution (36 mM), an aliquot was added directly to the culture at an OD600 of 0.05 to a calculated final concentration of 75.6 μM, and the same amount was added additionally at an OD600 of 0.9. The cells were collected immediately after the addition of AI-2 both times. Samples were fixed using RNA Protect bacterial reagent (Qiagen, Hilden, Germany). After centrifugation (10,000 rpm, 4°C, 10 min), the resulting cell pellets were stored at −70°C or applied for RNA extraction.
For cell lysis, lysozyme (2.5 mg/ml culture pellet) (Sigma-Aldrich, Taufkirchen, Germany) and mutanolysin (50 U/ml culture pellet) (Sigma-Aldrich, Taufkirchen, Germany) were added. After incubation at 37°C (15 min), sterile, acid-washed glass beads (diameter, 106 μm) were added and vortexed for 3 min. DNA-free RNA was extracted using the RNeasy minikit according to the manufacturer's protocol (Qiagen, Hilden, Germany). The quality and integrity of the total RNA was controlled by running all samples on an Agilent Technologies 2100 bioanalyzer (Agilent Technologies, Waldbronn, Germany). About 35 to 75 μg RNA was obtained per culture pellet.
For cDNA synthesis, the RNA was transcribed using SuperScript II reverse transcriptase and random hexamers (1.125 μg per reaction; Invitrogen). The cDNA products were then fragmented by DNase I and labeled with terminal transferase in the presence of biotin-ddUTP to biotinylate cDNA at the 3′ termini as described previously (http://www.affymetrix.com/support/technical/manual/expression_manual.affx, section 3: Prokaryotic sample and array processing).
Samples were hybridized to an individual S. mutans UA159 chip for 16 h at 42°C using a Lucidea Slidepro instrument (Amersham Biosciences). After hybridization, the microarrays were washed as recommended by the manufacturers (CodeLink expression bioarray system, Amersham Biosciences), stained with Cy5-streptavidin (Amersham Biosciences), and read using an array WoRx biochip reader (Applied Precision, LLC, Washington, DC).
Data analysis.
Raw signal intensities were quantified by means of Imagene software v5.5.2 (BioDiscovery, Inc., Los Angeles, CA). Raw signal intensities were further analyzed using the Array Assist 4.0 software package (Stratagene Europe, Amsterdam, The Netherlands). Before normalization, each individual array spot was pruned to exclude bad and empty spots flagged by the Imagene software. Normalization was performed using the LOWESS algorithm. For each time point, two biological and two technical replicates were measured, resulting in a maximum of 12 gene expression values per gene per time point. Signal intensities were averaged among the technical replicates. Two types of data analysis were performed. To identify density-dependent changes in gene expression, single averaged normalized signal intensities for each time point were compared to the averaged normalized signal intensity at time zero (OD600 = 0.05). Genes which showed a relative signal log2 ratio value above 1.0 at any time point during growth were selected for further analysis. k-means clustering was applied to organize and visualize expression profiles of the selected data set. In a second analysis, genes which were differentially expressed between the wild type and the luxS mutant and between the luxS mutant with and the luxS mutant without addition of AI-2 were detected using two-class paired significance analysis of microarrays (TCPSAM) for OD600s of 0.1, 0.7, 1.0, and 1.6 for the wild type and OD600s of 0.1, 0.7, 0.9, and 2.0 for the luxS mutant with and without AI-2 using the software program SAM 3.0 (71), downloaded from http://www-stat-class.stanford.edu/∼tibs/clickwrap/sam.html. The entire data set in a MIAME (minimal information about a microarray experiment)-compliant format and an array description file are available at the public GEO database under http://www.ncbi.nlm.nih.gov/geo/under accession number GSE5451.
The functions of the genes of interest were extracted from various databases, such as NCBI (http://www.ncbi.nlm.nih.gov), TIGR (http://www.tigr.org), European Bioinformatics Institute (http://www.ebi.ac.uk), ExPASy Proteomics Server (http://www.expasy.org), KEGG (http://www.genome.jp), and Los Alamos Oral Pathogens Database (http://www.oralgen.lanl.gov).
Real-time PCR.
Real-time PCR was performed using the Rotor-Gene cycler system (Roche) and a Quantitect SYBR green PCR kit (Qiagen). Primers were designed by using Primer 3 software (http://frodo.wi.mit.edu) to generate amplicons ranging from 100 to 170 bp in size (see Table S1 in the supplemental material). Real-time PCR was performed in triplicate in 20 μl of the reaction mixture containing 10 μl of 2× Quanti Tec SYBR green PCR master mix, 1 μl of primers (0.5 μM), and 5 ng cDNA. The reaction mixture without a template was run as a control. The cycling conditions were as follows: 95°C for 15 min, followed by 40 cycles of three steps consisting of denaturation at 94°C for 15 s, primer annealing at the optimal temperature for 30 s, and the primer extension at 72°C for 30 s. For each set of primers, the cycle threshold values (crossing point [CP]), defined as the first cycle showing an amount of PCR product above the background level, was determined. The relative expression based on the expression ratio between the target gene and a reference gene was calculated. Using a mathematical model, the target gene expression was normalized to the gene expression of a nonregulated reference gene using the software tool REST (relative expression software tool) (53). As a reference gene, SMU.1114 (gyrA) was chosen, which showed a constant low level of expression throughout growth and has also been used by other authors (5). The mean CP of the genes, the CP variation, and the coefficient of variation (CV) were calculated to determine reproducibility and variation. Linearity and amplification efficiency were determined for each primer pair, and only values corresponding to high amplification efficiency in the exponential range were used. Every real-time PCR was performed at least three times. The expression values determined in the microarray were confirmed by quantitative PCR for several genes, which were selected to represent both up- and downregulated genes from all three investigated cultures (see Table S2 in the supplemental material).
RESULTS
Phenotype of the luxS mutant.
When cultivated under the conditions used in the microarray analysis (planktonic culture in BM, 0.5% sucrose, anaerobic incubation at 37°C without shaking), wild-type S. mutans UA159 started to aggregate and to form flocs after reaching an OD600 between 0.1 and 0.2. By contrast, the luxS-deficient strain did not visibly aggregate. It produced only small flocs which were easily resuspended. When grown on glass slides, it formed a thinner and more diffuse biofilm than the wild type (see Fig. S2 in the supplemental material). AI-2 was not produced by the luxS mutant. In the wild type, the luxS gene, responsible for AI-2 synthesis, and the pfs gene, coding for the nucleosidase enzyme performing the preceding step in the activated methyl cycle (74), were expressed at a low but constant rate throughout growth (Fig. 1); thus, DPD, the precursor of AI-2, must have been formed. However, AI-2 activity detected in the V. harveyi bioassay was at background level (around 1% of positive controls) using cell supernatants of anaerobic or aerobic cultures of S. mutans during the entire growth curve in the following media: BHI, THBY, LB, BHI containing 1% (wt/vol) sucrose, and BMS (0.5% or 1% of sucrose) (data not shown). When cell culture supernatants from S. mutans were added to positive controls containing chemically synthesized AI-2, the recovery of AI-2 activity was strongly reduced compared to results for controls without added S. mutans culture supernatants (see Fig. S3 in the supplemental material). This effect increased with the OD of the culture under both aerobic and anaerobic conditions. It was not caused by a change in pH, since the medium was buffered and thus during growth the pH decreased only slightly, to pH 6.5. Since AI-2 was also not detected in media which did not contain glucose, which might be produced by S. mutans from sucrose during growth and is known to be a strong inhibitor of V. harveyi luminescence (14, 70), we conclude that glucose was not responsible for the lack of detection of AI-2. For our experiment, it is important that the luxS gene was expressed at a constant rate in the wild type and was absent in the mutant. Since AI-2 is hypothesized to be an interspecies signaling molecule, the chemically synthesized AI-2 added to the luxS mutant culture mimicked the signal produced by diverse luxS-harboring microorganisms in the oral cavity.
FIG. 1.
Expression of luxS and pfs in Streptococcus mutans UA159 during anaerobic growth on BMS.
Overview of microarray analysis.
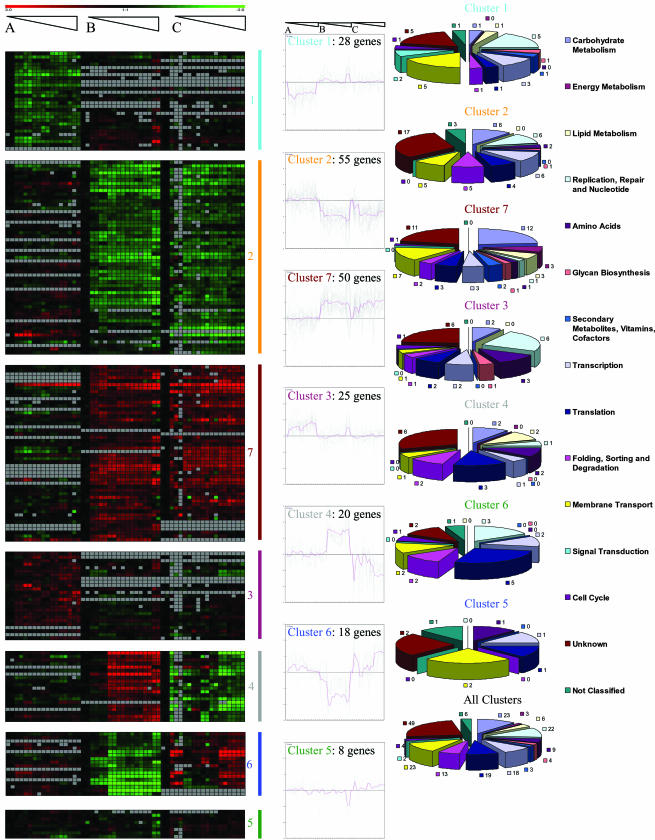
The total transcriptome of S. mutans UA159 was analyzed for the wild type, a luxS mutant, and a luxS mutant complemented with synthetic AI-2 in a density-dependent manner to identify possible quorum-sensing-related genes. To this end, the expression of each gene was normalized to its expression at time zero, corresponding to an OD600 of 0.05. With a cutoff value of 2 for the n-fold change of expression (P < 0.05), we found 204 genes of all 1,952 genes present in the microarray to be differentially regulated in a density-dependent way, 10.4% of the total genome. The regulated genes were then sorted into seven clusters according to their transcription pattern in the three treatments (Fig. 2,left and center) and finally assigned to their main functional categories (Fig. 2, right). Transcription patterns in clusters 1, 2, 3, and 7 show 158 genes which were up- or downregulated during growth in the luxS mutant but which were unaffected by addition of AI-2 and retained their luxS type expression level. Thus, these genes can be assumed to be responding to the metabolic changes resulting from the knockout of the luxS gene (see Table S3 in the supplemental material). Clusters 4 and 6 show 38 genes which were up- or downregulated in the luxS mutant during growth but which upon addition of AI-2 resumed their wild-type pattern of expression. Since AI-2 is not known to be an intermediate metabolite, these genes can therefore be assumed to be responding to the signaling molecule AI-2 (Table 1). Finally, cluster 7 shows 8 genes whose density-dependent regulation was very small in all three cases studied (see Table S3 in the supplemental material).
FIG. 2.
Density-dependent cluster analysis of significantly changed genes in S. mutans UA159. Left column: A, wild type; B, luxS null mutant; C, luxS null mutant complemented with AI-2; each square represents one time point. The change of gene expression compared to that at an OD600 of 0.05 is indicated by color: green, downregulated; red, upregulated; black; below cutoff; gray, expression at background level (not detectable [ND]). Middle column: gene expression for clusters 1 to 7 throughout growth for cultures A, B, and C. Right column: pathway assignment of the genes in each cluster and of all clusters taken together.
TABLE 1.
AI-2-regulated genes (density-dependent analysis)a
| Cluster | Gene | Description | Fold change in expressionb
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
S. mutans wild type
|
luxS mutant
|
luxS mutant with Al-2 added
|
|||||||||
| Avg | Minimum | Maximum | Avg | Minimum | Maximum | Avg | Minimum | Maximum | |||
| 4 | SMU.53 | Conserved hypothetical protein | 1,471 | −1,000 | 1,879 | 1,954 | −1,424 | 3,941 | −1,030 | −2,466 | 2,063 |
| SMU.74 | Conserved hypothetical protein | −1,075 | −1,807 | 1,409 | 5,724 | −1,087 | 24,991 | −3,562 | −16,502 | 1,098 | |
| SMU.82 | Heat shock protein, DnaK (HSP-70), dnaK | −1,373 | −2,052 | 1,122 | 1,458 | −1,179 | 2,559 | −2,914 | −5,675 | 1,014 | |
| SMU.153 | Hypothetical protein | −1,216 | −1,881 | 1,135 | 1,792 | −1,132 | 3,606 | −1,260 | −2,180 | 1,439 | |
| SMU.170 | 30S ribosomal protein S9 | NDc | ND | ND | 2,591 | −1,117 | 7,716 | −1,033 | −3,187 | 4,252 | |
| SMU.188c | Putative 33-kDa chaperonin (heat shock protein 33) | 1,144 | −1,593 | 1,815 | ND | ND | ND | −2,491 | −12,862 | 1,055 | |
| SMU.197c | Hypothetical protein | −1,432 | −1,916 | 1,093 | 1,679 | −1,117 | 3,978 | −2,544 | −7,726 | 1,148 | |
| SMU.357 | 30S ribosomal protein S12 | ND | ND | ND | 5,549 | −1,000 | 15,947 | −2,465 | −9,457 | 1,121 | |
| SMU.422 | Ribosome binding factor A | −1,237 | −2,059 | 1,244 | 1,643 | −1,404 | 3,045 | −2,520 | −6,866 | 1,219 | |
| SMU.436c | Putative transposase, ISSmu1 | ND | ND | ND | 1,558 | −1,347 | 4,292 | −2,204 | −7,655 | 3,710 | |
| SMU.522 | Conserved hypothetical protein | −1,290 | −1,796 | 1,214 | 1,662 | −0,998 | 2,719 | −1,175 | −2,188 | 1,927 | |
| SMU.643 | Putative esterase | 1,173 | −1,098 | 1,409 | 1,441 | −1,275 | 2,921 | −1,297 | −2,356 | 1,517 | |
| SMU.750c | Hypothetical protein | 1,543 | −1,007 | 2,615 | 2,359 | −0,999 | 5,337 | −1,595 | −3,799 | 1,360 | |
| SMU.798c | Hypothetical protein | 1,120 | −1,153 | 1,583 | 1,924 | −1,376 | 4,867 | −1,503 | −3,287 | 1,207 | |
| SMU.1589c | Putative hexosyltransferase | −1,043 | −1,532 | 1,309 | 2,363 | −1,240 | 7,787 | −1,325 | −2,887 | 1,357 | |
| SMU.1667 | Putative branched chain amino acid ABC transporter, permease protein, livM | −1,221 | −2,266 | 1,202 | 1,784 | −1,903 | 2,893 | −1,616 | −8,290 | 1,114 | |
| SMU.1741 | Putative malonyl-CoA (acyl-carrier-protein) transacylase, fabD | −1,046 | −1,269 | 1,149 | 2,535 | −1,176 | 4,645 | −1,416 | −2,342 | 1,480 | |
| SMU.2114c | Putative transcriptional regulator | 1,136 | −1,042 | 1,647 | 1,806 | −1,414 | 5,562 | −1,604 | −4,275 | 1,290 | |
| SMU.2153c | Putative peptidase | −1,197 | −1,736 | 1,231 | 1,569 | −1,101 | 2,481 | −1,329 | −2,074 | 1,110 | |
| SMU.2154c | Putative peptidase | −1,267 | −2,842 | 1,220 | 1,824 | −1,206 | 4,450 | −1,524 | −4,096 | 1,399 | |
| 6 | SMU.15 | Putative cell division protein FtsH, ftsH | 1,010 | −1,275 | 1,257 | −6,112 | −17,995 | 1,055 | 1,692 | −2,080 | 9,377 |
| SMU.31 | Hypothetical protein | −2,056 | −3,010 | 1,000 | −1,802 | −2,881 | 1,000 | 1,272 | −1,312 | 2,091 | |
| SMU.44 | Conserved hypothetical protein; possible DNA mismatch repair protein | 1,288 | −1,107 | 1,958 | −1,529 | −2,146 | 1,275 | 1,836 | −1,000 | 3,865 | |
| SMU.48 | Putative phosphoribosyl glycinamide synthetase, purD | ND | ND | ND | −2,225 | −5,274 | 1,434 | 3,975 | −1,076 | 20,928 | |
| SMU.97 | ATP synthetase (UTP-ammonia lyase), pyrG | 1,271 | −1,079 | 1,974 | −11,305 | −57,739 | 1,919 | ND | ND | ND | |
| SMU.135 | Putative transcriptional regulator, mleR | 1,371 | −1,072 | 2,331 | −2,795 | −4,568 | 1,041 | 1,864 | −1,709 | 9,661 | |
| SMU.184 | Putative ABC transporter, metal binding lipoprotein; (Lral family), sloC | 1,346 | −1,329 | 2,895 | −2,045 | −4,889 | 1,647 | ND | ND | ND | |
| SMU.341 | Putative deoxyribonuclease | 1,345 | −1,079 | 2,395 | −2,407 | −3,948 | 1,011 | 1,009 | −2,112 | 2,000 | |
| SMU.391c | Conserved hypothetical protein | −1,521 | −2,253 | 1,068 | −1,861 | −4,504 | 1,729 | 2,408 | −1,097 | 7,508 | |
| SMU.408 | Putative permease | 1,389 | −1,035 | 2,042 | −3,746 | −6,949 | 1,119 | 1,809 | −1,271 | 7,328 | |
| SMU.818 | 30S ribosomal protein S21 | 1,066 | −1,741 | 1,752 | −1,711 | −3,182 | −1,008 | 1,631 | −1,549 | 3,470 | |
| SMU.1168 | Putative transcriptional regulator | −1,152 | −2,803 | 1,652 | −2,180 | −9,190 | −1,001 | 1,605 | −1,731 | 3,337 | |
| SMU.1646c | Conserved hypothetical protein, possible hemolysis-inducing protein | −1,247 | −2,211 | 1,187 | −1,709 | −4,734 | 1,093 | 1,445 | −1,999 | 2,961 | |
| SMU.1847 | Putative translation elongation factor P, efp | 1,058 | −1,287 | 1,490 | −1,462 | −3,052 | 1,042 | 1,186 | −1,442 | 1,682 | |
| SMU.1954 | Putative chaperonin GroEL, groEL | ND | ND | ND | −2,671 | −5,386 | 1,000 | 1,154 | −2,264 | 2,230 | |
| SMU.2007 | 50S ribosomal protein L15, rl15 | ND | ND | ND | −1,423 | −2,458 | −1,003 | 1,614 | −1,947 | 3,127 | |
| SMU.2011 | 50S ribosomal protein L6 (BL10), rl6 | 1,211 | −1,094 | 2,282 | −1,614 | −2,571 | −1,001 | 1,274 | −3,334 | 2,329 | |
Thirty-eight genes were found to be significantly up- or downregulated (Fig. 2, clusters 4 and 6) in the luxS mutant during growth and reverted to the wild-type pattern of expression upon addition of AI-2. A gene appears in this list if its expression was changed more than twofold for at least one time point during growth in any of the three experimental treatments.
Avg, average value for up to six technical replicates. Genes which were also detected in the TCPSAM analysis (see Table 2) and n-fold change in values above the cutoff of 2.0 are shown in bold.
ND, not detectable (expression of gene at background level).
A second analysis was carried out to detect genes which were differentially expressed in the luxS mutant compared to the wild type or in the luxS mutant with and without addition of AI-2 but whose rate of expression was not necessarily changed during growth. These genes would therefore escape the density-dependent analysis described above. TCPSAM was used to this end, using a cutoff value for the n-fold change of expression of 2.0 and a Δ value of 0.25 (<1 false positives; q < 0.0001). A total of 453 genes of all 1,952 genes present in the microarray (25.0%) were found to be differentially expressed in the luxS mutant compared to the expression in the wild type and did not respond to AI-2 (see Table S4 in the supplemental material). In addition, 35 genes were significantly changed between the luxS mutant and the luxS mutant complemented with AI-2 and thus were regulated by AI-2. Of these, six genes displayed their wild-type level of expression upon addition of AI-2. For the remaining 29 genes, their expression in the luxS mutant was similar to that in the wild type or greater and was increased further by addition of AI-2 (Table 2).
TABLE 2.
AI-2-regulated genes (TCPSAM analysis)a
| Genec | Description | Fold change in expressionb |
|---|---|---|
| SMU.96 | Putative DNA-directed RNA polymerase, delta subunit, rpoE | 147.42 |
| SMU.234 | Threonine dehydratase, livA | 8.21 |
| SMU.2032 | 30S ribosomal protein S2, rs2 | 4.40 |
| SMU.1665* | Putative branched-chain amino acid ABC transporter, ATP-binding protein, livF | 4.04 |
| SMU.2020* | 50S ribosomal protein L16, rl16 | 3.86 |
| SMU.1168* | Putative transcriptional regulator | 3.39 |
| SMU.2001 | DNA-directed RNA polymerase, alpha subunit, rpoA | 3.33 |
| SMU.1816c | Putative maturase-related protein | 3.07 |
| SMU.848 | Conserved hypothetical protein | 3.06 |
| SMU.188c | Putative 33-kDa chaperonin (heat shock protein 33) | 3.00 |
| SMU.358 | 30S ribosomal protein S7 | 2.98 |
| SMU.2003 | 30S ribosomal protein S13, rs13 | 2.80 |
| SMU.05 | Conserved hypothetical protein | 2.79 |
| SMU.2026c | 30S ribosomal protein S10 | 2.79 |
| SMU.1667* | Putative branched-chain amino acid ABC transporter, permease protein, livM | 2.77 |
| SMU.1858 | 30S ribosomal protein S18, rs18 | 2.67 |
| SMU.2002 | 30S ribosomal protein S11, rs11 | 2.63 |
| SMU.1615c | Conserved hypothetical protein | 2.55 |
| SMU.2000 | 50S ribosomal protein L17, rl17 | 2.48 |
| SMU.103* | Putative PTS system, IIA component | 2.46 |
| SMU.958 | Hypothetical protein | 2.41 |
| SMU.361 | Phosphoglycerate kinase, pgk | 2.36 |
| SMU.2016 | 50S ribosomal protein L24, rl24 | 2.27 |
| SMU.1860 | 30S ribosomal protein S6, rs6 | 2.26 |
| SMU.2008 | 50S ribosomal protein L30, rl30 | 2.24 |
| SMU.697 | Putative translation initiation factor IF3 | 2.19 |
| SMU.2009 | 30S ribosomal protein S5, rs5 | 2.16 |
| SMU.990* | Putative dihydrodipicolinate synthase, dapA | 2.12 |
| SMU.2153c | Putative peptidase | 2.08 |
| SMU.1955* | Putative cochaperonin GroES, groES | 2.06 |
| SMU.741 | Conserved hypothetical protein | 2.06 |
| SMU.2014 | 30S ribosomal protein S14, rs14 | 2.05 |
| SMU.357 | 30S ribosomal protein S12 | 2.01 |
| SMU.1511c | Putative acetyltransferase | −2.02 |
| SMU.750c | Hypothetical protein | −2.11 |
Thirty-five genes were found to have significantly changed expression in the luxS mutant complemented with AI-1 from those in the luxS mutant without AI-2 (cutoff for the n-fold change of expression of 2.0, Δ = 0.25; rate of false positives, 0.8 per 100 genes).
Average n-fold change in expression for up to six technical replicates.
Asterisks indicate genes in which AI-2 restored the wild-type level of expression. Genes displayed in bold were also detected in the density-dependent analysis (see Table 1).
If the two analyses are taken together, a total of 585 genes were found to be affected by the luxS mutation (30.0% of the genome), i.e., they were differentially expressed in the luxS mutant culture and did not revert to the wild type in the AI-2-complemented culture. Of these, 107 genes were detected in both analyses. Genes affected by the luxS mutation were distributed in all metabolic pathways (Fig. 3). Unknown or unclassified genes contributed the largest fraction, followed by genes required for membrane transport, carbohydrate metabolism, folding, sorting and degradation, translation, transcription, amino acid synthesis, replication, and repair. Important effects were also observed for signal transduction-, cell cycle-, and glycan biosynthesis-related genes. The density-related analyses revealed both up- and downregulated genes, while in the TCPSAM analysis the majority of luxS-affected genes were downregulated.
FIG. 3.
Breakdown of genes affected by the luxS mutation according to functional categories. “luxS influenced genes” refers to genes whose expression was not restored by added AI-2; “AI-2 influenced genes” refers to genes whose expression was restored upon addition of AI-2. For each group, both TCPSAM analysis and density-dependent analysis are shown.
A total of 59 genes (3% of the genome) were found which were differentially expressed in the luxS mutant but whose expression responded to the addition of AI-2. They can thus be assumed to be regulated by the signaling molecule AI-2. Of these, 12 genes were found in both analyses. In summary, the luxS mutation induced deep changes in the transcription of many genes, and after addition of AI-2, the majority of them did not restore the transcription level observed in the wild-type strain. However, a number of genes were discovered which were regulated by AI-2.
LuxS-dependent changes in gene expression in S. mutans UA159.
Table S3 in the supplemental material shows the list of 166 genes which were up- or downregulated in the luxS mutant during growth and did not restore the wild-type gene expression pattern upon addition of AI-2 (abbreviated LuxS-affected genes, density-dependent cluster analysis). Table S4 in the supplemental material shows the list of 453 genes which were differentially expressed in the luxS mutant compared to wild-type expression and did not restore the wild-type expression level upon addition of AI-2 (LuxS-affected genes, TCPSAM analysis). We will here focus on the most strongly affected genes or on those which are known to be related to biofilm growth and virulence properties of S. mutans.
(i) Membrane transport.
ABC transporters form the largest group of paralogous genes in bacterial genomes, and they were most strongly affected by the luxS mutation. Changes in the transcription of more than 40 ABC transporters were induced which could not be restored by addition of AI-2. The most strongly affected transporter was SMU.238c, which was downregulated 108-fold. It is predicted to encode an ATP binding protein, the most conserved component of an ABC transport system with unknown function. The membrane component of this ABC transporter, SMU.237c, was recently found to be significantly downregulated in strain UA159 grown as biofilm compared to results with planktonic growth (65).
(ii) Bacteriocins and bacteriocin immunity.
The second most strongly affected gene was SMU.2035, encoding a putative bacteriocin immunity protein, which was downregulated 101.2-fold and was also discovered in the density-dependent analysis. The genome sequence of S. mutans UA159 encodes at least 12 putative bacteriocins (4, 47, 73), of which the nlmB gene (SMU.151), which together with nlmA forms the two-component mutacin IV (54), was downregulated 25.8-fold and is most active against Streptococcus sanguinis and Streptococcus mitis (36). In addition, the newly described mutacin V (SMU.1914c, nlmC), which is active against nonstreptococcal species (24), was down regulated 2.0-fold. Moreover, the bsmI gene (SMU.1895c) and the gene encoding the CSP (SMU.1915) were downregulated 2.0- and 2.3-fold, respectively. The nlmA gene, which showed the greatest antimicrobial activity of S. mutans UA159, even without nlmB (24), was below our cutoff criteria. Only two putative bacteriocins, SMU.1896c (bsmH; +2.8-fold) and SMU.1905 (bsmL) were upregulated in the luxS mutant.
(iii) Acid tolerance.
Strongly downregulated was aguA (SMU.264; −72.97-fold), encoding the agmatine deiminase enzyme, which is used for generating ammonia to reduce the intracellular pH in less acid tolerant streptococci (22).
(iv) Cell envelope.
The gene pbp2b, encoding a penicillin binding protein, was also strongly downregulated (SMU.597; −38.48). Since these proteins are responsible for the assembly, maintenance, and regulation of the cell wall component peptidoglycan, disruption of their expression can affect osmoadaptation, which is important during sessile growth (42). Mutation of pbp2b in Streptococcus gordonii resulted in reduced biofilm formation (9).
(v) Local and global transcriptional regulators.
Genes belonging to the transcriptional regulation category composed a big fraction of the luxS-affected genes, with 39 transcriptional regulators changed due to the lack of luxS. In addition, several genes connected with global transcriptional control were found: the luxS mutant showed a 24-fold downregulation of the rpoD gene (SMU.822) encoding the sigma factor σA. Catabolite control protein A (SMU.1591) was downregulated 2.4-fold.
(vi) Glucan biosynthesis.
Glucan plays a central role in the sucrose-dependent biofilm formation of S. mutans. Soluble and insoluble glucans are synthesized by glycosyltransferases (encoded by gtf genes) and form the main component of the sticky extracellular polysaccharide matrix. In the luxS mutant, the expression of the glycosyltransferase gene gtfD, responsible for the synthesis of soluble glucan, was slightly increased (SMU.910; +2.0-fold) while all other glycosyltransferases were unchanged. The biofilm architecture is strongly affected by the membrane-bound glucan binding proteins (encoded by gbp genes), which link the bacteria with extracellular glucan. The lack of any of them has been shown to reduce biofilm depth and even to influence cariogenicity (45, 46). In the luxS mutant, two of the four glucan binding proteins were expressed differentially. Glucan binding protein C, which binds soluble glucan, was upregulated (SMU.1396; gbpC; +4.2-fold), and glucan binding protein A was strongly downregulated (SMU.2112; gbpA; −7.8-fold). The repressor vicX, which is part of the VicRK signal transduction system (63), was downregulated (SMU.1515; vicX; −2.5-fold). A deletion mutant of vicX was shown to be more adhesive because of increased expression of gtfB and gtfC but was also impaired in competence and stress tolerance (64). Thus, contrasting changes in genes related to glucan synthesis and binding were observed.
(vii) Uptake and metabolism of sugars.
The phosphoenolpyruvate phosphotransferase system (PTS) is the most important sugar uptake mechanism in S. mutans (16, 72). Many components of PTSs were weakly downregulated, between twofold and threefold (SMU.1960c, SMU.313, SMU.271, SMU.1958c, and SMU.1600), while one mannose-specific component (SMU.1878, ptnC; +2.5-fold) and one fructose-specific component (SMU.872, +4.0-fold) was upregulated. The nonspecific energy coupling HPr component of PTSs was also strongly upregulated (SMU.674, ptsH; +8.6-fold). Internalized sugars are subsequently metabolized through glycolysis to pyruvate. A gene encoding a putative fructokinase enzyme initiating this metabolic route was strongly downregulated (SMU.1840, scrK; −12.5-fold), while SMU.1191 (pfkA; 6-phosphofructokinase) was upregulated 5.1-fold.
AI-2-dependent changes in gene expression in S. mutans UA159.
Genes which responded to the addition of AI-2 by a significant change in expression are listed in Table 1 (density-dependent analysis) and Table 2 (TCPSAM analysis). Genes in cluster 4 were upregulated in the luxS mutant during growth, while genes in cluster 6 were downregulated (Table 1). Addition of AI-2 restored the wild-type pattern of expression according to the clustering procedure, and thus, this molecule could induce both downregulation and upregulation. Of the AI-2 regulated genes found in the TCPSAM analysis (Table 2), 33 were upregulated in comparison to results for the luxS mutant and two were downregulated.
(i) Protein synthesis and stress-related genes.
Table 1 shows that except for a large fraction of hypothetical proteins, many of the AI-2-regulated genes were related to protein synthesis, protein degradation, DNA synthesis and repair, and general stress response. These effects might be indirect and result from an interaction of AI-2 with one or several regulatory elements responsible for these processes.
(ii) Global transcriptional regulators.
The largest change in gene expression which was observed in the whole study was for SMU.96, encoding the δ subunit of the RNA polymerase (RpoE), whose expression was stimulated 147-fold by AI-2 (Table 2). The wild type expressed this gene at a low level throughout growth, while the luxS mutant showed a pronounced peak of expression at the beginning of the exponential growth phase, followed by low-level transcription subsequently. In addition, the α subunit of the RNA polymerase (SMU.2001; RpoA) also was upregulated 3.3-fold in the AI-2-supplemented culture.
(iii) Transcription factors.
The density-dependent analysis of the array data revealed three different types of transcriptional regulators which responded directly to AI-2. Two of them were induced (SMU.135 and SMU.1168) (Table 1, cluster 6) and one was repressed (SMU.2114c) (Table 1, cluster 4) by AI-2; SMU.1168 was also detected by the TCPSAM analysis (Table 2).
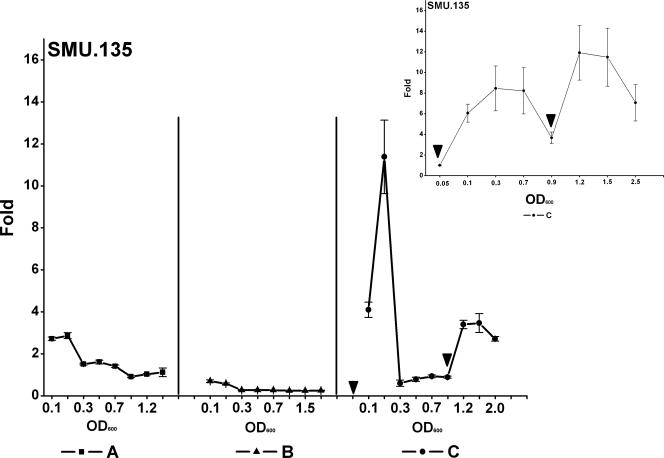
The transcriptional regulator SMU.135 showed an interesting density-dependent pattern of expression (Fig. 4). In the wild type, SMU.135 was upregulated at the beginning of growth, and during the logarithmic phase its expression decreased continuously. In the luxS mutant, the expression of the gene was very low and decreased right from the beginning of growth. Addition of AI-2 resulted in an immediate increase in the expression of SMU.135, both at an OD600 of 0.05 and at an OD600 of 0.9, suggesting that this transcription factor may have been regulated directly by AI-2. Real-time PCR confirmed the induction of SMU.135 expression after addition of AI-2 to the culture medium (Fig. 4).
FIG. 4.
Expression of gene SMU.135 in S. mutans UA159 during a complete growth curve. SMU.135 is a LysR-type transcriptional regulator which is induced by AI-2. Means and standard deviations of normalized spot intensities for up to six technical replicas are given. A, wild type; B, luxS mutant; C, luxS mutant complemented with AI-2. Arrows indicate addition of AI-2. Inset shows real-time PCR quantification of gene expression for treatment C.
The transcriptional regulator SMU.1168 was 3.4-fold upregulated in the AI-2-complemented luxS mutant culture (Table 2). Its expression was correlated to cell density, since it was also discovered in the time zero normalized data (Table 1). Maximum expression in the wild type was observed at the late logarithmic phase of growth.
The transcriptional regulator SMU.2114c was expressed at a constant rate throughout growth in the wild type and was strongly induced in the luxS mutant. Upon addition of AI-2, the wild-type pattern of expression was restored.
(iv) Membrane transport.
The expression of four transport proteins was induced by addition of AI-2, namely, SMU.103, SMU.408, SMU.1665, and SMU.1667. SMU.408 is a putative permease which was strongly upregulated well beyond the wild-type level of expression immediately after each addition of AI-2. Two other components of a membrane transporter which were regulated by AI-2 were SMU.1667 (livM), which was 2.8-fold upregulated, and SMU.1665 (livF), which was 4.0-fold upregulated. They are annotated to encode the ATP-binding component (encoded by livF) and the permease component (encoded by livM) of an ABC transporter for branched-chain amino acids. Finally, expression of SMU.103 encoding the IIA component of a putative PTS was upregulated 2.5-fold upon addition of AI-2. These data suggest that there are several transporters which are regulated by AI-2 and which might be specific for different variants of the AI-2 group of molecules.
DISCUSSION
Phenotype of the luxS mutant.
Four other luxS mutants of S. mutans have been previously studied, one in a UA159 background (78), two in a GS5 background (50, 82), and one in a UA140 background (49). All authors observed impaired biofilm formation, especially on chemically defined medium with sucrose as a carbon source. No effects or only small effects were observed with glucose as a carbon source or on complex media. Biofilms of the UA159 luxS null mutant growing on hydroxylapatite discs in BMS under CO2-enriched conditions (78) were “loose, hive-like, and had large gaps”, similar to our observations, which were, however, obtained with an oxygen-free atmosphere. The analysis reported here was performed with planktonic culture to determine density-dependent effects. However, the clearly different flocculation behavior of the wild type and the luxS mutant in planktonic culture indicates that traits relevant for biofilm formation were expressed. Since the mutant analyzed here is isogenic to the one constructed by Wen and Burne (78), it can be expected to also be more sensitive to acids and more tolerant to oxidative stress, although these traits were not tested by us.
We consistently found background-level AI-2 activity in sterile culture supernatants of S. mutans UA159, in spite of an active and functional LuxS enzyme, as judged by its constant expression during growth. Similar results were described by Merritt et al. (50) for S. mutans GS-5 and by Joyce et al. (30) for S. pneumoniae: culture supernatants of the wild type could not induce luminescence in V. harveyi BB170, although the respective LuxS enzyme was shown to be functional when expressed in Escherichia coli DH5α. The data suggest the presence of an inhibitor of V. harveyi luminescence or an enzyme which binds, modifies, or degrades AI-2 in the cell culture supernatants of S. mutans (and possibly S. pneumoniae). Candidates for such proteins have been detected in this analysis, e.g., the product of SMU.408, a permease whose transcription was strongly induced upon addition of AI-2, as well as those of SMU.103 and SMU.1667, which encode the IIA component of a PTS and the permease component of an ABC transporter, respectively.
Chemical detection of AI-2 would circumvent many of the problems inevitably associated with a bioassay. The published method has until now been applied only to the high concentrations present during chemical synthesis (13, 48, 62). Its sensitivity does not yet allow determination of in situ concentrations of biological samples reliably.
Genomewide changes in gene expression.
The transcriptome analysis showed that the luxS mutation is immensely pleiotropic, affecting 585 (roughly 30%) of the genes of S. mutans. In E. coli, DeLisa et al. (15) found 242 genes changed more than 2.3-fold in a luxS mutant (5.6% of the genome), and for enterohemorrhagic E. coli, Sperandio et al. reported that 404 genes were differentially expressed in a luxS mutant using a cutoff of 5 (66). The cutoff value of 2.0 is routinely used, e.g., by a recent microarray investigation of sugar transporters in S. mutans (5) or in a genomewide analysis of the role of luxS (30) or comX (52) in S. pneumoniae. The number of differentially expressed genes in the luxS mutant would have dropped to 201 had we used a cutoff value of 3.0. However, by using the cutoff of 2.0, it was possible to detect changes in key genes, e.g., comC or pleotropic regulators, which might be very important for the regulatory network of S. mutans.
Temporal pattern of changes in gene expression.
Quorum-sensing-controlled genes are expected to be expressed in a density-dependent manner, and therefore a growth-phase-dependent analysis of changes in gene expression was conducted. In such a way, Peterson et al. (52) showed that three temporal waves of gene expression were induced by the CSP, termed early, late, and delayed genes, over a time period of 22 min in S. pneumoniae. Our density-dependent analysis of the microarray data revealed no temporal pattern of gene expression of the luxS mutant. Most dysregulated genes responded already at the beginning of growth. These observations are similar to those obtained by Joyce et al. (30) with a luxS mutant of S. pneumoniae and led these authors to conclude that quorum sensing is not the mechanism mediating the differential gene expression seen in the luxS-deficient strain. However, since AI-2 is proposed to act as an interspecies signaling molecule, it could function to inform the recipient cell about the density of luxS-expressing bacteria in the immediate neighborhood rather than about its own cell density. This would be consistent with the constant expression of luxS during growth observed here but would imply that S. mutans can differentiate between its own excreted AI-2 (which is also rapidly taken up or modified) and AI-2 that is produced by neighboring bacteria.
Complementation experiments.
The majority of the differentially expressed genes did not respond to chemically synthesized DPD, among them many genes which are known to be important for biofilm formation. In accordance with the transcriptome data, DPD was not able to cure the altered floc formation behavior and biofilm growth of the luxS mutant (data not shown). Since it was reported that DPD restored interdigitated biofilm growth in a two-species system in a narrow concentration range between 0.08 and 0.8 nM while higher concentrations were inhibitory (57), we added DPD over a range of concentrations (0.04, 0.08, 8, 80, 800, and 8,000 nM at a starting OD600 of 0.05) to cultures of S. mutans, but wild-type floc formation was not obtained. Supplementation of the culture with wild-type sterile culture supernatant (20% or 80%) also did not restore wild-type floc formation.
The timing of added autoinducer might also influence the transcription of quorum-controlled genes. Thus, for Pseudomonas aeruginosa, transcriptome analysis revealed 315 quorum-induced genes and 38 quorum-repressed genes; their induction was strongly dependent on the timing of added autoinducer (59, 60). In our experiment, DPD was added at the beginning of growth (OD600 = 0.05) and at the end of the exponential growth phase (OD600 = 0.9), thus covering all growth stages. The used concentration of DPD is not inhibitory for V. harveyi but is relatively high to account for possible losses in the culture. The half-life of AI-2 in BMS (37°C, anaerobic incubation) is approximately 5 h (see Fig. S1 in the supplemental material), and the extracellular polysaccharides produced by S. mutans might bind and inactivate AI-2. In a microarray analysis of AI-2 regulation in enterohemorrhagic E. coli, Kendall et al. (31) added DPD to the culture in an even higher concentration (100 μM).
LuxS-dependent genes.
The luxS mutation resulted in changes in all cellular processes, and the affected genes were distributed throughout the genome (Fig. 5). Consistent transcriptional changes in complete operons (e.g., such as those seen for the CSP-regulated genes [52]) were rarely detected; in most cases the observed pattern resembled an uncoordinated response, similar to data reported for Neisseria meningitidis (18). Some of the affected genes are known to have pleiotropic effects themselves, e.g., transcriptional regulators (64, 77), sigma factors, iron acquisition systems (25), and PTS transporters (2). Below we will discuss several genes related to phenotypes described before for luxS mutants which were found to be differentially expressed in our analysis and did not respond to added DPD.
FIG. 5.
Localization of luxS-regulated genes in the genome of S. mutans UA159. The graph depicts 453 genes differentially expressed in the luxS mutant of S. mutans UA159 which did not respond to AI-2 (TCPSAM analysis). The two circles represent the two coding strands. Downregulated genes are shown in green and upregulated genes in red. The size of the genome is indicated in kbp. See Table S4 in the supplemental material for a complete list of the genes shown here.
(i) Bacteriocins and competence.
An effect of the luxS mutation on bacteriocin production has been described previously for S. mutans UA140 (49). This strain produces the lantibiotic mutacin I, which is active against a wide spectrum of gram-positive bacteria. Transcription of the mutacin I gene (mutA) and its transcriptional activator (mutR) was shown to be impaired in a luxS null mutant of UA140 due to the increased expression of the repressor irvA (SMU.1274). S. mutans UA159 does not produce mutacins I to III, which belong to the lantibiotics; they have complex biosynthesis pathways and regulatory circuits (69). In strain UA159, analyzed here, a homologue to the repressor irvA (SMU.1937c) and the positive regulator mutR (SMU.110) is present, but not a homologue to the mutacin-encoding gene mutA. In our analysis, irvA was downregulated 9.9-fold, in contrast to the observations of Merritt et al. (49). Its function in strain UA159, which does not produce mutacin I, remains to be elucidated.
However, a role of luxS for bacteriocin production was confirmed in our analysis, although it was not regulated by the signaling molecule AI-2. We found reduced transcription of 4 of the 12 putative bacteriocins that were identified in the genome sequence of strain UA159 (4, 73), including the CSP. The activities of the putative bacteriocins were analyzed, and mutacins IV and V, which were donwregulated in the luxS mutant analyzed here, were shown to have the strongest antimicrobial activity (23, 24, 54) whereas the putative bacteriocin-encoding genes SMU.281, -283, -423, -1892c, -1895c, -1896c, -1905c, and -1906c (two of which were upregulated in the luxS mutant) had no effect on 84 indicator strains tested (24). Thus, the luxS mutant was probably impaired in antibacterial activity. The comCDE regulatory circuit was shown to modulate the expression of some bacteriocin genes (35, 37, 47, 73). Downregulation of the comC gene 2.3-fold caused by the luxS mutation might therefore be responsible for an attenuated antimicrobial activity in the luxS mutant strain.
(ii) Biofilm formation and acid tolerance.
Biofilm formation is one of the main cariogenic properties of S. mutans, and it is impaired in luxS mutants (49, 50, 78, 82). The transcriptome analysis revealed changes in many biofilm-related genes which could not be restored by addition of DPD, including some of the most strongly dysregulated genes. Examples are a fructokinase gene (SMU.1840, −12.5-fold), pbp2b (SMU.597, −38.5-fold) (9), comC (SMU.1915, −2.3-fold) (41), and gbpA (SMU.2112, −7.8-fold) (45). Moreover, many of the membrane transporters which were downregulated (e.g., SMU.238c, −108.0-fold) may have an impact on biofilm formation (65). Aciduricity is considered another important virulence trait in S. mutans and was reported to be impaired in a luxS mutant (78), consistent with the strong downregulation of aguA (−73.0-fold) observed here.
These data show that luxS-related phenotypes may be caused by central metabolic defects rather than quorum-sensing-related signaling. Similar observations have been made by other authors. A luxS mutant of Salmonella enterica serovar Typhimurium has impaired biofilm growth, and this trait could not be restored by complementation with chemically synthesized AI-2 (13). The defects in growth and biofilm architecture of a luxS mutant of Lactobacillus rhamnosus were not affected by AI-2 (40).
AI-2-regulated genes.
While the majority of transcriptional changes observed in the luxS mutant were not significantly changed by addition of DPD, a number of genes clearly restored their wild-type expression level and thus were regulated directly by the signaling molecule AI-2. Here we will focus on regulatory genes which in turn could control several AI-2-dependent traits and on transport proteins likely to be important in uptake or excretion of AI-2.
(i) SMU.96.
The δ subunit (encoded by rpoE) of the RNA polymerase has a molecular mass of 21 kDa and is widespread in gram-positive bacteria, but there is no counterpart in gram-negative bacteria (29). It has been speculated that σA and δ together have activity similar to that of σ70 from E. coli (26). Spiegelman et al. (67) suggested a role for δ in maintaining transcriptional specificity in Bacillus subtilis. Thus, RpoE is a modulator of the specificity of RNA polymerase, and the 147-fold increase in its expression which was observed in this study likely affects many diverse genes. The expression of rpoE was reported to be maximal during mid-exponential growth in Streptococcus agalactiae and at the transition between logarithmic and stationary phases in B. subtilis (29, 44, 61), indicating that it might respond to cell density-related signals. The presence of a relatively high concentration of the AI-2 molecule might have mimicked a very high cell density and thus induced the strong upregulation of rpoE transcription in S. mutans. Further work is necessary to confirm this hypothesis.
(ii)SMU.135.
SMU.135 belongs to the LysR-type regulators, which are the most common positive transcriptional regulators (58). Most of them autoregulate their own transcription in a positive or negative way. LysR-type regulators are able to bind to their cognate DNA binding motif directly, but most of them require small ligands as coinducers for full activation of transcription of their target promoters. The binding cavity of the coinducer was located on the C-terminal domain, which exhibits large sequence divergence (6, 75). Studies based on the crystal structure of CysB from Klebsiella aerogenes and OxyR from E. coli revealed that they are structurally similar to the periplasmic ligand binding protein II superfamily. Both LLTR and periplasmic ligand binding protein were shown to bind many different classes of ligands, including sugars, amino acids, and inorganic ions (58, 83). The AI-2 molecule has a large structural similarity to d-ribose (11), thus making it a possible candidate to act as a coinducer for this LLTR. Addition of AI-2 immediately increased the transcription level of SMU.135; therefore, we suggest that SMU.135 is positively autoregulated. SMU.135 is a homologue of mleR from Lactococcus lactis (40% identity). In these organisms, it was shown to regulate malolactic fermentation (56). The mleR homologue in Oenococcus oeni is not involved in regulation of malolactic fermentation and could not be assigned a function yet (38). In H+-ATPase-deficient mutants, it was not transcribed (20), leading Galland et al. to the suggestion that a regulatory factor is needed which is presumably linked to metabolic energy.
(iii) SMU.1168.
SMU.1168 belongs to the TetR family of repressors. Ramos et al. (55) identified 16 TetR-type regulators in S. mutans, employing a bioinformatical approach. Transcriptional regulators of this family are known to act as repressors and to regulate diverse functions in bacteria, mainly related to multidrug resistance, biosynthesis of antibiotics, osmotic stress, and pathogenicity (55). The family has been named after its best-characterized member, TetR (tetracycline resistance), controlling expression of tetA, an efflux pump for tetracycline.
(iv) SMU.2114c.
SMU.2114c is a transcriptional regulator which binds DNA through a helix-turn-helix motif and belongs to the PFAM family PF00376 (merR). Members of this family are sensitive to metal ions, redox stress, and other chemical signals. They are positive regulators, which upon sensing their ligand release the DNA binding site and allow RNA polymerase to initiate transcription.
(v) SMU.103.
SMU.103 is part of a PTS, which catalyzes the phosphorylation of incoming sugar substrates concomitant with their translocation across the cell membrane. The EIIA domain is a membrane-bound protein which carries the first permease-specific phosphorylation site, a histidine which is phosphorylated by another component of the PTS, the phosphoryl carrier protein (HPr). SMU.103 belongs to the mannitol class of PTS. This EIIA domain has an average length of about 142 amino acids, and the three-dimensional structure has been determined (Prosite documentation PD0C00528). It remains to be tested if SMU.103 is able to phosphorylate and translocate AI-2 across the membrane.
(vi) SMU.408.
SMU.408 belongs to the major facilitator superfamily transporters, which are described to translocate diverse molecules, including amino acids, nucleotides, and sugars (1). Only a general function prediction is possible using the PFAM database. The COG2252 family includes permeases for diverse substrates, such as xanthine, uracil, and vitamin C, which have 10 predicted transmembrane helices. However, many members of this family are functionally uncharacterized, and the transported molecules are not known. The strong response of this transporter to the AI-2 molecule suggests that SMU.408 expression is regulated by AI-2 and that it may act as an uptake mechanism for AI-2. The strong induction of SMU.408 by AI-2 might also be responsible for the lack of detectable AI-2 in culture supernatants of S. mutans. SMU.408 has no similarity to the Lsr transporter of Salmonella enterica serovar Typhimurium (51, 68, 81). It is also not homologous to LsrR, the transcriptional regulator of Salmonella and E. coli, which binds phosphorylated AI-2 and activates transcription of the lsr operon. Finally, it is also not similar to the YdgG transporter of E. coli (27) and the RbsB ribose binding protein of Actinobacillus actinomycetemcomitans (28).
(vii) SMU.1667.
SMU.1667 is a bacterial inner membrane translocator which is part of a multicomponent branched-chain amino acid transport system. These are typically composed of a periplasmic substrate-binding protein, one or two reciprocally homologous integral inner-membrane proteins, and one or two peripheral membrane ATP-binding proteins that couple energy to the active transport system. SMU.1667c represents the permease component of this multicomponent transport system which translocates the substrate across the membrane. It has been shown that most of these proteins contain a conserved region located about 80 to 100 residues from their C-terminal extremity. This region seems to be located in a cytoplasmic loop between two transmembrane domains. Apart from the conserved region, the sequences of these proteins are quite divergent. Also found within this family are proteins from the galactose transport system permease and a ribose transport system (PF02653; http://pfam.sanger.ac.uk/family?acc=PF02653).
Conclusions.
In summary, the observed changes in the transcriptome of a luxS-deficient strain of S. mutans are consistent with the reported phenotypic changes. Many of them affected biofilm-related traits but also genes important for virulence, sugar metabolism, stress responses, acid tolerance, osmotolerance, protein synthesis, competence, and DNA repair. All functional categories were affected, but most of them in a subtle and apparently “uncoordinated” way. We found examples where certain genes responsible for a trait were upregulated while others were downregulated (e.g., glucan binding proteins). This suggests further that LuxS is a central metabolic enzyme with multiple and diverse roles in protein, RNA, and DNA synthesis, in such a way as to indirectly affect the majority of cellular processes. However, there is presently no evidence to ascribe these phenotypes to quorum-sensing-related signaling, since they could not be restored by addition a of chemically pure autoinducer.
A small subset of the genes disturbed in the luxS mutant of S. mutans responded clearly and repeatedly to an externally added autoinducer, which simulates a high density of actively growing cells in the immediate environment. Protein synthesis- and stress-related genes were among these genes. In addition, potential transcriptional regulators of AI-2 signaling distinct from any of the known quorum-sensing regulators were identified, as were AI-2-regulated membrane transport proteins. The delta subunit of the RNA polymerase was the most strongly upregulated gene in our analysis and might represent a mechanism for high-level regulatory effects of AI-2. Detailed studies of the properties of these genes and analyses of the regulatory networks will be required to clarify these questions.
Supplementary Material
Acknowledgments
We thank Pjotr Bielicki for support with real-time PCR. We gratefully acknowledge the constant supply of AI-2 from Verena Thiel. Georg Conrads is most gratefully acknowledged for his support in the manufacturing of the whole-genome microarray.
R.V. was supported by a grant from the University of Granada.
Footnotes
Published ahead of print on 2 November 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abramson, J., I. Smirnova, V. Kasho, G. Verner, H. R. Kaback, and S. Iwata. 2003. Structure and mechanism of the lactose permease of Escherichia coli. Science 301610-615. [DOI] [PubMed] [Google Scholar]
- 2.Abranches, J., M. M. Candella, Z. T. Wen, H. V. Baker, and R. A. Burne. 2006. Different roles of EIIABMan and EIIGlc in regulation of energy metabolism, biofilm development, and competence in Streptococcus mutans. J. Bacteriol. 1883748-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn, S. J., Z. T. Wen, and R. A. Burne. 2006. Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect. Immun. 741631-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 9914434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ajdic, D., and V. T. Pham. 2007. Global transcriptional analysis of Streptococcus mutans sugar transporters using microarrays. J. Bacteriol. 1895049-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnett, M. J., and S. R. Long. 1990. DNA sequence and translational product of a new nodulation-regulatory locus: SyrM has sequence similarity to NodD proteins. J. Bacteriol. 1723695-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bassler, B. L., E. P. Greenberg, and A. M. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 1794043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Battaglia, C., G. Salani, C. Consolandi, L. R. Bernardi, and G. De Bellis. 2000. Analysis of DNA microarrays by non-destructive fluorescent staining using SYBR green II. BioTechniques 2978-81. [DOI] [PubMed] [Google Scholar]
- 9.Bizzini, A., S. Beggah-Moller, P. Moreillon, and J. M. Entenza. 2006. Lack of in vitro biofilm formation does not attenuate the virulence of Streptococcus gordonii in experimental endocarditis. FEMS Immunol. Med. Microbiol. 48419-423. [DOI] [PubMed] [Google Scholar]
- 10.Camilli, A., and B. L. Bassler. 2006. Bacterial small-molecule signaling pathways. Science 3111113-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415545-549. [DOI] [PubMed] [Google Scholar]
- 12.Cvitkovitch, D. G., Y. H. Li, and R. P. Ellen. 2003. Quorum sensing and biofilm formation in Streptococcal infections. J. Clin. Investig. 1121626-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Keersmaecker, S. C., C. Varszegi, N. van Boxel, L. W. Habel, K. Metzger, R. Daniels, K. Marchal, D. De Vos, and J. Vanderleyden. 2005. Chemical synthesis of (S)-4,5-dihydroxy-2,3-pentanedione, a bacterial signal molecule precursor, and validation of its activity in Salmonella typhimurium. J. Biol. Chem. 28019563-19568. [DOI] [PubMed] [Google Scholar]
- 14.DeKeersmaecker, S. C., and J. Vanderleyden. 2003. Constraints on detection of autoinducer-2 (AI-2) signalling molecules using Vibrio harveyi as a reporter. Microbiology 1491953-1956. [DOI] [PubMed] [Google Scholar]
- 15.DeLisa, M. P., C. F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 1835239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deutscher, J., C. Francke, and P. W. Postma. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70939-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diaz, P. I., N. I. Chalmers, A. H. Rickard, C. Kong, C. L. Milburn, R. J. Palmer, Jr., and P. E. Kolenbrander. 2006. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl. Environ. Microbiol. 722837-2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dove, J. E., K. Yasukawa, C. R. Tinsley, and X. Nassif. 2003. Production of the signalling molecule, autoinducer-2, by Neisseria meningitidis: lack of evidence for a concerted transcriptional response. Microbiology 1491859-1869. [DOI] [PubMed] [Google Scholar]
- 19.Frias, J., E. Olle, and M. Alsina. 2001. Periodontal pathogens produce quorum sensing signal molecules. Infect. Immun. 693431-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galland, D., R. Tourdot-Marechal, M. Abraham, K. S. Chu, and J. Guzzo. 2003. Absence of malolactic activity is a characteristic of H+-ATPase-deficient mutants of the lactic acid bacterium Oenococcus oeni. Appl. Environ. Microbiol. 691973-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greenberg, E. P., J. W. Hastings, and S. Ulitzur. 1979. Induction of luciferase synthesis in Beneckea harveyi by other marine bacteria. Arch. Microbiol. 12087-91. [Google Scholar]
- 22.Griswold, A. R., M. Jameson-Lee, and R. A. Burne. 2006. Regulation and physiologic significance of the agmatine deiminase system of Streptococcus mutans UA159. J. Bacteriol. 188834-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hale, J. D., N. C. Heng, R. W. Jack, and J. R. Tagg. 2005. Identification of nlmTE, the locus encoding the ABC transport system required for export of nonlantibiotic mutacins in Streptococcus mutans. J. Bacteriol. 1875036-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hale, J. D., Y. T. Ting, R. W. Jack, J. R. Tagg, and N. C. Heng. 2005. Bacteriocin (mutacin) production by Streptococcus mutans genome sequence reference strain UA159: elucidation of the antimicrobial repertoire by genetic dissection. Appl. Environ. Microbiol. 717613-7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanks, T. S., M. Liu, M. J. McClure, and B. Lei. 2005. ABC transporter FtsABCD of Streptococcus pyogenes mediates uptake of ferric ferrichrome. BMC Microbiol. 562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helmann, J. D., L. M. Marquez, and M. J. Chamberlin. 1988. Cloning, sequencing, and disruption of the Bacillus subtilis sigma 28 gene. J. Bacteriol. 1701568-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herzberg, M., I. K. Kaye, W. Peti, and T. K. Wood. 2006. YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. J. Bacteriol. 188587-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.James, D., H. Shao, R. J. Lamont, and D. R. Demuth. 2006. The Actinobacillus actinomycetemcomitans ribose binding protein RbsB interacts with cognate and heterologous autoinducer 2 signals. Infect. Immun. 744021-4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones, A. L., R. H. Needham, and C. E. Rubens. 2003. The Delta subunit of RNA polymerase is required for virulence of Streptococcus agalactiae. Infect. Immun. 714011-4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joyce, E. A., A. Kawale, S. Censini, C. C. Kim, A. Covacci, and S. Falkow. 2004. LuxS is required for persistent pneumococcal carriage and expression of virulence and biosynthesis genes. Infect. Immun. 722964-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kendall, M. M., D. A. Rasko, and V. Sperandio. 2007. Global effects of the cell-to-cell signaling molecules autoinducer-2, autoinducer-3, and epinephrine in a luxS mutant of enterohemorrhagic Escherichia coli. Infect. Immun. 754875-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolenbrander, P. E. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54413-437. [DOI] [PubMed] [Google Scholar]
- 33.Kolenbrander, P. E., R. N. Andersen, D. S. Blehert, P. G. Egland, J. S. Foster, and R. J. Palmer, Jr. 2002. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66486-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolenbrander, P. E., P. G. Egland, P. I. Diaz, and R. J. Palmer, Jr. 2005. Genome-genome interactions: bacterial communities in initial dental plaque. Trends Microbiol. 1311-15. [DOI] [PubMed] [Google Scholar]
- 35.Kreth, J., J. Merritt, W. Shi, and F. Qi. 2005. Co-ordinated bacteriocin production and competence development: a possible mechanism for taking up DNA from neighbouring species. Mol. Microbiol. 57392-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kreth, J., J. Merritt, W. Shi, and F. Qi. 2005. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J. Bacteriol. 1877193-7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kreth, J., J. Merritt, L. Zhu, W. Shi, and F. Qi. 2006. Cell density- and ComE-dependent expression of a group of mutacin and mutacin-like genes in Streptococcus mutans. FEMS Microbiol. Lett. 26511-17. [DOI] [PubMed] [Google Scholar]
- 38.Labarre, C., C. Divies, and J. Guzzo. 1996. Genetic organization of the mle locus and identification of a mleR-like gene from Leuconostoc oenos. Appl. Environ. Microbiol. 624493-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lau, P. C., C. K. Sung, J. H. Lee, D. A. Morrison, and D. G. Cvitkovitch. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49193-205. [DOI] [PubMed] [Google Scholar]
- 40.Lebeer, S., S. C. De Keersmaecker, T. L. Verhoeven, A. A. Fadda, K. Marchal, and J. Vanderleyden. 2007. Functional analysis of luxS in the probiotic strain Lactobacillus rhamnosus GG reveals a central metabolic role important for growth and biofilm formation. J. Bacteriol. 189860-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, Y. H., N. Tang, M. B. Aspiras, P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 1842699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 1821374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loo, C. Y., K. Mitrakul, I. B. Voss, C. V. Hughes, and N. Ganeshkumar. 2003. Involvement of an inducible fructose phosphotransferase operon in Streptococcus gordonii biofilm formation. J. Bacteriol. 1856241-6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lopez de Saro, F. J., N. Yoshikawa, and J. D. Helmann. 1999. Expression, abundance, and RNA polymerase binding properties of the delta factor of Bacillus subtilis. J. Biol. Chem. 27415953-15958. [DOI] [PubMed] [Google Scholar]
- 45.Lynch, D. J., T. L. Fountain, J. E. Mazurkiewicz, and J. A. Banas. 2007. Glucan-binding proteins are essential for shaping Streptococcus mutans biofilm architecture. FEMS Microbiol. Lett. 268158-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsumoto, M., K. Fujita, and T. Ooshima. 2006. Binding of glucan-binding protein C to GTFD-synthesized soluble glucan in sucrose-dependent adhesion of Streptococcus mutans. Oral Microbiol. Immunol. 2142-46. [DOI] [PubMed] [Google Scholar]
- 47.Matsumoto-Nakano, M., and H. K. Kuramitsu. 2006. Role of bacteriocin immunity proteins in the antimicrobial sensitivity of Streptococcus mutans. J. Bacteriol. 1888095-8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meijler, M. M., L. G. Hom, G. F. Kaufmann, K. M. McKenzie, C. Sun, J. A. Moss, M. Matsushita, and K. D. Janda. 2004. Synthesis and biological validation of a ubiquitous quorum-sensing molecule. Angew. Chem. Int. Ed. Engl. 432106-2108. [DOI] [PubMed] [Google Scholar]
- 49.Merritt, J., J. Kreth, W. Shi, and F. Qi. 2005. LuxS controls bacteriocin production in Streptococcus mutans through a novel regulatory component. Mol. Microbiol. 57960-969. [DOI] [PubMed] [Google Scholar]
- 50.Merritt, J., F. Qi, S. D. Goodman, M. H. Anderson, and W. Shi. 2003. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 711972-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller, S. T., K. B. Xavier, S. R. Campagna, M. E. Taga, M. F. Semmelhack, B. L. Bassler, and F. M. Hughson. 2004. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol. Cell 15677-687. [DOI] [PubMed] [Google Scholar]
- 52.Peterson, S. N., C. K. Sung, R. Cline, B. V. Desai, E. C. Snesrud, P. Luo, J. Walling, H. Li, M. Mintz, G. Tsegaye, P. C. Burr, Y. Do, S. Ahn, J. Gilbert, R. D. Fleischmann, and D. A. Morrison. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 511051-1070. [DOI] [PubMed] [Google Scholar]
- 53.Pfaffl, M. W., G. W. Horgan, and L. Dempfle. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qi, F., P. Chen, and P. W. Caufield. 2001. The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV. Appl. Environ. Microbiol. 6715-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramos, J. L., M. Martinez-Bueno, A. J. Molina-Henares, W. Teran, K. Watanabe, X. Zhang, M. T. Gallegos, R. Brennan, and R. Tobes. 2005. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69326-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Renault, P., C. Gaillardin, and H. Heslot. 1989. Product of the Lactococcus lactis gene required for malolactic fermentation is homologous to a family of positive regulators. J. Bacteriol. 1713108-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rickard, A. H., R. J. Palmer, Jr., D. S. Blehert, S. R. Campagna, M. F. Semmelhack, P. G. Egland, B. L. Bassler, and P. E. Kolenbrander. 2006. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol. Microbiol. 601446-1456. [DOI] [PubMed] [Google Scholar]
- 58.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47597-626. [DOI] [PubMed] [Google Scholar]
- 59.Schuster, M., and E. P. Greenberg. 2006. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 29673-81. [DOI] [PubMed] [Google Scholar]
- 60.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 1852066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seepersaud, R., R. H. Needham, C. S. Kim, and A. L. Jones. 2006. Abundance of the delta subunit of RNA polymerase is linked to the virulence of Streptococcus agalactiae. J. Bacteriol. 1882096-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Semmelhack, M. F., S. R. Campagna, M. J. Federle, and B. L. Bassler. 2005. An expeditious synthesis of DPD and boron binding studies. Org. Lett. 7569-572. [DOI] [PubMed] [Google Scholar]
- 63.Senadheera, M. D., B. Guggenheim, G. A. Spatafora, Y. C. Huang, J. Choi, D. C. Hung, J. S. Treglown, S. D. Goodman, R. P. Ellen, and D. G. Cvitkovitch. 2005. A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J. Bacteriol. 1874064-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Senadheera, M. D., A. W. Lee, D. C. Hung, G. A. Spatafora, S. D. Goodman, and D. G. Cvitkovitch. 2007. The Streptococcus mutans vicX gene product modulates gtfB/C expression, biofilm formation, genetic competence, and oxidative stress tolerance. J. Bacteriol. 1891451-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shemesh, M., A. Tam, and D. Steinberg. 2007. Differential gene expression profiling of Streptococcus mutans cultured under biofilm and planktonic conditions. Microbiology 1531307-1317. [DOI] [PubMed] [Google Scholar]
- 66.Sperandio, V., A. G. Torres, J. A. Giron, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 1835187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spiegelman, G. B., W. R. Hiatt, and H. R. Whiteley. 1978. Role of the 21,000 molecular weight polypeptide of Bacillus subtilis RNA polymerase in RNA synthesis. J. Biol. Chem. 2531756-1765. [PubMed] [Google Scholar]
- 68.Taga, M. E., J. L. Semmelhack, and B. L. Bassler. 2001. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42777-793. [DOI] [PubMed] [Google Scholar]
- 69.Tsang, P., J. Merritt, W. Shi, and F. Qi. 2006. IrvA-dependent and IrvA-independent pathways for mutacin gene regulation in Streptococcus mutans. FEMS Microbiol. Lett. 261231-234. [DOI] [PubMed] [Google Scholar]
- 70.Turovskiy, Y., and M. L. Chikindas. 2006. Autoinducer-2 bioassay is a qualitative, not quantitative method influenced by glucose. J. Microbiol. Methods 66497-503. [DOI] [PubMed] [Google Scholar]
- 71.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 985116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vadeboncoeur, C., and M. Pelletier. 1997. The phosphoenolpyruvate:sugar phosphotransferase system of oral streptococci and its role in the control of sugar metabolism. FEMS Microbiol. Rev. 19187-207. [DOI] [PubMed] [Google Scholar]
- 73.van der Ploeg, J. R. 2005. Regulation of bacteriocin production in Streptococcus mutans by the quorum-sensing system required for development of genetic competence. J. Bacteriol. 1873980-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vendeville, A., K. Winzer, K. Heurlier, C. M. Tang, and K. R. Hardie. 2005. Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 3383-396. [DOI] [PubMed] [Google Scholar]
- 75.Viale, A. M., H. Kobayashi, T. Akazawa, and S. Henikoff. 1991. rbcR [sic], a gene coding for a member of the LysR family of transcriptional regulators, is located upstream of the expressed set of ribulose 1,5-bisphosphate carboxylase/oxygenase genes in the photosynthetic bacterium Chromatium vinosum. J. Bacteriol. 1735224-5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vilchez, R., A. Lemme, V. Thiel, S. Schulz, H. Sztajer, and I. Wagner-Dobler. 2007. Analysing traces of autoinducer-2 requires standardization of the Vibrio harveyi bioassay. Anal. Bioanal. Chem. 387489-496. [DOI] [PubMed] [Google Scholar]
- 77.Wang, B., and H. K. Kuramitsu. 2006. A pleiotropic regulator, Frp, affects exopolysaccharide synthesis, biofilm formation, and competence development in Streptococcus mutans. Infect. Immun. 744581-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wen, Z. T., and R. A. Burne. 2004. LuxS-mediated signaling in Streptococcus mutans is involved in regulation of acid and oxidative stress tolerance and biofilm formation. J. Bacteriol. 1862682-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Winzer, K., K. R. Hardie, and P. Williams. 2003. LuxS and autoinducer-2: their contribution to quorum sensing and metabolism in bacteria. Adv. Appl. Microbiol. 53291-396. [DOI] [PubMed] [Google Scholar]
- 80.Xavier, K. B., and B. L. Bassler. 2005. Interference with AI-2-mediated bacterial cell-cell communication. Nature 437750-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xavier, K. B., S. T. Miller, W. Lu, J. H. Kim, J. Rabinowitz, I. Pelczer, M. F. Semmelhack, and B. L. Bassler. 2007. Phosphorylation and processing of the quorum-sensing molecule autoinducer-2 in enteric bacteria. ACS Chem. Biol. 2128-136. [DOI] [PubMed] [Google Scholar]
- 82.Yoshida, A., T. Ansai, T. Takehara, and H. K. Kuramitsu. 2005. LuxS-based signaling affects Streptococcus mutans biofilm formation. Appl. Environ. Microbiol. 712372-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zaim, J., and A. M. Kierzek. 2003. The structure of full-length LysR-type transcriptional regulators. Modeling of the full-length OxyR transcription factor dimer. Nucleic Acids Res. 311444-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.