Abstract
We examined the activity of polynucleotide phosphorylase (PNPase) from Streptomyces coelicolor, Streptomyces antibioticus, and Escherichia coli in phosphorolysis using substrates derived from the rpsO-pnp operon of S. coelicolor. The Streptomyces and E. coli enzymes were both able to digest a substrate with a 3′ single-stranded tail although E. coli PNPase was more effective in digesting this substrate than were the Streptomyces enzymes. The kcat for the E. coli enzyme was ca. twofold higher than that observed with the S. coelicolor enzyme. S. coelicolor PNPase was more effective than its E. coli counterpart in digesting a substrate possessing a 3′ stem-loop structure, and the Km for the E. coli enzyme was ca. twice that of the S. coelicolor enzyme. Electrophoretic mobility shift assays revealed an increased affinity of S. coelicolor PNPase for the substrate possessing a 3′ stem-loop structure compared with the E. coli enzyme. We observed an effect of nucleoside diphosphates on the activity of the S. coelicolor PNPase but not the E. coli enzyme. In the presence of a mixture of 20 μM ADP, CDP, GDP, and UDP, the Km for the phosphorolysis of the substrate with the 3′ stem-loop was some fivefold lower than the value observed in the absence of nucleoside diphosphates. No effect of nucleoside diphosphates on the phosphorolytic activity of E. coli PNPase was observed. To our knowledge, this is the first demonstration of an effect of nucleoside diphosphates, the normal substrates for polymerization by PNPase, on the phosphorolytic activity of that enzyme.
Polynucleotide phosphorylase (PNPase) is a 3′-5′ exoribonuclease that functions in the phosphorolytic degradation of RNA molecules in bacteria and in eukaryotic organelles (20, 24). In Escherichia coli and other bacteria, PNPase plays an important role in the degradation of messenger RNAs. Thus, endonucleolytic cleavage of RNA molecules generates 3′ ends that are substrates for the action of PNPase and RNase II, an exonuclease that functions hydrolytically (9, 17, 31). PNPase plays another role in E. coli, at least under some circumstances. As is the case in eukaryotes, the 3′ ends of at least some RNA molecules in bacteria are polyadenylated (35, 36). Polyadenylation facilitates the degradation of RNAs in bacteria (26, 27, 30). While the major enzyme responsible for RNA polyadenylation in E. coli is poly(A) polymerase I (PAP I) (8), mutants of E. coli lacking PAP I still retain the ability to polyadenylate RNAs (29), indicating that there is at least one other polyadenylating enzyme in those cells. Mohanty and Kushner have presented evidence indicating that the second PAP in E. coli is none other than PNPase (28). They argue that under appropriate conditions in vivo, PNPase can serve to degrade RNAs or synthesize poly(A) tails and that this enzyme is responsible for the G, C, and U residues that are found at low frequency in the poly(A) tails of RNAs from wild-type E. coli (28).
PNPase structure and function have been studied extensively in Streptomyces (5, 6, 22, 40). Of particular relevance to the present study are the observations that Streptomyces species do not appear to contain a dedicated poly(A) polymerase like PAP I and that the polyadenylation of RNA 3′ ends in Streptomyces is catalyzed by PNPase (3, 37). Similar conclusions have been reached in studies of RNA polyadenylation in spinach chloroplasts and in a Synechocystis species (34, 41). Moreover, in all of these systems the tails associated with the cellular RNAs are heteropolymeric, like those observed in E. coli mutants lacking PAP I (3, 4, 28, 34, 41). Thus, PNPase plays a dual role in these systems, serving as both a 3′-5′ exoribonuclease and as an RNA 3′ polyribonucleotide polymerase, i.e., the analog of poly(A) polymerase in E. coli.
One question which emerges from consideration of these dual roles is how PNPase functions as both a nuclease and a polymerase in the same cell. To begin to answer this question, we describe here a kinetic comparison of PNPases from Streptomyces and E. coli using naturally occurring substrates for phosphorolysis. Most kinetic studies of PNPase have been performed using synthetic substrates, in particular, various forms of oligo(A) and poly(A) (13-15, 18, 19). Using transcripts of the rpsO-pnp intergenic region from Streptomyces coelicolor, we show that while the E. coli enzyme is more effective in digesting substrates with single-stranded 3′ tails, the Streptomyces enzyme is more effective than its E. coli counterpart with substrates containing a 3′ stem-loop (hairpin) structure. We show, moreover, that nucleoside diphosphates (NDPs) stimulate the phosphorolytic activity of the Streptomyces PNPase but not the E. coli enzyme.
MATERIALS AND METHODS
Synthesis and 32P labeling of in vitro transcripts of the rpsO-pnp intergenic region.
The templates used to synthesize the rpsO-pnp transcripts used in this study were derived from plasmids pJSE600 and pJSE5600, which we described previously (10). pJSE600 contains the entire S. coelicolor rspO-pnp operon, and pJSE5600 contains a 459-bp fragment from pJSE600 that includes the rpsO terminator and the intergenic hairpin targeted by RNase III (10). In the experiments reported here, pJSE5600 was linearized with SpeI. In vitro transcription of this linearized form of pJSE5600 generated a 507-base RNA transcript which ends at the pnp start site. That product is referred to below as the 5601 transcript. The pJSE5650 plasmid was created by PCR using primers rps1 and Sc Hairpin R1 and the plasmid pJSE600 as a template (Table 1). The PCR product was cloned into pCR2.1-TOPO (Invitrogen) following the manufacturer's protocol, and plasmids with the insert in the correct orientation relative to the T7 promoter were identified by restriction digestion and sequencing. The Sc Hairpin R1 primer contains a BamHI site which was used to linearize pJSE5650 for runoff transcription. The 420-base transcript from pJSE5650, called the 5650 transcript or RP3 (Fig. 1 and 2), contains the same sequences as the 5601 transcript except that the 5650 transcript terminates near the base of the 3′ end of the rpsO-pnp intergenic hairpin (Fig. 1, G420). Both transcripts include 72 bases of pCR2.1-TOPO vector sequence at the 5′ end.
TABLE 1.
Primers used for construction of plasmids and for cDNA cloning
| Primer | Sequence |
|---|---|
| rps1 | 5′-CACAAGCACGACCACCACTCC-3′ |
| Sc Hairpin R1 | 5′-GTGCAATGGATCCCGGTCTTC-3′ |
| pnpB1 | 5′-CTCGTCGCGGGATCCGACGTG-3′ |
| ScPNPF1N | 5′-GTACCCTGTTTTCATATGCCGAACGCC-3′ |
| AD20 | 5′-GGATCCGAATTCTCTAGAGC-3′ |
| AD20 complement | 5′-GCTCTAGAGAATTCGGATCC-3′ |
| TOPO R1 | 5′-CCACTAGTAACGGCCGCC-3′ |
FIG. 1.
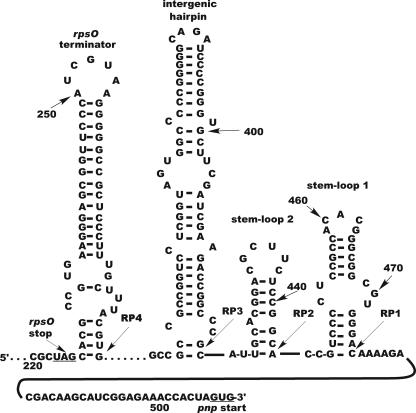
Mfold (42) model of the structure of the 5601 transcript. The numbers represent bases counted from the 5′ end of the transcript beginning with the initiation site for T7 RNA polymerase. RP1 to RP4 represent the approximate 3′ endpoints for the intermediates produced by PNPase digestion of the transcript. As noted in the text, PNPases produce heterogeneous 3′ ends at stall points, and the arrows indicate the most 5′ endpoint of the sequenced termini.
FIG. 2.
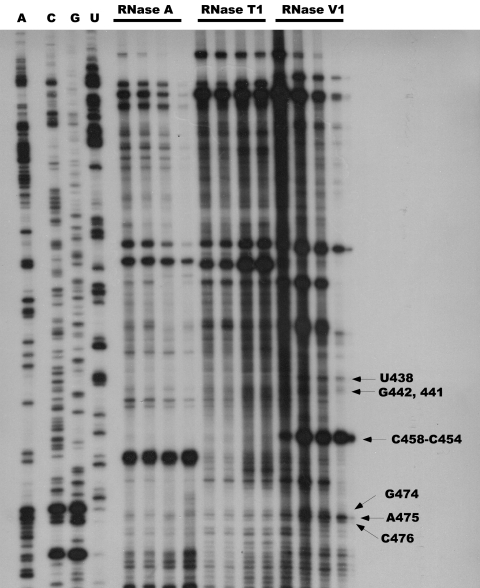
Autoradiogram of a gel showing RNase probing results. Only the portion of the gel corresponding to predicted stem-loops 1 and 2 (Fig. 1) is shown. The 5600 transcript was digested, and the digestion products were analyzed by primer extension as described in Materials and Methods. Lanes 1 to 4 show the sequencing ladder and the lanes are labeled A, C, G, and U, representing the RNA complement of the actual sequencing results. Other lanes represent (left to right in respective order) the results of digestion of the transcript with 0.3, 0.5, 1.0, and 3.0 ng of RNase A; with 0.05, 0.1, 0.3, and 0.5 units of RNase T1; and with 0.001, 0.005, 0.01, and 0.03 units of RNase V1. The arrows identify specific bases in the transcript.
Transcripts were synthesized using a His-tagged T7 RNA polymerase expressed from pBH161 (21) and purified as described previously (10) using the Talon immobilized metal affinity chromatography resin (BD Biosciences Clontech). In a few experiments, we used a commercially available preparation of T7 RNA polymerase (Fermentas). Large-scale transcription reactions were used to synthesize transcripts internally labeled with [α-32P]CTP (3,000 Ci/mmol; GE Healthcare), which were then purified as described previously (10). Transcripts were further purified using a modified protocol for the SV Total RNA Isolation kit (Promega Corp.), eluted with water, and stored at −20°C.
RNase probing of the structure of the rpsO-pnp transcript.
A preparation of nonradioactive 5600 transcript (10), synthesized as described above, was digested with RNases A, T1, and V1, essentially as described by Chen et al. (11). Briefly, 3 μg of RNA was heated to 80°C and allowed to cool slowly to room temperature. The RNA was digested with the amounts of RNase indicated in the legend to Fig. 2 in 30 mM Tris-HCl, pH 7.5, 3 mM Na2EDTA, 200 mM NaCl, 100 mM LiCl, and 10 mM MgCl2. Digestions were performed for 10 min at room temperature in 60-μl mixtures. The reactions were quenched by phenol-chloroform-isoamyl alcohol extraction of digestion mixtures, and products were recovered by ethanol precipitation. The reaction products were then analyzed by primer extension using 5′ end-labeled primer TOPO R1 (Table 1) as described previously (10). Extension products were displayed on a 5.2% sequencing polyacrylamide gel along with a sequencing ladder generated from plasmid pJSE5600 with the TOPO R1 primer.
Expression and purification of PNPases.
PNPases from Streptomyces antibioticus and E. coli were expressed and purified as described previously (23). The PNPase gene from S. coelicolor was amplified via PCR using primers pnpB1 and ScPNPF1N (Table 1) and S. coelicolor cosmid 3C3 DNA as template. The PCR product was digested with NdeI and BamHI and ligated to NdeI/BamHI-digested pET-11a (Novagen). The resulting plasmid, designated pJSE3512, was transformed into E. coli DH5α, and clones were screened by restriction digestion of plasmid DNAs. DNA from a positive clone was transformed into the E. coli expression host BL21 Gold(DE3) pLysS (Stratagene). The S. coelicolor PNPase was then expressed and purified as described previously (23). All enzymes were at least 95% pure as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Conditions for PNPase digestions.
Thirty-microliter phosphorolysis reaction mixtures containing 50 mM Tris (pH 8.0), 5 mM MgCl2, 50 μg/ml bovine serum albumin, 50 mM KCl, and 1 mM potassium phosphate (pH 7.5) were incubated with up to 500 ng of PNPase (ca. 70 nM) and 32P-labeled rpsO-pnp RNA, either the 5601 or 5650 transcript (see below for concentrations). It should be noted that 1 mM phosphate was found to be saturating under the conditions employed here. In some experiments, various amounts of a mixture of ADP, UDP, CDP, and GDP (Sigma Aldrich) were included in the reaction mixture. Reactions were incubated at 37°C for 5 min and terminated by the addition of 30 μl of Sequencing Stop Solution, containing formamide and gel tracking dyes (U.S. Biochemical Corp.). Duplicate samples were removed from the reaction mixtures, and phosphorolysis products were separated on a 7 M urea-5% polyacrylamide gel and visualized by autoradiography. Autoradiograms were scanned, and the densities of bands were quantified using the Scion Image for Windows software. The amounts of RNA in the intermediates produced by phosphorolysis were determined from standard curves generated using the intact 32P-labeled transcript and were converted to nanomoles of product using the calculated molecular weight of the relevant intermediate. For the 5601 transcript, the amount of the RP1 intermediate was determined, and for the 5650 transcript the amount of the RP4 intermediate was determined (Fig. 3). Preliminary assays (not shown) showed that product formation proceeded linearly at the protein concentrations and for the incubation times used in the experiments reported here.
FIG. 3.
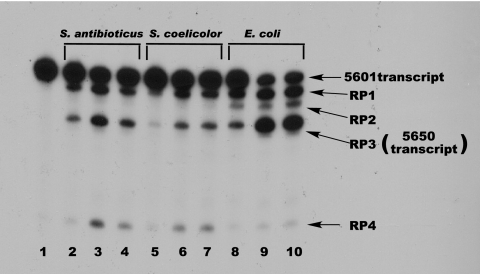
Gel electrophoresis of PNPase digests of the 5601 transcript. Digestion conditions were as described in Materials and Methods; products were separated on a denaturing polyacrylamide gel, and the gel was subjected to autoradiography. Lane 1, undigested 5601 transcript; lanes 2 to 4, digestion with 50, 250, and 500 ng of S. antibioticus PNPase; lanes 5 to 7, digestion with 50, 250, and 500 ng of S. coelicolor PNPase; lanes 8 to 10, digestion with 50, 250, and 500 ng of E. coli PNPase. The product designations at the right of the figure relate the bands produced with the 3′ endpoints mapped by cDNA cloning and sequencing (Fig. 1). The difference in intensities of the RP3 and RP4 bands in lanes 3 and 4 reflects a gel loading artifact.
For kinetic studies, substrate concentrations ranged from 0.2 to 3.3 μM for the 5601 transcript and from 0.2 to 2 μM for the 5650 transcript. Reaction mixtures contained 35 nM PNPase. Initial reaction velocities were calculated from the data obtained as described above, and Eadie-Hofstee plots were used to estimate kinetic parameters. In some kinetic experiments, reaction mixtures contained ADP, CDP, GDP, and UDP, each at 20 μM. The data of Table 2 were obtained from two separate assays in which each PNPase digestion was performed in duplicate. The data obtained from those assays were averaged to produce the results shown in Table 2. The same preparations of transcript and PNPase were used in both assays.
TABLE 2.
Kinetic parameters for polynucleotide phosphorylases
| Strain and condition | Value of kinetic parameter for the indicated transcripta
|
|||||
|---|---|---|---|---|---|---|
|
Km (μM)
|
kcat (min−1)
|
kcat/Km (mol−1 min−l × 106)
|
||||
| 5601 | 5650 | 5601 | 5650 | 5601 | 5650 | |
| S. coelicolor | ||||||
| No NDPs | 0.62 ± 0.04 | 3.12 ± 0.15 | 5.83 ± 0.20 | 5.34 ± 0.18 | 9.40 ± 0.16 | 1.71 ± 0.03 |
| Plus NDPs | 0.65 ± 0.17 | 4.74 ± 0.19 | 7.29 ± 0.02 | |||
| E. coli | ||||||
| No NDPs | 0.78 ± 0.02 | 6.33 ± 0.44 | 11.2 ± 0.20 | 5.55 ± 0.34 | 14.4 ± 0.18 | 0.88 ± 0.10 |
| Plus NDPs | 6.11 ± 0.42 | 6.07 ± 0.59 | 0.99 ± 0.17 | |||
Data are averages of two sets of duplicates ± standard errors of the means.
Incubations for electrophoretic mobility shift assays (EMSAs) were performed in 10-μl reaction mixtures under the same buffer and ionic conditions used for the kinetic assays except that mixtures contained 50 μg/ml of E. coli tRNA, and potassium phosphate was omitted. Reaction mixtures contained 7.4 nM 5650 transcript and 0 to 1.3 μM PNPase. Mixtures were incubated for 5 min at 37°; mixed with 10 μl of a solution containing 0.05% bromphenol blue, 0.05% xylene cyanol, and 20% glycerol; and applied to 5% polyacrylamide gels without urea. Radioactive bands were visualized by autoradiography.
cDNA cloning.
Large-scale phosphorolysis reactions were performed under the conditions described above except that 10 mM potassium phosphate (pH 7.5) was used and 20 μg of S. coelicolor or E. coli PNPase was used with 1.65 μg of unlabeled 5601 transcript. Reaction mixtures were incubated for 3 min at 37°C and extracted with phenol-chloroform-isoamyl alcohol, and the products were precipitated with ethanol. The 3′ ends of the intermediates resulting from PNPase digestion were determined by a modified RNA ligase-mediated amplification of cDNA ends (25). Briefly, the 5′ end of the AD20 primer (Table 1) was phosphorylated with T4 polynucleotide kinase (Promega Corp.) and then ligated to the ends of the PNPase phosphorolysis products with T4 RNA ligase (Ambion). Ligated products were reverse transcribed at 48°C using SuperScript II reverse transcriptase (Invitrogen) and the AD20 complement primer (Table 1). Reverse transcriptase products were PCR-amplified using the rps1 and AD20 complement primers (Table 1), cloned into pCR2.1-TOPO, and sequenced. The 3′ endpoints of the relevant phosphorolysis products were determined by comparing the results of cDNA sequencing with the known sequence of the 5601 transcript (Fig. 1).
Miscellaneous methods.
Protein concentrations were determined by the Bradford method (2) using the Bio-Rad protein reagent and bovine serum albumin as a standard. All PCRs were performed as follows: 95°C for 5 min and then 35 cycles at 95°C for 1 min, 50°C for 2 min, and 72°C for 1 min, with a final extension at 72°C for 10 min. The RNA folding program Mfold, version 3.2, was used to predict the secondary structure of the rpsO-pnp in vitro transcripts (42).
RESULTS
Structure of the 5601 transcript.
Several synthetic transcripts, based on the rpsO-pnp region of S. coelicolor, were used in this study. The 5601 transcript was synthesized by linearization of plasmid pJSE5600 with SpeI and transcription of the resulting template with T7 RNA polymerase. The 5601 transcript is 64 bases shorter than the 5600 transcript employed in the earlier studies (10) and contains no E. coli sequences at its 3′ end. The 5601 transcript includes 153 bases from the 3′ end of rpsO, including the rpsO terminator, the rpsO-pnp intergenic hairpin, and the pnp start codon. The secondary structure of this transcript was predicted using Mfold software (42), with the results shown in Fig. 1.
The rpsO terminator is a common feature of bacterial rspO-pnp operons (16, 32), and we demonstrated the presence of the intergenic hairpin in earlier studies using S. coelicolor RNase III (10). It remained to demonstrate the existence of the predicted stem-loops 1 and 2 in the 5601 transcript. RNase probing was used for that demonstration. Because predicted stem-loops 1 and 2 are very near the 3′ end of the 5601 transcript, it was necessary to use a primer for the primer extension analysis that was downstream of this region. This, in turn, required the use of the 5600 transcript (10) as the substrate for the RNase digestions with TOPO R1 (Table 1) as the primer extension and DNA sequencing primer. Mfold predicted the presence of stem-loops 1 and 2 in the 5600 transcript as well as the 5601 transcript.
Results of RNase A, T1, and V1 digestion of the end-labeled 5600 transcript are shown in Fig. 2. Of particular interest are the labeled regions in the figure, regions that show extension products that were obtained following RNase V1 cleavage. RNase V1 is specific for double-stranded RNA and cleaves base-paired nucleotides. Figure 2 shows RNase V1 cleavage at C476, A475, G474, C458-C454, G442 and G441, and U438, among others. These bases are predicted to occur in the double-stranded regions depicted in stem-loops 1 and 2 in Fig. 1. There is also a strong RNase A cleavage at A460 which is predicted to be present in the loop of stem-loop 1 (Fig. 1). Although the interpretation of the results is complicated somewhat by the high GC content of the 5601 transcript (the rpsO-pnp region of 5601 is 67% GC), these results do provide support for the existence of the predicted structures in that transcript.
Action of PNPases on the 5601 transcript.
We examined the activity of purified PNPases from S. antibioticus, S. coelicolor, and E. coli on the 5601 transcript, labeled internally with [32P]CTP, with the results shown in Fig. 3. The figure shows an autoradiogram of a denaturing polyacrylamide gel on which the digestion products were fractionated. PNPases are highly processive (38-40); thus, all three enzymes yielded discrete digestion products whose 3′ ends correspond to points at which the enzymes stalled during the digestion process. We designated the products produced by the PNPases RP1 to RP4. To identify the 3′ends of each species, and thus the point at which the enzymes stalled, transcription products were isolated from a PNPase digest of unlabeled 5601 transcript as described in Materials and Methods. The reverse transcription products were used for PCR with the adaptor primer and the gene-specific rps1 primer for RNA ligase-mediated amplification of cDNA ends. The sequence of the rsp1 primer corresponds to the first 21 bases of the rpsO-pnp intergenic region cloned in pJSE5600 (10). The PCR products were cloned in pCR2.1-TOPO, and the inserts were sequenced. We performed this analysis with products obtained from digests with all three PNPases, and the 3′ ends generated by the enzymes are indicated in Fig. 1. Thus, e.g., the 3′ end of RP1 is situated at or near C476 (Fig. 1) of the 5601 transcript. As PNPase digestions produce heterogeneous 3′ ends (see reference 38), it is likely that each of the product bands shown in Fig. 2 contains a mixture of products with different 3′ ends. We sequenced multiple products corresponding to each band and did indeed observe 3′-end heterogeneity. However, the various 3′ ends for a given product were never separated by more than 3 to 5 bases. Thus, this heterogeneity would have essentially no effect on the calculation of product molecular weights for the kinetic analyses described below.
As Fig. 3 demonstrates, all three PNPases produced digestion products RP1, RP3, and RP4. The E. coli enzyme also produced RP2. Once the 3′ ends of these products were determined by cDNA cloning, we compared those ends with the secondary structure of the 5601 transcript predicted by the Mfold software. In all cases, the 3′ ends observed in the cDNA clones corresponded to the 3′ end of a stem-loop structure predicted by Mfold. Thus, RP1 is predicted to contain the rpsO terminator, the rpsO-pnp intergenic hairpin, and stem-loops 1 and 2. The predicted folding free energies for these structures are −29.5 kcal/mol (rpsO terminator), −45.2 kcal/mol (intergenic hairpin), −7.5 kcal/mol (stem-loop 2), and −14.6 kcal/mol (stem-loop 1). RP2 is predicted to lack stem-loop 1, and Fig. 3 clearly shows that it does not represent a barrier to digestion by the Streptomyces PNPases. RP3 terminates at or near the base of the intergenic hairpin, and the 3′ end of RP4 is at or near the base of the rpsO terminator.
Action of PNPases on the 5650 transcript.
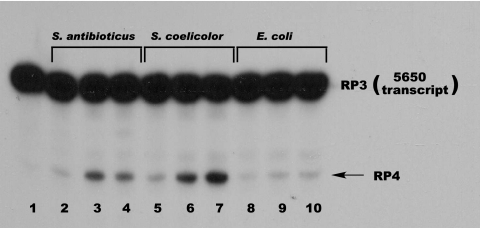
The results shown in Fig. 3 demonstrate that both the Streptomyces and E. coli PNPases are active against the 5601 transcript. However, Fig. 3 suggests that the Streptomyces enzymes are more effective than the E. coli enzyme in converting RP3 to RP4. To confirm this suggestion, we cloned the region corresponding to the RP3 sequence, containing both the rpsO terminator and the rpsO-pnp intergenic hairpin, in pCR2.1-TOPO and synthesized the relevant transcript from the linearized plasmid template. This RNA, designated the 5650 transcript, terminates at the G residue at the 3′ end of the intergenic stem-loop (Fig. 1, G420). We then used the 5650 transcript as a substrate for digestion by the Streptomyces and E. coli PNPases, with the results shown in Fig. 4. Again, it is apparent that increasing concentrations of the Streptomyces enzymes are capable of converting RP3 to RP4 whereas the E. coli PNPase is much less effective in catalyzing this reaction. We suggest reasons for this observation below.
FIG. 4.
Gel electrophoresis of PNPase digests of the 5650 transcript. Digestions conditions were as described in Materials and Methods. Products were separated on a denaturing polyacrylamide gel, and the gel was subjected to autoradiography. Lane 1, undigested 5650 transcript; lanes 2 to 4, digestion with 50, 250, and 500 ng of S. antibioticus PNPase; lanes 5 to 7, digestion with 50, 250, and 500 ng of S. coelicolor PNPase; lanes 8 to 10, digestion with 50, 250, and 500 ng of E. coli PNPase. The product designations at the right of the figure relate the bands produced with the 3′ endpoints mapped by cDNA cloning and sequencing.
Kinetic analysis of the PNPases with the 5601 and 5650 transcripts.
A number of kinetic analyses have been performed using E. coli and Micrococcus PNPases (13-15, 18, 19). However, these studies were all performed with synthetic substrates, usually oligo(A) or poly(A). As we have characterized the discrete products obtained by digestion of substrates derived from the rpsO-pnp intergenic region and as those RNAs may serve as substrates for PNPase digestion in vivo, it was of interest to examine the kinetics of PNPase action on those transcripts. Since the S. antibioticus and S. coelicolor PNPases behaved in an essentially identical fashion in all our initial analyses, subsequent experiments were performed using the S. coelicolor PNPase as the Streptomyces representative.
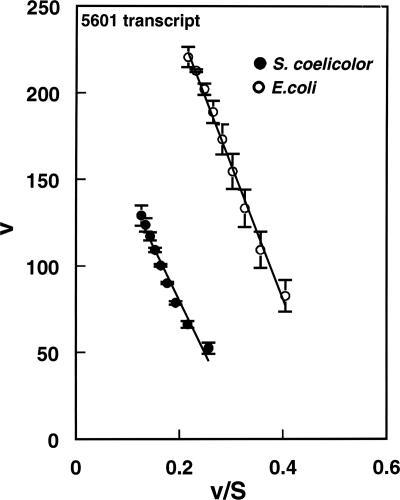
For the kinetic analysis, increasing amounts of the 5601 and 5650 transcripts were used in reactions containing ca. 35 nM PNPase. Substrate concentrations from ca. 0.2 to 3.3 μM were used in studies with the 5601 transcript and from ca. 0.2 to 2 μM in studies with the 5650 transcript. Eadie-Hofstee plots of typical data obtained for the S. coelicolor and E. coli PNPases with the 5601 transcript are shown in Fig. 5, and the kinetic data obtained from such analyses are shown in Table 2. It can be seen that the Km values for the S. coelicolor and E. coli enzymes with the 5601 transcript are quite similar, i.e., 0.62 and 0.78 μM, respectively. The kcat value for the E. coli enzyme is about twice the corresponding value obtained for the S. coelicolor PNPase; thus, the kcat/Km value for the E. coli enzyme is ca. 1.5 times the corresponding value for the S. coelicolor enzyme. The kinetic data are consistent with the gel patterns shown in Fig. 3, which indicate more effective conversion of the 5601 transcript to RP3 by the E. coli enzyme than by the S. coelicolor PNPase. Caveats related to the interpretation of these results will be discussed below.
FIG. 5.
Eadie-Hofstee plot of kinetic data for S. coelicolor and E. coli PNPases with the 5601 transcript. Kinetic assays were performed as described in Materials and Methods. The amounts of RP1 were determined and used to calculate initial velocities. Velocities (v) are expressed as nmol liter−1 min−1, and substrate concentrations are in μM. Data in the figure are averages of two sets of duplicates ± standard errors of the means. Kinetic parameters were calculated from the regression statistics corresponding to the slopes and intercepts of the plots.
Table 2 also shows the results of the kinetic analyses performed with the 5650 transcript. A Km value of ca. 3 μM was observed for the S. coelicolor PNPase while the corresponding value for the E. coli enzyme was ca. 6 μM. The kcat values for both enzymes with this substrate were essentially identical, ca. 5.5 min−1. The lower Km for the S. coelicolor enzyme results in a kcat/Km value that is twice that observed for E. coli PNPase. Again, the kinetic data are consistent with the gel patterns shown in Fig. 4, which shows clearly that the Streptomyces enzymes are more effective than the E. coli PNPase in digesting the 5650 transcript.
Effects of NDPs on the digestion of the 5650 transcript by Streptomyces and E. coli PNPases.
We have shown that, as is the case in E. coli, streptomycete RNAs possess 3′ tails (4, 7). Unlike the situation in E. coli, the RNA tails associated with streptomycete RNAs are highly heteropolymeric (4). Both Sohlberg et al. (37) and studies from our laboratory (3) have shown that these tails are probably synthesized by PNPase rather than by a dedicated PAP like E. coli PAP I. As poly(A) tails facilitate the degradation of RNAs by exonucleases like PNPase, it seemed possible that the Streptomyces PNPases might add 3′ tails to RNAs transiently during the process of phosphorolysis and that those tails might increase the ability of the enzyme to degrade the RNAs. To test this possibility, we examined the effects of a mixture of all four NDPs, the usual substrates for the polymerization reactions catalyzed by PNPase, on the phosphorolysis of the 5650 transcript by PNPases from S. coelicolor and E. coli. We utilized all four NDPs because the RNA tails associated with streptomycete RNAs are heteropolymeric (4), and it seemed possible, therefore, that all four would be necessary to observe an effect on PNPase activity.
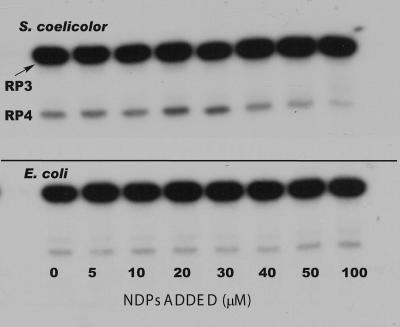
Results of a typical experiment are shown in Fig. 6. The concentration of the NDP mixture was varied between 0 and 100 μM, under conditions that were optimal for phosphorolysis. There was no observed effect of the NDPs on the formation of RP4 from RP3 (the 5650 transcript) when the E. coli PNPase was used. In marked contrast, Fig. 6 shows a significant effect of 20 to 30 μM NDPs on the activity of the S. coelicolor PNPase. Densitometry and quantification of the RP4 bands in the autoradiogram shown in Fig. 6 indicated an approximately twofold increase in the amount of RP4 produced by digestion of RP3 with S. coelicolor PNPase compared with the amount of RP3 produced in the absence of NDPs. At the highest NDP concentrations used, e.g., 50 to 100 μM, phosphorolysis by the S. coelicolor PNPase appeared to be inhibited somewhat, as indicated by a decrease in the amount of the RP4 band (Fig. 6). The cause of this inhibition is unknown at this point.
FIG. 6.
Effects of NDPs on the phosphorolysis of the 5650 transcript. Reaction conditions were as described in Materials and Methods. Products were separated by gel electrophoresis. The top panel shows the results obtained with S. coelicolor PNPase, and the bottom panel shows results using E. coli PNPase. Reactions were conducted in the presence of increasing concentrations of a mixture of ADP, CDP, UDP, and GDP as indicated.
To explore the effects of NDPs on the phosphorolysis of the 5650 transcript by the PNPases, kinetic analyses were performed, as described in Materials and Methods and in the preceding section, in the presence of 20 μM NDPs. Kinetic constants are shown in Table 2. The kcat value for S. coelicolor PNPase observed in a typical experiment in the presence of NDPs was similar to that measured in the absence of NDPs, and the same was true for the E. coli enzyme. However, the Km observed for the S. coelicolor enzyme in the presence of NDPs was approximately fivefold lower than the value observed in the absence of NDPs. No effect of NDPs on the Km for the E. coli enzyme was observed (Table 2). Thus, kcat/Km did not change appreciably when reactions were performed with the E. coli enzyme and 20 μM NDPs, but the catalytic activity of the S. coelicolor enzyme in the phosphorolysis of the 5650 transcript was ca. four times greater in the presence of NDPs than in their absence. The kcat/Km value for the S. coelicolor enzyme in the presence of NDPs was seven times greater than the corresponding value for the E. coli enzyme. It should be noted that we observed a similar effect of NDPs on the phosphorolysis of the 5650 transcript by S. antibioticus PNPase (data not shown).
EMSAs.
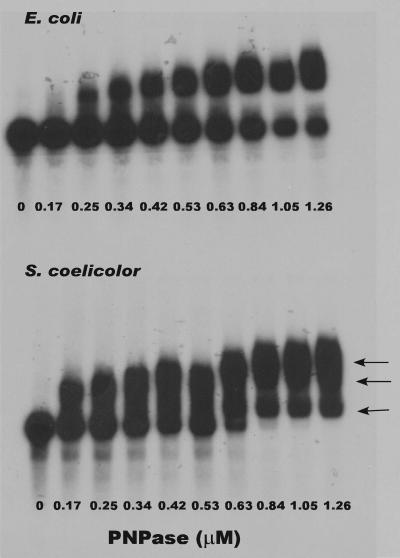
The observation that the Km for the S. coelicolor enzyme in the digestion of the 5650 transcript was half that observed for E. coli PNPase indicated that the former enzyme had a greater affinity for that transcript. Further support for this observation was obtained from EMSAs performed as described in Materials and Methods. Results of a typical set of assays are shown in Fig. 7. Binding of S. coelicolor and E. coli PNPases to the 5650 transcript was readily observed. Only a single complex was observed with E. coli PNPase under the conditions employed. However, multiple complexes (arrows) were observed with S. coelicolor PNPase, indicating the possible presence of multiple binding sites for the enzyme on the 5650 transcript (1). The addition of 100 μg/ml of unlabeled 5650 transcript completely abolished PNPase binding (data not shown). This observation, coupled with the fact that reaction mixtures contained a nonspecific competitor RNA, indicates that the binding demonstrated in Fig. 7 is specific for the 5650 transcript. As we expected, it was not possible to observe an effect of NDPs in the binding assays (see further below).
FIG. 7.
EMSA of the binding of the 5650 transcript by E. coli and S. coelicolor PNPases. Reaction and electrophoretic conditions were as described in Materials and Methods. The top panel shows results obtained using E. coli PNPase, and the bottom panel shows results using S. coelicolor PNPase. The concentrations of enzyme in each reaction mixture are shown at the bottom of each lane. Arrows in the bottom panel show the positions of three different complexes formed by the S. coelicolor PNPase with the 5650 transcript.
DISCUSSION
The data presented above demonstrate that PNPases from Streptomyces and E. coli are capable of digesting structured substrates derived from the rpsO-pnp region of S. coelicolor. The data of Fig. 3 suggest that E. coli PNPase is more effective than the Streptomyces enzymes in digesting the 5601 transcript to RP1. This observation is confirmed by the kinetic data which show a ca. 1.5-fold higher kcat/Km for E. coli PNPase. This difference may reflect the fact that E. coli PNPase digests single-stranded RNAs with high efficiency (12-15, 18, 19, 38). There is an important caveat that applies to the data obtained for the 5601 transcript. It was not possible in the kinetic analyses to utilize enzyme concentrations that were sufficiently low or incubation times that were sufficiently short to prevent the conversion of RP1 to smaller products while still allowing us to quantify the product bands by densitometry. Thus, there was some conversion of RP1 to smaller products in the kinetic analyses, and our kinetic measurements reflect only the amounts of RP1 present when the reactions were quenched and do not account for any conversion of RP1 to those products. Nevertheless, the kinetic data do appear to be internally consistent and demonstrate, as noted above, that the E. coli PNPase functions more effectively in digesting the 5601 transcript than the S. coelicolor PNPase. As Km is a measure of the affinity of an enzyme for its substrate, the kinetic data suggest that the Streptomyces and E. coli PNPases have about the same affinity for the 5601 transcript (Table 2). The difference in kcat is responsible for the higher catalytic efficiency of the E. coli enzyme with this substrate. It should also be noted that the Streptomyces and E. coli enzymes produce identical digestion products from the 5601 transcript with one exception. RP2 was never observed among the products obtained with the Streptomyces enzymes. We suspect that the Streptomyces enzymes do not find the small stem-loop whose 3′ end is predicted to represent the terminus of RP2 to be a barrier to digestion.
Although both the Streptomyces and E. coli PNPases can efficiently digest the small stem-loops present in the region of the 5601 transcript that is 3′ to the intergenic stem-loop (Fig. 1), the S. coelicolor PNPase catalyzes the digestion of the intergenic stem-loop itself (digesting RP3 to RP4) twice as efficiently as does the E. coli enzyme (Table 2). We speculate that the Streptomyces enzymes have evolved to digest RNAs derived from their GC-rich genomes efficiently. Although the rpsO-pnp region of E. coli contains an intergenic hairpin (32, 33), secondary structure modeling predicts a folding free energy for that structure of −30.5 kcal/mole, whereas the predicted folding free energy for the intergenic hairpin in the 5650 transcript is −45.2 kcal/mol. This difference in stability may manifest itself as a decreased ability of the E. coli enzyme, compared with the Streptomyces PNPases, to digest the hairpin.
As noted above, most kinetic studies of PNPases have been performed using synthetic substrates, so it is not possible to compare in a detailed and meaningful way the kinetic constants we have obtained with those that have been published previously. Nevertheless, it is relevant to review some of the data. In particular, it is noteworthy that Km values have been shown by several authors to decrease with increasing chain length. For example, Km values of 0.033 and 0.067 mM were observed in studies of the Micrococcus luteus PNPase with (Ap)5A and (Ap)4A, respectively (14). Similarly, Chou and Singer reported values ranging from 2.5 to 0.033 mM as substrate chain length increased from 2 to 8 nucleotides (13). Reported Km values for poly(A) range from 1 to 10 nM for chain lengths between 300 and 500 nucleotides (15, 19). As shown in Table 2, the Km values observed with our substrates ranged from 0.6 to 6 μM.
Perhaps the most significant observation deriving from the studies presented here is the stimulation of phosphorolysis by the Streptomyces PNPases by NDPs. To our knowledge, this is the first demonstration of a stimulatory effect of NDPs on phosphorolysis by PNPase. Although our data do not reveal the mechanism for this stimulation, we believe that the most straightforward explanation for the effect involves known properties of PNPase. The reaction catalyzed by PNPase is reversible, and the enzyme can polymerize NDPs under appropriate conditions (20, 24). We posit that the Streptomyces enzymes have evolved with the ability to carry out polymerization under conditions that also support phosphorolysis and that short 3′ tails are added to the ends of RNA substrates under these conditions. These tails enhance the binding of the RNA substrate to the enzyme, manifested as the decrease in Km observed in the presence of NDPs, leading to an increase in the efficiency of phosphorolysis. This hypothesis is supported by the observation that the Km for the digestion of the 5650 transcript in the presence of NDPs is identical to the Km for the digestion of the 5601 transcript, which possesses a single-stranded 3′ tail, in the absence of NDPs. Our EMSA studies are consistent with this hypothesis as the data of Fig. 7 demonstrate that S. coelicolor PNPase binds the 5650 transcript more effectively than the E. coli PNPase. Thus, the NDPs “convert” the 5650 transcript to a form for which the S. coelicolor PNPase has a higher affinity. Since multiple complexes are observed for the S. coelicolor PNPase, it was not a simple matter to calculate dissociation constants from the data of Fig. 7. Nevertheless, inspection of the figure shows that essentially all of the substrate in the reaction mixture was bound by the S. coelicolor PNPase when the enzyme was present at a concentration of 0.6 μM while a significant amount of unbound substrate was observed in reaction mixtures containing 1.3 μM E. coli PNPase. As we expected, it was not possible to observe an effect of NDPs on substrate binding, since in the absence of potassium phosphate the added NDPs were polymerized by the PNPases (data not shown). We attempted to demonstrate the synthesis of short 3′ tails in phosphorolysis reactions in the presence of NDPs using cDNA cloning but obtained no clones with such tails. We suspect that such tails, if they are synthesized as we envision during phosphorolysis in the presence of NDPs, are too short-lived to be isolated by cDNA cloning. We note here that we cannot at this point eliminate other explanations for the effects of NDPs on phosphorolysis, such as an allosteric effect on PNPase. There are three potential active sites in the full trimer, only one of which can be occupied by RNA. However, NDPs could presumably bind to any or all of the sites to exert an allosteric effect on PNPase activity.
The question remains, since PNPases clearly prefer single-stranded RNAs as substrates, how are the E. coli and particularly the Streptomyces enzymes able to digest the stem-loop structures found in the substrates we used in the experiments described here? Spickler and Mackie have suggested a reasonable explanation for this observation (38). They argue that the balance between PNPase stalling and digestion at stem-loops is determined by a corresponding balance between the rate of enzyme dissociation at stem-loop stall points and the melting of those stem-loops, making them accessible to PNPase cleavage. One possibility is that either the core or the peripheral RNA binding sites in Streptomyces PNPases are slightly stronger, resulting in retention of “stalled substrates,” rather than their release. Retention would allow time for breathing of the stem, permitting the stalled enzyme to continue phosphorolysis. Put differently, the Streptomyces enzymes appear to be slightly more processive, and this must reflect RNA binding.
Acknowledgments
This work was supported by a grant from the Emory University Research Committee, by National Science Foundation grant MCB-0133520 to G.H.J., and by grant MT5396 from the Canadian Institutes of Health Research to G.A.M.
Footnotes
Published ahead of print on 26 October 2007.
REFERENCES
- 1.Black, D. L., R. Chan, H. Min, J. Wang, and L. Bell. 1998. The electrophoretic mobility shift assay for RNA binding proteins, p. 109-136. In C. W. H. Smith (ed.), RNA: protein interactions. Oxford University Press, Oxford, United Kingdom.
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 3.Bralley, P., B. Gust, S. A. Chang, K. F. Chater, and G. H. Jones. 2006. RNA 3′-tail synthesis in Streptomyces: in vitro and in vivo activities of RNase PH, the SCO3896 gene product and PNPase. Microbiology 152627-636. [DOI] [PubMed] [Google Scholar]
- 4.Bralley, P., and G. H. Jones. 2002. cDNA cloning confirms the polyadenylation of RNA decay intermediates in Streptomyces coelicolor. Microbiology 1481421-1425. [DOI] [PubMed] [Google Scholar]
- 5.Bralley, P., and G. H. Jones. 2004. Organization and expression of the polynucleotide phosphorylase gene (pnp) of Streptomyces: Processing of pnp transcripts in Streptomyces antibioticus. J. Bacteriol. 1863160-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bralley, P., and G. H. Jones. 2003. Overexpression of the polynucleotide phosphorylase gene (pnp) of Streptomyces antibioticus affects mRNA stability and poly(A) tail length but not ppGpp levels. Microbiology 1492173-2182. [DOI] [PubMed] [Google Scholar]
- 7.Bralley, P., and G. H. Jones. 2001. Poly(A) polymerase activity and RNA polyadenylation in Streptomyces coelicolor A3(2). Mol. Microbiol. 401155-1164. [DOI] [PubMed] [Google Scholar]
- 8.Cao, G. J., and N. Sarkar. 1992. Identification of the gene for an Escherichia coli poly(A) polymerase. Proc. Natl. Acad. Sci. USA 8910380-10384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpousis, A. J., N. F. Vanzo, and L. C. Raynal. 1999. mRNA degradation: a tale of poly (A) and multiprotein machines. Trends Genet. 1524-28. [DOI] [PubMed] [Google Scholar]
- 10.Chang, S. A., P. Bralley, and G. H. Jones. 2005. The absB gene encodes a double-strand specific endoribonuclease that cleaves the readthrough transcript of the rspO-pnp operon in Streptomyces coelicolor. J. Biol. Chem. 28033213-33219. [DOI] [PubMed] [Google Scholar]
- 11.Chen, H.-C., Y. H. Hsu, and N.-S. Lin. 2007. Downregulation of Bamboo mosaic virus replication requires the 5′ apical hairpin stem loop structure and sequence of satellite RNA. Virology. doi: 10.1016/j.virol. 2007.03.050. [DOI] [PubMed]
- 12.Cheng, Z. F., and M. P. Deutscher. 2005. An important role for RNase R in mRNA decay. Mol. Cell 17313-318. [DOI] [PubMed] [Google Scholar]
- 13.Chou, J. Y., and M. Singer. 1970. The effect of chain length on the phosphorolysis of oligonucleotides by polynucleotide phosphorylase. J. Biol. Chem. 2451005-1011. [PubMed] [Google Scholar]
- 14.Chou, J. Y., and M. Singer. 1970. A kinetic analysis of the phosphorolysis of oligonucleotides by polynucleotide phosphorylase. J. Biol. Chem. 245995-1004. [PubMed] [Google Scholar]
- 15.Chou, J. Y., M. Singer, and P. McPhie. 1975. Kinetic studies on the phosphorolysis of polynucleotides by polynucleotide phosphorylase. J. Biol. Chem. 250508-514. [PubMed] [Google Scholar]
- 16.Clarke, D. J., and B. C. Dowds. 1994. The gene coding for polynucleotide phosphorylase in Photorhabdus sp. strain K122 is induced at low temperatures. J. Bacteriol. 1763775-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coburn, G. A., and G. A. Mackie. 1999. Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog. Nucleic Acids Res. Mol. Biol. 6255-105. [DOI] [PubMed] [Google Scholar]
- 18.Godefroy, T. 1970. Kinetics of polymerization and phosphorolysis reactions of Escherichia coli polynucleotide phosphorylase-evidence for multiple binding of polynucleotide in phosphorolysis. Eur. J. Biochem. 14222-231. [DOI] [PubMed] [Google Scholar]
- 19.Godefroy, T., M. Cohn, and M. Grunberg-Manago. 1970. Kinetics of polymerization and phosphorolysis reactions of E. coli polynucleotide phosphorylase-role of oligonucleotides in polymerization. Eur. J. Biochem. 12236-249. [DOI] [PubMed] [Google Scholar]
- 20.Godefroy-Colburn, T., and M. Grunberg-Manago. 1972. Polynucleotide phosphorylase. Enzymes 7533-574. [Google Scholar]
- 21.He, B., M. Rong, D. Lyakhov, H. Gartenstein, G. Diaz, R. Castagna, W. T. McAllister, and R. K. Durbin. 1997. Rapid mutagenesis and purification of phage RNA polymerases. Protein Expr. Purif. 9142-151. [DOI] [PubMed] [Google Scholar]
- 22.Jones, G. H., and M. J. Bibb. 1996. Guanosine pentaphosphate synthetase from Streptomyces antibioticus is also a polynucleotide phosphorylase. J. Bacteriol. 1784281-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones, G. H., M. F. Symmons, J. S. Hankins, and G. A. Mackie. 2003. Overexpression and purification of untagged polynucleotide phosphorylases. Protein Expr. Purif. 32202-209. [DOI] [PubMed] [Google Scholar]
- 24.Littauer, U. Z., and H. Soreq. 1982. Polynucleotide phosphorylase. Enzymes 15517-553. [Google Scholar]
- 25.Liu, X., and M. A. Gorovsky. 1993. Mapping the 5′ and 3′ ends of Tetrahymena thermophila mRNAs using RNA ligase mediated amplification of cDNA ends (RLM-RACE). Nucleic Acids Res. 214954-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohanty, B. K., and S. R. Kushner. 1999. Analysis of the function of Escherichia coli poly(A) polymerase in RNA metabolism. Mol. Microbiol. 341094-1108. [DOI] [PubMed] [Google Scholar]
- 27.Mohanty, B. K., and S. R. Kushner. 2002. Polyadenylation of Escherichia coli transcripts plays an integral role in regulating intracellular levels of polynucleotide phosphorylase and RNase E. Mol. Microbiol. 451315-1324. [DOI] [PubMed] [Google Scholar]
- 28.Mohanty, B. K., and S. R. Kushner. 2000. Polynucleotide phosphorylase functions both as a 3′-5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl. Acad. Sci. USA 9711966-11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohanty, B. K., and S. R. Kushner. 1999. Residual polyadenylation in poly(A) polymerase I (pcnB) mutants of Escherichia coli does not result from the activity encoded by the f310 gene. Mol. Microbiol. 341109-1119. [DOI] [PubMed] [Google Scholar]
- 30.O'Hara, E. B., J. A. Chekanova, C. A. Ingle, Z. R. Kushner, E. Peters, and S. R. Kushner. 1995. Polyadenylation helps regulate mRNA decay in Escherichia coli. Proc. Natl. Acad. Sci. USA 921807-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rauhut, R., and G. Klug. 1999. mRNA degradation in bacteria. FEMS Microbiol. Rev. 23353-370. [DOI] [PubMed] [Google Scholar]
- 32.Régnier, P., and C. Portier. 1986. Initiation, attenuation and RNase III processing of transcripts from the Escherichia coli operon encoding ribosomal protein S15 and polynucleotide phosphorylase. J. Mol. Biol. 18723-32. [DOI] [PubMed] [Google Scholar]
- 33.Robert-Le Meur, M., and C. Portier. 1992. E. coli polynucleotide phosphorylase expression is autoregulated through an RNase III-dependent mechanism. EMBO J. 112633-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rott, R., G. Zipor, V. Portnoy, V. Liveanu, and G. Schuster. 2003. RNA polyadenylation and degradation in cyanobacteria are similar to the chloroplast but different from E. coli. J. Biol. Chem. 27815771-15777. [DOI] [PubMed] [Google Scholar]
- 35.Sarkar, N. 1996. Polyadenylation of mRNA in bacteria. Microbiology 1423125-3133. [DOI] [PubMed] [Google Scholar]
- 36.Sarkar, N. 1997. Polyadenylation of mRNA in prokaryotes. Annu. Rev. Biochem. 66173-197. [DOI] [PubMed] [Google Scholar]
- 37.Sohlberg, B., J. Huang, and S. N. Cohen. 2003. The Streptomyces coelicolor polynucleotide phosphorylase homologue, and not the putative poly(A) polymerase can polyadenylate RNA. J. Bacteriol. 1857273-7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spickler, C., and G. A. Mackie. 2000. Action of RNase II and polynucleotide phosphorylase against RNAs containing stem-loops of defined structure. J. Bacteriol. 1822422-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sulewiski, M., S. P. Marchese-Ragona, K. A. Johnson, and S. J. Benkovic. 1989. Mechanism of PNPase. Biochemistry 285855-5864. [DOI] [PubMed] [Google Scholar]
- 40.Symmons, M., G. H. Jones, and B. Luisi. 2000. A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity and regulation. Structure 81215-1226. [DOI] [PubMed] [Google Scholar]
- 41.Yehudai-Resheff, S., M. Hirsh, and G. Schuster. 2001. Polynucleotide phosphorylase functions as both an exonuclease and a poly(A) polymerase in spinach chloroplasts. Mol. Cell. Biol. 215408-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 313406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]