Abstract
Chorismate mutase (CM) catalyzes the rearrangement of chorismate to prephenate in the biosynthetic pathway that forms phenylalanine and tyrosine in bacteria, fungi, plants, and apicomplexan parasites. Since this enzyme is absent from mammals, it represents a promising target for the development of new antimycobacterial drugs, which are needed to combat Mycobacterium tuberculosis, the causative agent of tuberculosis. Until recently, two putative open reading frames (ORFs), Rv0948c and Rv1885c, showing low sequence similarity to CMs have been described as “conserved hypothetical proteins” in the M. tuberculosis genome. However, we and others demonstrated that these ORFs are in fact monofunctional CMs of the AroQ structural class and that they are differentially localized in the mycobacterial cell. Since homologues to the M. tuberculosis enzymes are also present in Mycobacterium smegmatis, we cloned the coding sequences corresponding to ORFs MSMEG5513 and MSMEG2114 from the latter. The CM activities of both ORFs was determined, as well as their translational start sites. In addition, we analyzed the promoter activities of three M. tuberculosis loci related to phenylalanine and tyrosine biosynthesis under a variety of conditions using M. smegmatis as a surrogate host. Our results indicate that the aroQ (Rv0948c), *aroQ (Rv1885c), and fbpB (Rv1886c) genes from M. tuberculosis are constitutively expressed or subjected to minor regulation by aromatic amino acids levels, especially tryptophan.
Tuberculosis (TB), a serious infectious disease caused by Mycobacterium tuberculosis, still remains a public health problem in the world. It has been estimated that one-third of the world's population is latently infected with M. tuberculosis and that almost 9 million new cases and 2 million deaths occur each year (19). The low efficacy of the currently available TB vaccine (Mycobacterium bovis BCG), the emergence of multidrug-resistant strains of M. tuberculosis, and the global spread of human immunodeficiency virus (which increases the risk of TB development) are among the factors underlying the resurgence of TB (24). New drugs and second-generation vaccines are urgently required to control TB, but the complex biology of M. tuberculosis has hindered the development of novel therapeutic tools.
M. tuberculosis is a sophisticated intracellular pathogen that can persist for months or years within the human host, and different mechanisms related to its entry, survival, and replication in macrophages are involved in the infection process (51, 56). The bacilli are notably able to interfere with normal macrophage functions, such as antigen presentation and phagosome maturation, residing in a specialized, nonacidified compartment (44). Of particular interest is the fact that important nutrients are restricted or unavailable inside macrophages, so that M. tuberculosis cells are probably subjected to long-term starvation during the early and latent stages of infection (6, 36, 50). Since many anabolic and catabolic pathways should be specifically activated or repressed in response to these challenging conditions encountered by the bacilli, a better understanding of the general metabolism of M. tuberculosis is crucial to identifying good targets for rational drug design.
In this context, a possible approach to selective antimycobacterial chemotherapy is the characterization of enzymes not only unique to the pathogen but also vital to its growth. The shikimate pathway, for instance, constitutes an attractive target for the development of antimicrobial agents and herbicides, since it is present in bacteria, fungi, plants, and apicomplexan parasites but is absent from animals (5). In this pathway, erythrose 4-phosphate and phosphoenolpyruvate are converted to chorismate through seven catalytic steps. Chorismate is a key intermediate in the biosynthesis of a wide range of compounds, including aromatic amino acids, folate cofactors, menaquinones, ubiquinones, and siderophores (17). In addition, it was shown that the shikimate pathway is essential for the in vitro viability of M. tuberculosis cells (39), thus demonstrating that the enzymes of this pathway are promising targets for the search of inhibitors with potential antimycobacterial activity.
Although many shikimate pathway enzymes have been studied (20, 35, 52) and a tryptophan auxotroph has been generated (37), very little is known about aromatic amino acid metabolism in M. tuberculosis. The conversion of chorismate to prephenate, catalyzed by chorismate mutase (CM), is the first committed step in the biosynthesis of phenylalanine and tyrosine (40). As a rare example of an enzyme-catalyzed pericyclic process known as Claisen rearrangement, this strongly exergonic reaction provides rate enhancements of more than 106 and has become a model for the study of enzyme catalysis (23). Moreover, CMs are extremely versatile enzymes and can be structurally divided into two major groups: the type I or AroH class, which comprises CMs characterized by a trimeric pseudo α/β-barrel structure (12), and the type II or AroQ class, which comprises CMs characterized by a dimeric α-helical structure (29). CMs can also be monofunctional, bifunctional (generally fused to another shikimate pathway member), exported, and/or allosterically regulated (9, 17, 54).
Despite the structural diversity, all CMs perform the same basic reaction, and most active-site residues are highly conserved in both AroH and AroQ classes. On the other hand, these proteins are noted for their low sequence similarity at the amino acid level, a feature that often leads to the dubious assignment of open reading frames (ORFs) during the annotation phase of genomes (9). Accordingly, in the original genome annotation of M. tuberculosis, two ORFs, Rv0948c and Rv1885c, exhibiting CM motifs were classified as “conserved hypothetical proteins” (13). We show in the present study that these are both monofunctional CMs of the AroQ class. It should be pointed out that Rv1885c has been recently characterized in detail by others (27, 41, 45). We also show that homologues of each CM from M. tuberculosis are found in Mycobacterium smegmatis, a nonpathogenic species. These homologues (which correspond to ORFs MSMEG5513 and MSMEG2114, respectively) were cloned, expressed in Escherichia coli, and demonstrated to possess CM activity as their M. tuberculosis counterparts. Finally, we analyzed the promoter region of each CM gene and the fbpB operon from M. tuberculosis under a variety of conditions using a β-galactosidase (β-Gal)-based reporter system. Our results indicate that the expression of these promoters is constitutive or regulated in response to variable levels of aromatic amino acids in M. tuberculosis.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
E. coli DH5α, DH10B and BL21(DE3) (Novagen) strains were grown in Luria-Bertani (LB) medium (Difco) containing 50 μg of carbenicillin ml−1, 50 μg of kanamycin ml−1, 20 μg of streptomycin ml−1, or 20 μg of tetracycline ml−1, where appropriate. M. smegmatis mc2155 (47) was grown in Lemco medium (Lemco powder [5 g liter−1], NaCl [5 g liter−1], Bacto peptone [10 g liter−1]) with 0.05% (wt/vol) Tween 80 (liquid) or 15 g of Bacto agar (solid) liter−1. M. smegmatis was also grown in minimal medium (31) containing 0.05% (wt/vol) Tween 80 and glucose at 0.2% (wt/vol). l-Amino acids (phenylalanine, tyrosine, or tryptophan; Sigma) were added to minimal medium at the specified concentrations. Kanamycin or streptomycin were added to M. smegmatis cultures at a concentration of 20 μg ml−1, where required.
Bioinformatics.
Sequence data for M. tuberculosis H37Rv and Mycobacterium leprae TN were obtained from the TubercuList (http://genolist.pasteur.fr/TubercuList/) and Leproma (http://genolist.pasteur.fr/Leproma/) databases, respectively. Sequence data for M. smegmatis mc2155 and other bacteria were obtained from the Comprehensive Microbial Resource (http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi/) database at The Institute for Genomic Research (TIGR; http://www.tigr.org/).
Determination of translational start site.
In order to identify the translational start site of ORFs MSMEG5513 and MSMEG2114 from M. smegmatis, approximately 250 bp of DNA comprising sequences upstream and downstream of the −10 region of each putative CM gene were PCR amplified using the primers TSS1 (5′-TACCGCGCGTGGTACCCATCCTTTGCG-3′) and TSS2 (5′-CACGGCACTGCAGGTCTCAGGCATGTG-3′) for MSMEG5513 and the primers TSS3 (5′-GAGACGGTGCGGTACCCGCTGAACGG-3′) and TSS4 (5′-GGCGAGCCTGCAGGAAGCGAGCACCG-3′) for MSMEG2114. Each amplified product was ligated into the KpnI and PstI restriction sites (underlined in the primer sequences) of the pMV261 expression vector (48) in place of the hsp60 promoter, as described by Jain et al. (26), resulting in pSP1 and pSP2 (listed in Table 1) . A lacZ version from pMC1871 (11) was adapted for cloning into the PstI restriction site of pSP1 and pSP2, creating translational fusions of the M. smegmatis sequences to the E. coli β-Gal gene and resulting in pTASS1 and pTASS2, respectively. These plasmids were used as controls and also as templates for introducing three (pTASS1) and two (pTASS2) frameshift mutations downstream of the first start codon predicted for each M. smegmatis CM sequence (see Fig. 3A and B). Site-specific mutagenesis was carried out by using the QuikChange II site-directed mutagenesis kit (Stratagene) and appropriate primer pairs. Pfu Ultra DNA polymerase and 5 or 10% dimethyl sulfoxide were used in all of the amplifications. In outline, the PCR conditions were, following an initial denaturation step, 15 cycles of 95°C for 1 min, 55°C for 1 min, and 68°C for 15 min, with a final extension step of 68°C for 20 min. DpnI digestion of templates was performed at 37°C for 1 h, and 10 μl of each reaction was used to transform E. coli. After DNA sequencing to confirm correct translational fusion of the wild-type and mutant constructions to lacZ, control and mutant plasmids were separately transformed into electrocompetent M. smegmatis cells. Transformants were selected on Lemco medium containing kanamycin at 20 μg ml−1. Cell extracts from three individual M. smegmatis transformants (grown at 37°C for 20 h in 5 ml of Lemco medium) were prepared, and each extract was assayed in duplicate for β-Gal activity (32).
TABLE 1.
Plasmids used in this study
| Plasmid | Descriptiona | Source or reference |
|---|---|---|
| pCAN1 | Predicted aroQMt promoter region (C1, 250 bp) in pSM128 | This study |
| pET-23a(+) | Expression vector; Apr | Novagen |
| pET-23a(+)-Msm aroQ | pET-23a(+) plus aroQMs coding sequence (315 bp) | This study |
| pET-23a(+)-Msm *aroQ | pET-23a(+) plus *aroQMs coding sequence without signal peptide (489 bp) | This study |
| pET-23a(+)-Mtb aroQ | pET-23a(+) plus aroQMt coding sequence (318 bp) | This study |
| pET-23a(+)-Mtb *aroQ | pET-23a(+) plus *aroQMt coding sequence without signal peptide (504 bp) | This study |
| pGEM-T Easy | Cloning vector; Apr | Promega |
| pMC1871 | Cloning vector, lacZ; Tcr | 11 |
| pMV261 | E. coli-mycobacterial shuttle vector, oriE, oriM, Phsp60; Kmr | 48 |
| pPAN1 | Predicted M. tuberculosis fbpB promoter region (P1, 215 bp) in pSM128 | This study |
| pPAN2 | Predicted *aroQMt promoter region (P2, 267 bp) in pSM128 | This study |
| pSM128 | Promoter-probe vector, lacZ reporter gene; Smr | 18 |
| pSP1 | 250 bp of aroQMs promoter and 5′ end in place of Phsp60 from pMV261 | This study |
| pSP2 | 250 bp of *aroQMs promoter and 5′ end in place of Phsp60 from pMV261 | This study |
| pTASS1 | pSP1 plus lacZ gene from pMC1871 | This study |
| pTASS2 | pSP2 plus lacZ gene from pMC1871 | This study |
Tcr, tetracycline resistance; Apr, ampicillin resistance; Smr, streptomycin resistance; Kmr, kanamycin resistance.
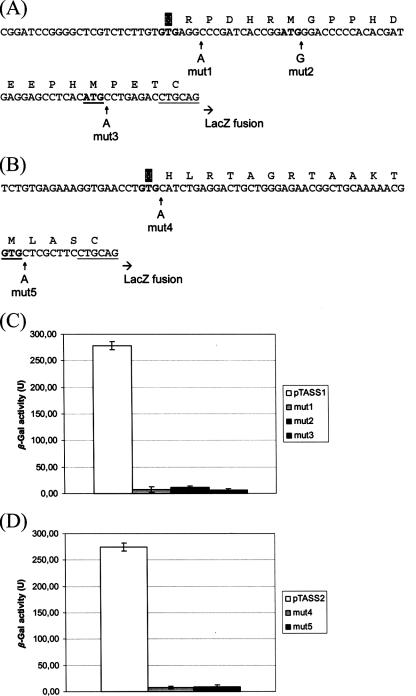
FIG. 3.
Determination of translational start site for ORFs MSMEG5513 and MSMEG2114 from M. smegmatis. The DNA sequences of the 5′ end and the region immediately upstream of the aroQMs (MSMEG5513) and *aroQMs (MSMEG2114) genes are shown (panels A and B, respectively). Potential translational start sites are indicated in boldface. Underlined start codons indicate the originally predicted start sites, and methionines in white on a black background correspond to the experimentally identified translational start sites. The introduced frameshift mutations are marked with arrows in each sequence. The PstI restriction site to which a lacZ fusion was made for each construction is underlined. (C) Quantitative analysis of β-Gal activity in M. smegmatis cells transformed with the wild-type (pTASS1) or mutant constructions (mut1, mut2, and mut3) for identifying the MSMEG5513 translational start site. (D) Quantitative analysis of β-Gal activity in M. smegmatis cells transformed with the wild-type (pTASS2) or mutant constructions (mut4 and mut5) for identifying the MSMEG2114 translational start site. The results are the means ± the standard deviations of three individual transformants, each assayed in duplicate. Units are given in nanomoles of o-nitrophenyl galactoside produced per minute per milligram of total protein.
Construction of CM expression plasmids.
The predicted M. tuberculosis aroQ (Rv0948) and *aroQ (Rv1885c) coding sequences were PCR amplified from genomic DNA with the primers MTB AROQC1 (forward, 5′-ACCATATGAGACCAGAACCCCCACATCACG-3′) and MTB AROQC2 (reverse, 5′-GTGGATCCTCAGTGACCGAGGCGGCCCC-3′), which amplified a 318-bp product, and the primers MTB AROQP2a (forward, 5′-CGCATATGGACGGCACCAGCCAGTTAGCC-3′) and MTB AROQP2b (reverse, 5′-CGGGATCCTCAGGCCGGCGGTAGGGCC-3′), which amplified a 504-bp product (corresponding to the predicted mature M. tuberculosis *AroQ protein), respectively. The predicted M. smegmatis aroQ (MSMEG5513) and *aroQ (MSMEG2114) coding sequences were PCR amplified from genomic DNA with the primers MSM CMC3 (forward, 5′-TCTCATATGAGGCCCGATCACCGGATG-3′) and MSM CMC4 (reverse, 5′-GCGGATCCTCAGTACCCGAGGCGG-3′), which amplified a 315-bp product, and the primers MSM CMP1 (forward, 5′-CCCATATGGACGACGCGAGCCCGCTG-3′) and MSM CMP4 (reverse, 5′-TGTGGATCCTCATCGGCAGTACGAATGCGT-3′), which amplified a 489-bp product (corresponding to the predicted mature M. smegmatis *AroQ protein), respectively. NdeI and BamHI restriction sites are underlined in the forward and reverse primer sequences, respectively. Pfu DNA polymerase (Stratagene) was used in all of the reactions under standard PCR conditions. Each amplified product was gel purified, cleaved with NdeI and BamHI, and ligated into the pET-23a(+) expression vector (Novagen) to construct the related CM expression plasmid (listed in Table 1). DNA sequences of the cloned fragments were determined by using a Thermo Sequenase radiolabeled terminator cycle sequencing kit (USB).
Expression of CM in E. coli cells.
A single colony of E. coli BL21(DE3) previously transformed with each CM expression plasmid [pET-23a(+)-Mtb aroQ, pET-23a(+)-Mtb *aroQ, pET-23a(+)-Msm aroQ, or pET-23a(+)-Msm *aroQ] was inoculated into 50 ml of LB medium containing carbenicillin at 50 μg ml−1. Control experiments were performed under the same conditions, except that transformed E. coli cells carried the pET-23a(+) expression vector. Cells were incubated (180 rpm) at 37°C up to a value of 0.4 for absorbance at 600 nm and either induced by supplementation with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) or not induced and grown at 37°C for additional 24 h. Culture aliquots were sampled at specific time intervals, centrifuged at 14,000 rpm for 3 min, and stored at −20°C. Pellets were suspended in 500 μl of 50 mM Tris-Cl (pH 7.8), disrupted by sonication, and centrifuged at 14,000 rpm for 30 min at 4°C. The soluble and insoluble fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with Coomassie brilliant blue staining. The CM activity of crude extracts was measured immediately after cell disruption.
CM activity assay.
CM activity was assayed by monitoring the conversion of chorismate to prephenate plus phenylpyruvate according to the method of Cotton and Gibson (15) and as adapted by Ahmad and Jensen (1). Briefly, the reaction mixture in a final volume of 200 μl contained modified buffer A (50 mM potassium phosphate buffer [pH 7.5], containing 1 mM dithiothreitol), 1 mM chorismic acid (barium salt; Sigma), and variable volumes (5 to 20 μl) of crude extract. After incubation at 37°C for 20 min, 100 μl of 1 N HCl was added to terminate the first reaction (the enzymatic formation of prephenate from chorismate). After a further incubation at 37°C for 15 min, 700 μl of 2.5 N NaOH was added to terminate the second reaction (the nonenzymatic formation of phenylpyruvate from prephenate), and the absorbance at 320 nm was measured to determine the phenylpyruvate concentration. Blanks (without crude extract) were included, since chorismate undergoes spontaneous conversion to prephenate at 37°C (21).
The protein concentration in crude extracts was determined by the method of Bradford (7) using a Bio-Rad protein assay kit and bovine serum albumin as a standard.
Construction of mycobacterial reporter plasmids.
The predicted aroQ (Rv0948c), *aroQ (Rv1885c), and fbpB (Rv1886c) promoter regions from M. tuberculosis were PCR amplified by using the following primers: AROQCAN1 (5′-TCCCCCGGGCCAATTTTGGGTGCACTTTT-3′) and AROQCAN2 (5′-TCCCCCGGGGGTTCATGGCTGCTAACTCC-3′) for a 250-bp region (C1) upstream of the aroQ gene; AROQPAN1 (5′-CCGCCCGGGGATTCTGAGAAATCCGCGATA-3′) and AROQPAN2 (5′-CCGCCCGGGGAATCTTTCGGCTCACGTCT-3′) for a 215-bp region (P1) upstream of the fbpB operon; and AROQPAN3 (5′-GTTCCCGGGGAACTTCGTTCGTAGCAGCA-3′) and AROQPAN4 (5′-CAGCCCGGGGCCTAGTGGTGCAATCAGC-3′) for a 267-bp product (P2) covering the 3′ end of the fbpB gene, the intergenic region fbpB-*aroQ, and the 5′ end of the *aroQ gene (see Fig. 5 for details). The amplified products were cloned into pGEM-T Easy (Promega) and subsequently excised as SmaI (the restriction site is underlined in primer sequences) fragments. These were gel purified and subcloned into the ScaI restriction site of the integrative promoter-probe vector pSM128 (18) to construct each mycobacterial reporter plasmid (listed in Table 1). The pSM128 vector carries a cII-lacZ fusion, the mycobacteriophage L5 integrase gene, attachment sites, and a streptomycin/spectinomycin resistance cassette. This vector is present in only one copy per cell. DNA sequences of the cloned fragments were determined by automated sequencing (MWG Biotech) to both confirm the identity of the amplified products and check the in-frame transcriptional fusion of inserts to the promoterless lacZ gene.
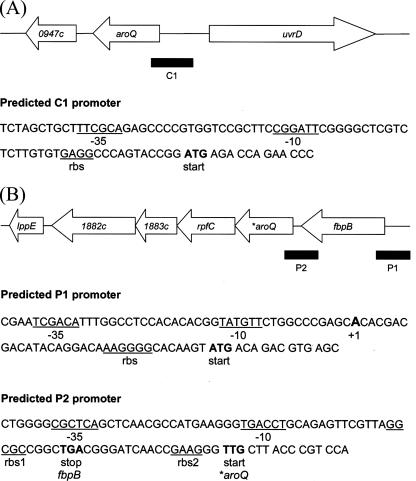
FIG. 5.
Genetic organization of the aroQ gene and the fbpB operon (which contains the *aroQ gene) on the M. tuberculosis H37Rv chromosome. (A) The genome region containing the M. tuberculosis aroQ gene and its predicted promoter (C1) is shown. The potential −35/−10 consensus sequence and ribosomal binding site (rbs) are underlined, and the aroQ start codon is indicated in boldface. The C1 sequence comprises approximately 250 bp covering the 5′ end and the upstream sequence of aroQ and was cloned in pSM128 to make pCAN1. (B) The fbp operon from M. tuberculosis. The predicted fbpB (P1) and *aroQ (P2) promoters are shown. The potential −35/−10 consensus sequences and ribosomal binding sequences for both genes are underlined (rbs1 and rbs2 refer to an annotated and proposed ribosomal binding site ] for the *aroQMt gene, respectively); start and stop codons are indicated in boldface. The fbpB transcriptional start site (+1) was determined by Harth et al. (25). The P1 (215 bp) and P2 (267 bp) sequences were PCR amplified and cloned into pSM128, yielding pPAN1 and pPAN2, respectively.
Promoter activity assays.
M. smegmatis electrocompetent cells were prepared and transformed as described by Snapper et al. (47) and Parish and Stoker (38). Each mycobacterial reporter plasmid (pCAN1, pPAN1, or pPAN2) was electroporated into M. smegmatis cells, and transformants were selected on Lemco medium containing 20 μg of streptomycin ml−1. For each construction, three independent transformants were assayed in duplicate for the determination of promoter activity in Lemco medium (liquid) or minimal medium (liquid) containing different concentrations of phenylalanine, tyrosine, or tryptophan (1 or 5 mM each). M. smegmatis cells transformed with the pSM128 vector were grown under the same conditions and used as controls. Cell extracts were prepared and assayed for β-Gal activity as described previously (32).
RESULTS AND DISCUSSION
Identification of CMs in mycobacteria.
We are interested in the chorismate biosynthesis pathway of mycobacteria. In particular, we wanted to identify the CM enzymes, which were not precisely annotated in the original genome sequence of M. tuberculosis H37Rv (13) and the subsequent reannotation (10). In the annotation, two ORFs (Rv0948c and Rv1885) with some similarity to CMs, including the well-known monofunctional periplasmic CM from Erwinia herbicola (54), are found, although these ORFs are described as “conserved hypothetical proteins.” In addition, genes coding for prephenate dehydratase (PheA) and prephenate dehydrogenase (TyrA), enzymes that utilize the prephenate formed by CM from chorismate, were promptly identified: they correspond to ORFs Rv3838c and Rv3754, respectively, and do not possess any associated CM domain (13). Thus, at least one CM should be present in the M. tuberculosis genome. The unclear description of CMs in M. tuberculosis was probably due to the short length of many CM genes, the low levels of sequence similarity among these proteins, and the fact that monofunctional CMs, which are becoming increasingly apparent (9), were little studied at that time.
Nevertheless, previous work provided evidence that Rv1885c constitutes a monofunctional CM of the AroQ structural class and that it is probably exported (9, 30). This was confirmed by others (27, 41, 45). Prakash et al. (41) and Sasso et al. (45) experimentally demonstrated that Rv1885c codes for an exported monofunctional CM in M. tuberculosis; these authors also determined basic structural and kinetic characteristics of the enzyme. Kim et al. (27) detected CM activity due to secretion of Rv1885c in the culture filtrate of bacilli in different stages of growth and identified CM active-site residues by site-directed mutagenesis and crystallographic studies.
In contrast to the studies in M. tuberculosis, very little is known of CMs in M. smegmatis. Homologues of Rv1885c and Rv0948c are present in the M. smegmatis genome, where ORFs MSMEG2114 and MSMEG5513 have been recently annotated as potential CMs (http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi/). A reasonable degree of identity (43.2%) was found between ORF Rv1885c and the M. smegmatis homologue MSMEG2114. Moreover, this protein has a cleavable signal peptide in the amino acid sequence (Fig. 1A). Compared to the prototypal CM domain of the bifunctional protein CM-prephenate dehydratase from E. coli (EcCM) (29), ORF MSMEG2114 shares a conserved active-site glutamate residue (equivalent to Glu52 in the, E. coli sequence), whereas ORF Rv1885c has a valine in the corresponding position. Ökvist et al. (34) proposed that Glu106 in the M. tuberculosis sequence performs the role of E. coli Glu52 in catalytic site formation.
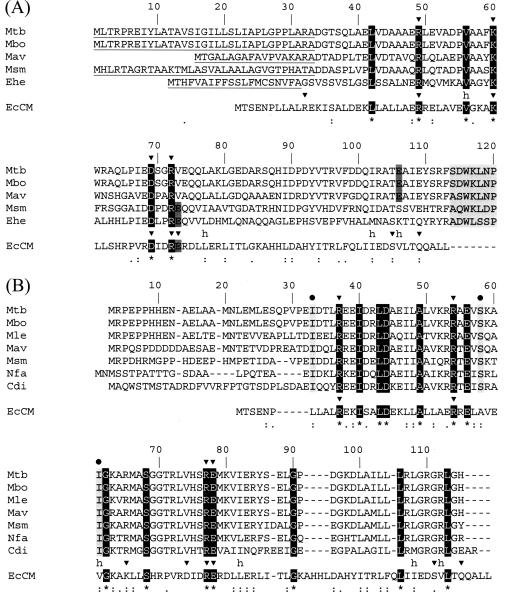
FIG. 1.
Multiple sequence alignments of bacterial CMs with the CM domain of the bifunctional protein CM-prephenate dehydratase from E. coli (EcCM). Sequence data were obtained as described in Materials and Methods, and the program CLUSTAL W (49) was used in both alignments. (A) The putative exported CMs for M. tuberculosis (Mtb), Mycobacterium bovis (Mbo), Mycobacterium avium (Mav), and M. smegmatis (Msm) were aligned with EcCM and the monofunctional *AroQ protein from Erwinia herbicola (Ehe), the first characterized periplasmic CM (54). The signal peptides of *AroQ proteins (as predicted by the program SignalP 3.0 [4]) are underlined. Identical residues are shown in white on a black background and are also indicated by asterisks below the alignment. Strongly similar and weakly similar residues are identified by colons and periods, respectively. The first residues of the C-terminal portion of *AroQ proteins (which displays no homology to known CMs [9]) are shown in light gray. Active-site residues of EcCM are marked with arrowheads, and each “h” represents a hydrophobic residue (29). Glu52 in EcCM has conserved residues in the M. smegmatis and E. herbicola sequences (in dark gray), and Ökvist et al. (34) propose that Glu106 in the M. tuberculosis sequence is equivalent (also in dark gray). We extrapolated their analysis to the M. bovis and M. avium sequences. (B) Multiple sequence alignment of actinomycete AroQ proteins with EcCM. Besides the previously mentioned species (except E. herbicola), Mycobacterium leprae (Mle), Nocardia farcinica (Nfa), and Corynebacterium diphtheriae (Cdi) were included in the alignment. Colors and symbols are the same as described for panel A. Black circles indicate substitutions that naturally occur in all analyzed sequences from actinomycetes (also shown in light gray) and that were demonstrated to maintain or increase CM catalytic efficiency when the corresponding E. coli residues (Leu7, Ala32, and Val35) are mutated in EcCM (28).
Multiple sequence alignment of EcCM with ORFs Rv0948c from M. tuberculosis and MSMEG5513 from M. smegmatis, as well as ORFs from other actinomycetes, indicates that most of the active-site residues are conserved in mycobacteria (Fig. 1B). In a recent work, computational protein design techniques were used to predict variants of EcCM that should maintain catalytic activity in comparison with the wild type (28). These variants were constructed, and determination of steady-state kinetic parameters showed that three mutants (Leu7Ile, Val35Ile, and Ile81Leu/Val85Ile) had similar catalytic efficiency and a mutant (Ala32Ser) had increased catalytic efficiency compared to wild-type EcCM (28). Interestingly, some of these substitutions (Leu7Ile, Ala32Ser, and Val35Ile) occur precisely in identical positions in actinomycete sequences and in both M. tuberculosis and M. smegmatis ORFs (Fig. 1B). Consequently, it appears to be plausible that the mycobacterial sequences use a set of alternative residues in their composition in such a way that final CM catalytic activity should not be markedly affected, pointing to a case of sequence divergence. Identity between Rv0948c and MSMEG5513 is 66.4% at the amino acid level.
In this way, according to the enzyme nomenclature proposed by Jensen and coworkers (9), ORFs Rv0948c and Rv1885c from M. tuberculosis were designated M. tuberculosis AroQMt and *AroQMt, and ORFs MSMEG5513 and MSMEG2114 from M. smegmatis were designated AroQMs and *AroQMs, respectively (asterisks indicate that the proteins possess a signal peptide and are exported, as predicted by the program SignalP 3.0 [4]; http://www.cbs.dtu.dk/services/SignalP/).
Genomic comparison of CM genes.
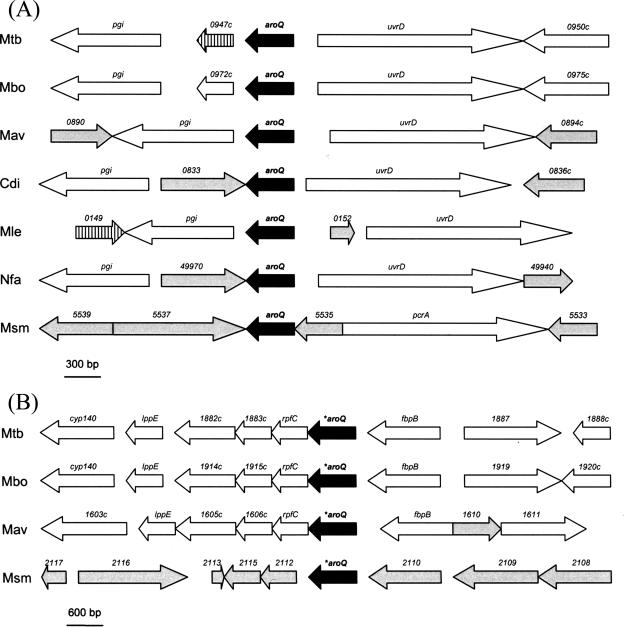
A brief comparison with other mycobacterial genomes reveals that the AroQMt and *AroQMt proteins (as well as the two M. smegmatis homologues) have highly similar counterparts in the related species Mycobacterium bovis, Mycobacterium avium, and Mycobacterium leprae, besides other actinomycetes (Fig. 2). Conservation of gene synteny is particularly significant for the M. tuberculosis aroQ gene and adjacent regions, where genes coding for a probable ATP-dependent DNA helicase and glucose 6-phosphate isomerase (corresponding to, respectively, uvrD/Rv0949 and pgi/Rv0946c in M. tuberculosis) are present in most species and in an almost unchanged pattern (Fig. 2A). Another interesting point is that all actinomycete aroQ genes seem to be monocistronic (in M. tuberculosis, for example, there is an intergenic gap of nearly 300 bp between uvrD and aroQ), while *aroQ genes are in principle part of operons (Fig. 2B). In the latter case, this may suggest a functional relationship among *aroQ and the neighboring genes, which presumably share one or more transcription signals.
FIG. 2.
Genome region comparison of actinomycete CM genes. (A) Genomic organization of aroQ genes in actinomycetes: M. tuberculosis (Mtb), M. bovis (Mbo), M. avium (Mav), C. diphtheriae (Cdi), M. leprae (Mle), N. farcinica (Nfa), and M. smegmatis (Msm). Black arrows indicate the conserved aroQ genes. White and gray arrows indicate genes homologous and nonhomologous to the M. tuberculosis pgi-Rv0950c region, respectively. Hatched arrows indicate pseudogenes. Gene names and numbers refer to the original ORF assignments or as defined by multiple sequence alignments. The numbering of genes is abbreviated, so 0947c represents Rv0947c, 0972c represents Mb0972c, 0890 represents MAP0890, 0833 represents DIP0833, 0149 represents ML0149, 49970 represents nfa49970, and 5539 represents MSMEG5539. (B) Genomic organization of *aroQ genes in mycobacteria. Black arrows indicate the conserved *aroQ genes. White and gray arrows indicate, respectively, genes homologous and nonhomologous to the M. tuberculosis cyp140-Rv1888c region. Gene names and numbers and the numbering of genes are as described for panel A.
However, *aroQ genes are not present in species such as Corynebacterium diphtheriae and Nocardia farcinica, as well as M. leprae. Hence, ML0151c (homologous to aroQMt) from M. leprae (which is supposed to contain the “minimal” mycobacterial gene set) codes for the unique CM present in this Mycobacterium sp., since ML2029 (homologous to *aroQMt) is a pseudogene (14). It is also noteworthy that genes coding for mycolyltransferases (enzymes required for the biosynthesis of trehalose dimycolate, an important molecule in the assembly of the mycobacterial cell envelope) occur surrounding both aroQ and *aroQ genes in M. tuberculosis, M. bovis, and M. avium but not M. smegmatis (Fig. 2). In M. tuberculosis these mycolyltransferases correspond to the Rv0947c and Rv1886c (fbpB) genes. Although Rv0947c codes for a putative mycolyltransferase which is in fact a pseudogene, fbpB codes for the secreted antigen 85B, one of the three components of the antigen 85 complex, a structure responsible for the high-affinity binding of mycobacteria to fibronectin (3).
Translational start site determination.
An inspection of the annotated CMs in the M. smegmatis genome (http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi/) revealed more than one potential translational start site for both aroQ and *aroQ genes (Fig. 3). Specifically, two and one additional start codons could act as sites for translation initiation of the AroQMs (Fig. 3A) and *AroQMs (Fig. 3B) proteins, respectively. Furthermore, multiple sequence alignments of the M. smegmatis enzymes with other mycobacterial CMs showed that homology extended some conserved amino acids upstream of the originally annotated initiation codons, suggesting that the start sites had been misannotated (Fig. 1).
To determine the correct translational start sites for MSMEG5513 (AroQMs) and MSMEG2114 (*AroQMs), we cloned approximately 250 bp of DNA covering the promoter region and the 5′ end of each putative CM gene from M. smegmatis into the pMV261 vector in place of the hsp60 promoter. A translational fusion to lacZ was made for each construction, giving rise to pTASS1 and pTASS2, respectively. These plasmids were used as controls (since they carry only wild-type sequences) and also as templates for introducing specific frameshift mutations (Fig. 3A and B). Each control and mutant plasmid was electroporated into M. smegmatis, cell extracts prepared and β-Gal activity measured in duplicate for three independent transformants. The results for MSMEG5513 and MSMEG2114 are shown in Fig. 3C and D, respectively.
Introduction of three distinct frameshift mutations downstream of the first start codon predicted for MSMEG5513 practically abolished β-Gal activity in M. smegmatis cells (Fig. 3C). Considering that the insertions will have a direct effect on translation, these data indicate that the upstream GTG in the aroQMs sequence represents the actual translational start site (Fig. 3A). Accordingly, the aroQMs coding sequence consists of 315 bp (including stop codon) and gives rise to a protein of 104 amino acids. Likewise, introduction of two different frameshift mutations downstream of the first start codon predicted for MSMEG2114 resulted in very low levels of β-Gal activity in M. smegmatis (Fig. 3D), supporting our hypothesis that the upstream GTG in the *aroQMs sequence is the actual translational start site (Fig. 3B). Thus, the *AroQMs coding sequence consists of 585 bp (including stop codon) and gives rise to a protein of 194 amino acids and to a predicted mature protein of 162 amino acids (Fig. 1A).
We therefore propose that the MSMEG5513 and MSMEG2114 loci are not correctly annotated in the M. smegmatis genome regarding their translational start sites. Since alternative start codons such as GTG frequently occur in mycobacteria, as well as “atypical” ribosomal binding sites, these factors may have influenced proper ORF allocation in the genome regions containing the MSMEG5513 and MSMEG2114 sequences. Moreover, our results indicate that the DNA sequences immediately upstream of both aroQ and *aroQ genes from M. smegmatis constitute functional promoters, since they were able to drive expression of lacZ under the conditions tested.
Expression of mycobacterial CMs in E. coli.
In order to determine whether Rv0948c, Rv1885c, MSMEG5513, and MSMEG2114 were bona fide CMs, we cloned the coding sequences corresponding to each ORF into the pET-23a(+) expression vector. We then conducted assays for CM activity on extracts generated from E. coli producing each recombinant protein. The expression of mycobacterial CMs was observed in the soluble fraction of crude extracts prepared from E. coli BL21(DE3) cells (Fig. 4). It should be pointed out that extensive enzymological and crystallographic data for the Rv1885c gene product have been reported by others (27, 34, 41, 45) and that in the present study this protein was used as a control for the experiments describing Rv0948c and the two M. smegmatis homologues.
FIG. 4.
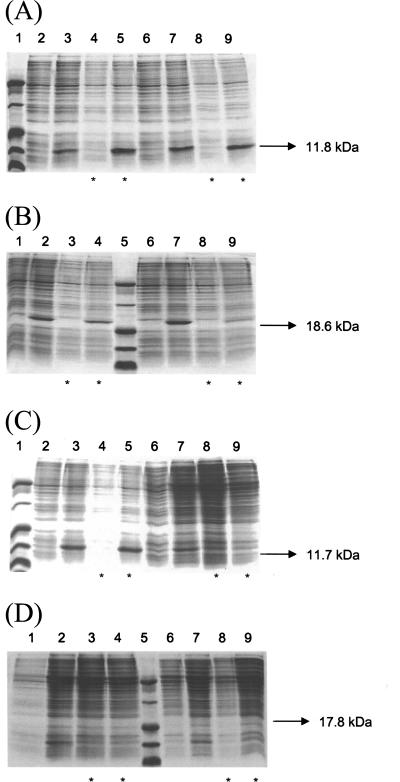
SDS-PAGE analysis of expression of M. tuberculosis and M. smegmatis CMs in the soluble fraction of crude extracts from E. coli BL21(DE3) cells. All samples were grown in LB medium, and asterisks indicate the addition of 1 mM IPTG to cultures reaching an OD600 of 0.4. E. coli transformed with the pET-23a(+) expression vector was used as negative control. (A) Expression of the AroQMt protein (11.8 kDa) in E. coli after 2 h (lanes 2 to 5) and 4 h (lanes 6 to 9) of cell growth. Lanes: 1, molecular mass marker; 2, 4, 6, and 8, host cell extract (negative controls); 3, 5, 7, and 9, E. coli transformed with pET-23a(+)-Mtb aroQ. (B) Expression of the *AroQMt protein (18.6 kDa) in E. coli after 3 h (lanes 1 to 4) and 6 h (lanes 6 to 9) of cell growth. Lanes: 1, 3, 6, and 8, host cell extract (negative controls); 5, molecular mass marker; 2, 4, 7, and 9, E. coli transformed with pET-23a(+)-Mtb *aroQ. (C) Expression of the AroQMs protein (11.7 kDa) in E. coli after 3 h (lanes 2 to 5) and 6 h (lanes 6 to 9) of cell growth. Lanes: 1, molecular mass marker; 2, 4, 6, and 8, host cell extract (negative controls); 3, 5, 7, and 9, E. coli transformed with pET-23a(+)-Msm aroQ. (D) Expression of the *AroQMs protein (17.8 kDa) in E. coli after 3 h (lanes 1 to 4) and 6 h (lanes 6 to 9) of cell growth. Lanes: 1, 3, 6, and 8, host cell extract (negative controls); 5, molecular mass marker; 2, 4, 7, and 9, E. coli transformed with pET-23a(+)-Msm *aroQ.
After 2 and 4 h of cell growth, with or without the addition of 1 mM IPTG to cultures reaching an optical density at 600 nm (OD600) of 0.4, expression of a recombinant protein in agreement with the theoretical molecular mass for AroQMt (11,770.64 Da) was observed in E. coli (Fig. 4A). Analysis by SDS-PAGE indicated that the mature *AroQMt protein (no signal peptide, calculated molecular mass of 18,605.75 Da) was produced in E. coli after 3 and 6 h of cell growth and that IPTG addition was dispensable for the best results (Fig. 4B). Although the expression of AroQMt in E. coli was nearly constant up to 24 h of cell growth, the protein band corresponding to *AroQMt was very faint at 6 h postinduction (Fig. 4B), disappearing completely after 9 h (data not shown).
Maximal expression of the recombinant protein AroQMs (calculated molecular mass of 11,753.54 Da) in soluble form was achieved after 3 h of cell growth, with or without the addition of 1 mM IPTG to E. coli cultures reaching an OD600 of 0.4 (Fig. 4C). Expression of this protein in E. coli decreased after 12 h of cell growth without IPTG supplementation and was very low from 6 h (Fig. 4C) to 24 h (data not shown) postinduction. The mature *AroQMs protein (no signal peptide, calculated molecular mass of 17,817.76 Da) was produced in low levels in E. coli after 3 and 6 h of cell growth, without IPTG supplementation (Fig. 4D).
Leaky protein expression due to cyclic AMP-dependent derepression of the lac operon has been shown to occur in the pET system (22). This may explain why moderate levels of expression were obtained for mycobacterial CMs in the absence of IPTG induction. Furthermore, it is possible that the mycobacterial *AroQ proteins have some toxic effects on E. coli after prolonged incubation at 37°C, since cell growth was diminished and expression of these enzymes disappeared by 6 h (*AroQMs) or 9 h (*AroQMt) postinduction.
Characterization of Rv0948c, Rv1885c, MSMEG5513, and MSMEG2114.
CM catalyzes the intramolecular rearrangement of chorismate to form prephenate. Although it is well established that the nonenzymatic conversion of chorismate to prephenate is significant at 37°C (21), the reaction rate is more than a million times faster at the active site of CMs than free in solution (2). We thus used blanks containing only chorismate in the absence of CM to account for nonenzymatic (chemical) conversion to prephenate. CM activity assays were carried out measuring, under acidic conditions, the formation of phenylpyruvate from prephenate, which was in turn formed from chorismate by the CM activity of each recombinant protein.
To characterize ORFs Rv0948c, MSMEG5513, and MSMEG2114 as CMs (and to confirm the role of Rv1885c), we measured the total CM enzyme activity in the soluble fraction of E. coli (transformed with the corresponding CM expression plasmid) cell extracts. E. coli cells grown under the same conditions and transformed with the pET-23a(+) expression vector were used as controls. CM specific activity in E. coli cells expressing either the AroQMt or the *AroQMt protein was determined to be, respectively, 143- and 106-fold higher than the enzyme activity obtained for E. coli cells carrying the pET-23a(+) expression vector (Table 2). Analysis of CM specific activity in E. coli cells producing the recombinant protein AroQMs showed a 155-fold increase compared to the E. coli control cell extract, whereas a 131-fold increase was detected in E. coli cells producing *AroQMs. Moreover, CM enzyme activity was linearly dependent on the volume of crude extract added to the reaction mixture (data not shown).
TABLE 2.
CM specific activity in crude extracts of E. coli cells producing the recombinant proteins AroQMt, *AroQMt, AroQMs, and *AroQMs
| Cell extracta | Sp act (U/mg)b | Cloned extract/control |
|---|---|---|
| Control (M. tuberculosis) | 0.889 | 1 |
| AroQMt (Rv0948c) | 127.38 | 143.28 |
| *AroQMt (Rv1885c) | 94.23 | 105.99 |
| Control (M. smegmatis) | 1.204 | 1 |
| AroQMs (MSMEG5513) | 186.31 | 154.74 |
| *AroQMs (MSMEG2114) | 158.29 | 131.47 |
Soluble fraction of crude extracts from E. coli BL21(DE3) cells transformed with the pET-23a(+) vector only (control) or each respective mycobacterial expression plasmid (cloned extract). Measurements of CM enzyme activity for the M. tuberculosis and M. smegmatis recombinant proteins were done separately.
Units are given in U ml−1/mg ml−1. CM enzyme activity was assayed in duplicate for each cell extract.
As demonstrated by SDS-PAGE analysis and CM enzyme activity measurement, products of each mycobacterial aroQ and *aroQ gene were expressed in soluble and active form in E. coli, indicating that protein folding was correctly achieved. Taken together, these data confirm that ORFs Rv0948c, Rv1885c, MSMEG5513, and MSMEG2114 code for functional CMs. In agreement with our results, the predicted PheA (Rv3838c) and TyrA (Rv3754) proteins from M. tuberculosis have been recently purified (42, 55), and no associated CM activity was detected. Similarly, the annotated PheA (MSMEG6381) and TyrA (MSMEG6289) proteins from M. smegmatis have no fused CM domain to their sequences (http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi/). We thus conclude that there are two monofunctional CMs of the AroQ structural class in M. tuberculosis and M. smegmatis and that they are probably directed to distinct cellular locations in mycobacteria.
Although Rv0948c and MSMEG5513 are cytoplasmic enzymes, the presence of a cleavable signal peptide in both Rv1885c and MSMEG2114 strongly suggests that they are exported (Fig. 1A). Kim et al. (27) detected CM activity due to the secretion of *AroQMt into the culture filtrate of bacilli in the early and late exponential phases of growth. However, the existence of a secreted CM in M. tuberculosis is an enigmatic question, since no genes coding for potentially exported cyclohexadienyl dehydratases and aromatic aminotransferases (enzymes that act downstream of prephenate and phenylpyruvate, respectively) have been identified in the genome (10, 13), despite extensive similarity searches in databases (data not shown). Thus, the biological role of *AroQMt is not obvious at the moment.
On the other hand, a different scenario may be present in M. smegmatis. Calhoun et al. (9) found a complete biosynthetic pathway for phenylalanine in the periplasm of Pseudomonas aeruginosa, where a cyclohexadienyl dehydratase (*PheC) and an aromatic aminotransferase occur, besides an exported CM. When we performed similarity searches using *PheC (PA3475) from P. aeruginosa as a query, an ORF (MSMEG3450) showing 49.1% identity (67.7% similarity) to that enzyme was found in the M. smegmatis genome. In addition, ORF MSMEG3450 clearly codes for an exported protein, as predicted by the program SignalP 3.0 (data not shown). Therefore, it is possible that an extracellular biosynthetic pathway from chorismate to phenylalanine exists in M. smegmatis, even if the function of such putative route is presently obscure.
Organization of Rv0948c and Rv1885c promoter regions.
As depicted in Fig. 2, in silico analyses (10, 13) indicate that Rv0948c (aroQ) forms a single transcriptional unit in M. tuberculosis, whereas Rv1885c (*aroQ) is probably the second gene in the fbpB operon (which in total contains six genes). The chromosomal regions comprising the aroQ gene and the fbpB operon are detailed in Fig. 5A and B, respectively, where predicted promoter regions are also shown.
The aroQMt gene is most probably monocistronic, since the intergenic region Rv0948c-uvrD is considerably large (almost 300 bp) and the two genes are transcribed in opposite directions (Fig. 5A). In addition, the nearby Rv0947c is a pseudogene. A potential RBS (GAGG) is predicted to occur close to the start codon, and putative −10 and −35 promoter consensus sequences were also identified.
The *aroQMt gene lies in the fbpB operon (Fig. 5B), which includes rpfC (resuscitation-promoting factor RpfC), Rv1883c (“conserved hypothetical protein”), Rv1882c (probable short-chain type dehydrogenase) and possibly lppE (conserved lipoprotein LppE). There are short intergenic regions between the genes, ranging from only 11 bp (*aroQ-rpfC) to 40 bp (Rv1882c-lppE). The intergenic region fbpB-*aroQ is 17 bp long and probably constitutes an active promoter with part of the 5′ upstream sequence, as we discuss next. A potential RBS (rbs1, GGCGC) for *aroQ was predicted in previous genome annotations (10, 13), but we suggest that there may be a second downstream element (rbs2, GAAG) that performs the same function. This requires further experimental investigation. The transcriptional start site of fbpB was mapped by Harth et al. (25), and the ribosomal binding site and the promoter region were assigned.
Analysis of promoter activity.
Considering the existence of two monofunctional CMs in M. tuberculosis, we wondered whether there might be a possible differential regulation of the aroQ and *aroQ genes at the transcriptional level. To evaluate this hypothesis, which could also give us clues about the biological role, we cloned the predicted promoter regions of the M. tuberculosis aroQ gene (C1) and the fbpB operon (P1) into the integrative promoter-probe vector pSM128 (see Fig. 5 for details). Since there is a short intergenic region between fbpB and *aroQ (and two potential ribosomal binding sites), we also tested the pertinent surrounding sequences (P2) for promoter activity (Fig. 5B). In this way, using the β-Gal reporter gene and M. smegmatis as a surrogate host, the promoter activity of the two M. tuberculosis CM loci and the fbpB operon was assessed under different growth conditions.
β-Gal assays from M. smegmatis cells grown in either enriched (Lemco medium) or defined (minimal medium) media detected the presence of low promoter activity levels in transformants harboring each construction (Table 3). In an arbitrary scale, promoter activity was classified as very weak (e.g., 5.76 ± 0.62 U of β-Gal for P1 in Lemco) to weak (e.g., 11.66 ± 0.19 U of β-Gal for C1 in Lemco) in the conditions tested. Although β-Gal activity values were lower in minimal medium (compared to Lemco), the general pattern among promoters from each group was maintained, and no significant difference was observed (in contrast, there were differences between P1 and P2 activities under the same condition, for instance). It should be noted that cell growth is restricted in minimal medium, a situation that could lead to great variations in the activity of each promoter. However, this does not appear to be the case, since the expression levels from all promoters were roughly similar to the observed for M. smegmatis cells grown in Lemco. An interesting result was that the intergenic region fbpB-*aroQ exhibited promoter activity levels higher than the fbp operon and comparable to the aroQ gene, showing that the P2 sequence probably contains regulatory signals (see Fig. 5B). Therefore, the M. tuberculosis *aroQ gene might be subjected to dual promoter control.
TABLE 3.
Promoter activity analysis of the M. tuberculosis C1, P1, and P2 sequences in M. smegmatis cells grown in different media
| Straina | Promoter activity (mean ± SD)b on:
|
|||
|---|---|---|---|---|
| Lemco | MM | MM + 1 mM Phe | MM + 1 mM Trp | |
| pSM128 (control) | 2.78 ± 0.12 | 1.52 ± 0.22 | 1.69 ± 0.23 | 1.80 ± 0.35 |
| pCAN1 (aroQ) | 11.66 ± 0.19 | 7.67 ± 0.51 | 8.58 ± 0.55 | 19.77 ± 0.97 |
| pPAN1 (fbpB) | 5.76 ± 0.62 | 3.69 ± 0.31 | 4.12 ± 0.32 | 6.32 ± 0.94 |
| pPAN2 (*aroQ) | 10.47 ± 0.44 | 5.82 ± 0.56 | 6.78 ± 1.13 | 20.96 ± 1.32 |
Each mycobacterial reporter plasmid was electroporated into M. smegmatis cells. Transformants were isolated, grown in Lemco or minimal medium, and assayed for β-Gal activity.
Results are the means for three independent transformants, each assayed in duplicate. Units are given in nmol of o-nitrophenyl galactoside produced per min per mg of total protein. MM, minimal medium.
To analyze the activity of these promoter regions in more detail, transformants carrying each mycobacterial reporter plasmid were grown in minimal medium supplemented with 0.2% glucose and 1 mM phenylalanine, 1 mM tyrosine, or 1 mM tryptophan. Our specific goal was to evaluate whether transcription of the aroQ and *aroQ genes (and the fbpB operon) is controlled by the presence of aromatic amino acids. The promoter activity levels of M. smegmatis cells grown in minimal medium supplemented with 1 mM phenylalanine were comparable to those observed when the same transformants were grown in minimal medium without amino acid supplementation (Table 3). No significant difference was found between the two conditions. Analogous results were obtained for M. smegmatis cells grown in minimal medium supplemented with 1 mM tyrosine (data not shown). However, when we changed to minimal medium supplemented with 1 mM tryptophan, there was a significant increase in promoter activity of the C1 and P2 sequences (Table 3), suggesting that the aroQ and *aroQ genes from M. tuberculosis may be subjected to a certain level of regulation under the conditions tested. The promoter activity of the P1 sequence was 6.32 ± 0.94 U of β-Gal in minimal medium supplemented with 1 mM tryptophan, indicating that expression of the fbpB gene (and indirectly the whole operon) is probably neither activated nor repressed in response to aromatic amino acids levels in M. tuberculosis in this condition.
In other experimental conditions tested, M. smegmatis cells harboring the pCAN1, pPAN1, or pPAN2 constructions were grown in minimal medium supplemented with 0.2% glucose and 5 mM phenylalanine, 5 mM tyrosine, or 5 mM tryptophan. Our specific goal was to evaluate whether a significant level of repression could be detected. Under the conditions tested, no repression of the promoter activity of the M. tuberculosis aroQ and *aroQ genes was observed when 5 mM phenylalanine or 5 mM tyrosine was added to the culture medium (data not shown). In fact, there was a very slight increase in promoter activity (in comparison to the nonsupplemented minimal medium condition), but this was not significant and can be explained as a result of increased cell growth upon amino acid addition, as previously reported (36). When 5 mM tryptophan was added to the culture medium, promoter activity values similar to those obtained for supplementation with 1 mM tryptophan were found: 21.71 ± 0.83, 7.29 ± 2.30, and 22.85 ± 0.44 U of β-Gal for the C1, P1, and P2 sequences, respectively. This demonstrates that the intergenic region fbpB-*aroQ definitely exhibits promoter activity and that this sequence (P2) and the M. tuberculosis aroQ promoter region (C1) show a certain level of regulation by tryptophan.
In conclusion, we established that the sequences immediately upstream of the aroQ gene, the fbpB operon and the *aroQ gene are functional promoter regions in M. tuberculosis using M. smegmatis as a model system. Although expression of these promoters seems to be constitutive in response to variable concentrations of phenylalanine and tyrosine in minimal medium, tryptophan is possibly a minor transcriptional inducer of the CM genes. As mentioned above, it could be argued that the apparent induction of aroQ and *aroQ simply results from a better amino acid supplementation to the culture medium. However, increased (and statistically significant) promoter activity of these two loci is not detected when M. smegmatis cells are grown in minimal medium containing either 1 or 5 mM phenylalanine or tyrosine, reinforcing a probable, though limited, regulation of aroQ and *aroQ by tryptophan. In addition, it should be observed that, in general, M. tuberculosis transcriptional signals are efficiently recognized by the transcription apparatus of the closely related M. smegmatis, which is extensively utilized as a surrogate host in mycobacterial genetics and biochemistry. We provided evidence that, although modest, the aroQ, *aroQ, and fbpB promoter regions were active in M. smegmatis under the conditions tested. However, it should be pointed out that the promoters in question may be differentially regulated in M. smegmatis and M. tuberculosis, and caution should thus be exercised when extrapolating the results to the pathogenic bacillus. Nevertheless, this is a first step toward understanding the promoter activity of CM genes in mycobacteria.
Involvement of *AroQMt (Rv1885c) in pathogenesis.
Although the presence of two monofunctional CMs in M. tuberculosis is still intriguing, it has been recently suggested that the *AroQMt protein, besides its CM activity, holds an additional unknown function that may be related to virulence (27, 45). This proposal is mainly based on three observations. First, *AroQ proteins are basically found in bacterial pathogens, such as E. herbicola, P. aeruginosa, Salmonella enterica serovar Typhimurium, Xylella fastidiosa, Yersinia pestis, and others (http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi/). Second, the *aroQMt gene is situated in an operon that contains fbpB and rpfC, whose products are considered to be important for the pathogenicity of the mycobacterial cell envelope (8) and reactivation of bacilli from a dormancy state (33), respectively. A third reason would be the long C-terminal portion of the exported CMs, which apparently shows no sequence similarity to known catalytic or regulatory motifs and could hence display an extra role yet to be unveiled (9).
This is further complicated because it is not yet known whether there is in vivo functional redundancy between the two CMs from M. tuberculosis. In addition, as previously discussed, no exported prephenate-utilizing enzymes were found in the M. tuberculosis genome, thus making unclear the biological role of *AroQMt. However, here we provided experimental evidence that M. smegmatis, a saprophytic species, possesses two monofunctional CMs of the AroQ class, one of which is probably exported (based on homology to the *AroQMt protein and the presence of a putative cleavable signal peptide) and may participate in an extracellular pathway for aromatic amino acids. To our knowledge, this is the first description of occurrence of an *AroQ protein in a nonpathogenic bacterium. On the other hand, *aroQ is a pseudogene in M. leprae, the etiological agent of leprosy (14). So, as proposed by Calhoun et al. (9), it is possible that the *AroQMt protein represents a remnant of an extinct periplasmic pathway for phenylalanine and tyrosine biosynthesis. Alternatively, it could have evolved to interact with a new pathway or even host proteins.
Although M. tuberculosis AroQ and *AroQ may exhibit in vivo overlapping functions, it is likely that these enzymes have differential spatial and/or temporal roles. TB is a complex, progressive infection, and M. tuberculosis uses many elaborated mechanisms and specific metabolic pathways in order to evade the host immune system (56). In our experiments, it is suggestive that tryptophan is a slight inducer of promoter activity of the M. tuberculosis aroQ and *aroQ genes, since availability of this amino acid is thought to be very limited within the intracellular compartment in which the bacilli reside. It is also remarkable that the *aroQ gene is probably subjected to dual promoter control in M. tuberculosis. Thus, it is tempting to speculate that the aroQ gene codes for the biologically relevant CM in this pathogen and that production of the *AroQMt protein is more “tightly” controlled or only necessary under defined conditions (for instance, nutrient restriction). Nevertheless, this hypothesis needs further examination, since expression of at least the fbpB gene is moderate to substantial in different in vitro and in vivo conditions (16, 43, 46, 53) and antigen 85B is one of the major secreted products in M. tuberculosis (3). These facts may have indirect implications on transcription of other genes from the fbpB operon, including *aroQ.
To conclude, the possible involvement of the *AroQMt protein in TB pathogenesis is not clear at the moment. Novel experimental approaches and a more comprehensive analysis of the aroQ and *aroQ promoter regions are necessary to both understand the exact contribution of each gene to biologically relevant CM activity and evaluate whether *AroQ actually plays a different functional role in M. tuberculosis. In addition, since CMs are known for a high degree of sequence divergence at the amino acid level without impairment of enzyme catalysis, these appear to be plausible avenues of research to be pursued to unveil the role, if any, of the exported CM in M. tuberculosis virulence mechanisms.
Acknowledgments
C.Z.S. was supported by postgraduate research studentships from the National Council for Scientific and Technological Development of Brazil (CNPq) and the CAPES Foundation. This study was funded by the Millennium Initiative Program MCT-CNPq, Ministry of Health-Department of Science and Technology-UNESCO (Brazil) to, D.S.S. and L.A.B. D.S.S. and L.A.B. also acknowledge grants awarded by PADCT and FINEP. D.S.S. (CNPq, 304051/1975-06) and L.A.B. (CNPq, 520182/99-5) are research career awardees from CNPq.
C.Z.S. thanks Giancarlo Pasquali for DNA sequencing; Michael S. Glickman for kindly providing pMV261; and Amanda C. Brown, Paul Carroll, and Jade N. James of the TB Group at CID, QMUL, London, United Kingdom, for help with M. smegmatis cultures.
Footnotes
Published ahead of print on 26 October 2007.
REFERENCES
- 1.Ahmad, S., and R. A. Jensen. 1988. Phylogenetic distribution of components of the overflow pathway to l-phenylalanine within the enteric lineage of bacteria. Curr. Microbiol. 16295-302. [Google Scholar]
- 2.Andrews, P. R., G. D. Smith, and I. G. Young. 1973. Transition-state stabilization and enzymic catalysis: kinetic and molecular orbital studies of the rearrangement of chorismate to prephenate. Biochemistry 123492-3498. [DOI] [PubMed] [Google Scholar]
- 3.Belisle, J. T., V. D. Vissa, T. Sievert, K. Takayama, P. J. Brennan, and G. S. Besra. 1997. Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science 2761420-1422. [DOI] [PubMed] [Google Scholar]
- 4.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340783-795. [DOI] [PubMed] [Google Scholar]
- 5.Bentley, R. 1990. The shikimate pathway: a metabolic tree with many branches. Crit. Rev. Biochem. Mol. Biol. 25307-384. [DOI] [PubMed] [Google Scholar]
- 6.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43717-731. [DOI] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 8.Brennan, P. J. 2003. Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis 8391-97. [DOI] [PubMed] [Google Scholar]
- 9.Calhoun, D. H., C. A. Bonner, W. Gu, G. Xie, and R. A. Jensen. 2001. The emerging periplasm-localized subclass of AroQ chorismate mutases, exemplified by those from Salmonella typhimurium and Pseudomonas aeruginosa. Genome Biol. 20030.1-0030.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camus, J.-C., M. J. Pryor, C. Médigue, and S. T. Cole. 2002. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology 1482967-2973. [DOI] [PubMed] [Google Scholar]
- 11.Casadaban, M. J., A. Martinez-Arias, S. K. Shapira, and J. Chou. 1983. Beta-galactosidase gene fusions for analyzing gene expression in Escherichia coli and yeast. Methods Enzymol. 100293-308. [DOI] [PubMed] [Google Scholar]
- 12.Chook, Y. M., H. Ke, and W. N. Lipscomb. 1993. Crystal structures of the monofunctional chorismate mutase from Bacillus subtilis and its complex with a transition state analog. Proc. Natl. Acad. Sci. USA 908600-8603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry, III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393537-544. [DOI] [PubMed] [Google Scholar]
- 14.Cole, S. T., K. Eiglmeier, J. Parkhill, K. D. James, N. R. Thomson, P. R. Wheeler, N. Honore, T. Garnier, C. Churcher, D. Harris, K. Mungall, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. M. Davies, K. Devlin, S. Duthoy, T. Feltwell, A. Fraser, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, C. Lacroix, J. Maclean, S. Moule, L. Murphy, K. Oliver, M. A. Quail, M. A. Rajandream, K. M. Rutherford, S. Rutter, K. Seeger, S. Simon, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, K. Taylor, S. Whitehead, J. R. Woodward, and B. G. Barrell. 2001. Massive gene decay in the leprosy bacillus. Nature 4091007-1011. [DOI] [PubMed] [Google Scholar]
- 15.Cotton, R. G. H., and F. Gibson. 1965. The biosynthesis of phenylalanine and tyrosine: enzymes converting chorismic acid into prephenic acid and their relationships to prephenate dehydratase and prephenate dehydrogenase. Biochim. Biophys. Acta 10076-88. [DOI] [PubMed] [Google Scholar]
- 16.DesJardin, L. E., L. G. Hayes, C. D. Sohaskey, L. G. Wayne, and K. D. Eisenach. 2001. Microaerophilic induction of the alpha-crystallin chaperone protein homologue (hspX) mRNA of Mycobacterium tuberculosis. J. Bacteriol. 1835311-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dosselaere, F., and J. Vanderleyden. 2001. A metabolic node in action: chorismate-utilizing enzymes in microorganisms. Crit. Rev. Microbiol. 2775-131. [DOI] [PubMed] [Google Scholar]
- 18.Dussurget, O., J. Timm, M. Gomez, B. Gold, S. Yu, S. Z. Sabol, R. K. Holmes, W. R. Jacobs, Jr., and I. Smith. 1999. Transcriptional control of the iron-responsive fxbA gene by the mycobaterial regulator IdeR. J. Bacteriol. 1813402-3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dye, C. 2006. Global epidemiology of tuberculosis. Lancet 367938-940. [DOI] [PubMed] [Google Scholar]
- 20.Fonseca, I. O., M. L. B. Magalhães, J. S. Oliveira, R. G. Silva, M. A. Mendes, M. S. Palma, D. S. Santos, and L. A. Basso. 2006. Functional shikimate dehydrogenase from Mycobacterium tuberculosis H37Rv: purification and characterization. Protein Exp. Purif. 46429-437. [DOI] [PubMed] [Google Scholar]
- 21.Gibson, F. 1964. Chorismic acid: purification and some chemical and physical studies. Biochem. J. 90256-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grossman, T. H., E. S. Kawasaki, S. R. Punreddy, and M. S. Osburne. 1998. Spontaneous cAMP-dependent derepression of gene expression in stationary phase plays a role in recombinant expression instability. Gene 20995-103. [DOI] [PubMed] [Google Scholar]
- 23.Guilford, W. J., S. D. Copley, and J. R. Knowles. 1987. On the mechanism of the chorismate mutase reaction. J. Am. Chem. Soc. 1095013-5019. [Google Scholar]
- 24.Harries, A. D., and C. Dye. 2006. Tuberculosis. Ann. Trop. Med. Parasitol. 100415-431. [DOI] [PubMed] [Google Scholar]
- 25.Harth, G., B.-Y. Lee, J. Wang, D. L. Clemens, and M. A. Horwitz. 1996. Novel insights into the genetics, biochemistry, and immunocytochemistry of the 30-kilodalton major extracellular protein of Mycobacterium tuberculosis. Infect. Immun. 643038-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain, V., S. Sujatha, A. K. Ojha, and D. Chatterji. 2005. Identification and characterization of rel promoter element of Mycobacterium tuberculosis. Gene 351149-157. [DOI] [PubMed] [Google Scholar]
- 27.Kim, S.-K., S. K. Reddy, B. C. Nelson, G. B. Vasquez, A. Davis, A. J. Howard, S. Patterson, G. L. Gilliland, J. E. Ladner, and P. T. Reddy. 2006. Biochemical and structural characterization of the secreted chorismate mutase (Rv1885c) from Mycobacterium tuberculosis H37Rv: an *AroQ enzyme not regulated by the aromatic amino acids. J. Bacteriol. 1888638-8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lassila, J. K., J. R. Keeffe, P. Oelschlaeger, and S. L. Mayo. 2005. Computationally designed variants of Escherichia coli chorismate mutase show altered catalytic activity. Protein Eng. Des. Sel. 18161-163. [DOI] [PubMed] [Google Scholar]
- 29.Lee, A. Y., P. A. Karplus, B. Ganem, and J. Clardy. 1995. Atomic structure of the buried catalytic pocket of Escherichia coli chorismate mutase. J. Am. Chem. Soc. 1173627-3628. [Google Scholar]
- 30.MacBeath, G., P. Kast, and D. Hilvert. 1998. A small, thermostable, and monofunctional chorismate mutase from the archeon Methanococcus jannaschii. Biochemistry 3710062-10073. [DOI] [PubMed] [Google Scholar]
- 31.Mahenthiralingam, E., P. Draper, E. O. Davis, and M. J. Colston. 1993. Cloning and sequencing of the gene which encodes the highly inducible acetamidase of Mycobacterium smegmatis. J. Gen. Microbiol. 139575-583. [DOI] [PubMed] [Google Scholar]
- 32.Miller, J. H. 1972. Assay of beta-galactosidase, p. 352-355. In J. H. Miller (ed.), Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Mukamolova, G. V., O. A. Turapov, D. I. Young, A. S. Kaprelyants, D. B. Kell, and M. Young. 2002. A family of autocrine growth factors in Mycobacterium tuberculosis. Mol. Microbiol. 46623-635. [DOI] [PubMed] [Google Scholar]
- 34.Ökvist, M., R. Dey, S. Sasso, E. Grahn, P. Kast, and U. Krengel. 2006. 1.6 Å crystal structure of the secreted chorismate mutase from Mycobacterium tuberculosis: novel fold topology revealed. J. Mol. Biol. 3571483-1499. [DOI] [PubMed] [Google Scholar]
- 35.Oliveira, J. S., C. A. Pinto, L. A. Basso, and D. S. Santos. 2001. Cloning and overexpression in soluble form of functional shikimate kinase and 5-enolpyruvylshikimate 3-phosphate synthase enzymes from Mycobacterium tuberculosis. Protein Expr. Purif. 22430-435. [DOI] [PubMed] [Google Scholar]
- 36.Parish, T. 2003. Starvation survival response of Mycobacterium tuberculosis. J. Bacteriol. 1856702-6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parish, T., B. G. Gordhan, R. A. McAdam, K. Duncan, V. Mizrahi, and N. G. Stoker. 1999. Production of mutants in amino acid biosynthesis genes of Mycobacterium tuberculosis by homologous recombination. Microbiology 1453497-3503. [DOI] [PubMed] [Google Scholar]
- 38.Parish, T., and N. G. Stoker. 1998. Electroporation of mycobacteria, p. 129-144. In T. Parish and N. G. Stoker (ed.), Methods in molecular biology, vol. 101. Mycobacteria protocols. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 39.Parish, T., and N. G. Stoker. 2002. The common aromatic amino acid biosynthesis pathway is essential in Mycobacterium tuberculosis. Microbiology 1483069-3077. [DOI] [PubMed] [Google Scholar]
- 40.Pittard, A. J. 1996. Biosynthesis of the aromatic amino acids, p. 458-484. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 41.Prakash, P., B. Aruna, A. A. Sardesai, and S. E. Hasnain. 2005. Purified recombinant hypothetical protein coded by open reading frame Rv1885c of Mycobacterium tuberculosis exhibits a monofunctional AroQ class of periplasmic chorismate mutase activity. J. Biol. Chem. 28019641-19648. [DOI] [PubMed] [Google Scholar]
- 42.Prakash, P., N. Pathak, and S. E. Hasnain. 2005. pheA (Rv3838c) of Mycobacterium tuberculosis encodes an allosterically regulated monofunctional prephenate dehydratase that requires both catalytic and regulatory domains for optimum activity. J. Biol. Chem. 28020666-20671. [DOI] [PubMed] [Google Scholar]
- 43.Rogerson, B. J., Y.-J. Jung, R. LaCourse, L. Ryan, N. Enright, and R. J. North. 2006. Expression levels of Mycobacterium tuberculosis antigen-encoding genes versus production levels of antigen-specific T cells during stationary level lung infection in mice. Immunology 118195-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Russell, D. G. 2001. Mycobacterium tuberculosis: here today, and here tomorrow. Nat. Rev. Mol. Cell. Biol. 2569-577. [DOI] [PubMed] [Google Scholar]
- 45.Sasso, S., C. Ramakrishnan, M. Gamper, D. Hilvert, and P. Kast. 2005. Characterization of the secreted chorismate mutase from Mycobacterium tuberculosis. FEBS J. 272375-389. [DOI] [PubMed] [Google Scholar]
- 46.Shi, L., Y.-J. Jung, S. Tyagi, M. L. Gennaro, and R. J. North. 2003. Expression of Th1-mediated immunity in mouse lungs induces a Mycobacterium tuberculosis transcription pattern characteristic of nonreplicating persistence. Proc. Natl. Acad. Sci. USA 100241-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 41911-1919. [DOI] [PubMed] [Google Scholar]
- 48.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, G. F. Hatfull, S. B. Snapper, R. G. Barletta, W. R. Jacobs, Jr., and B. R. Bloom. 1991. New use of BCG for recombinant vaccines. Nature 351456-460. [DOI] [PubMed] [Google Scholar]
- 49.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner, D., J. Maser, B. Lai, Z. Cai, C. E. Barry, III, K. Höner zu Bentrup, D. G. Russell, and L. E. Bermudez. 2005. Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell's endosomal system. J. Immunol. 1741491-1500. [DOI] [PubMed] [Google Scholar]
- 51.Warner, D. F., and V. Mizrahi. 2007. The survival kit of Mycobacterium tuberculosis. Nat. Med. 13282-284. [DOI] [PubMed] [Google Scholar]
- 52.Webby, C. J., H. M. Baker, J. S. Lott, E. N. Baker, and E. J. Parker. 2005. The structure of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase from Mycobacterium tuberculosis reveals a common catalytic scaffold and ancestry for type I and type II enzymes. J. Mol. Biol. 354927-939. [DOI] [PubMed] [Google Scholar]
- 53.Wilkinson, R. J., L. E. DesJardin, N. Islam, B. M. Gibson, R. A. Kanost, K. A. Wilkinson, D. Poelman, K. D. Eisenach, and Z. Toossi. 2001. An increase in expression of a Mycobacterium tuberculosis mycolyl transferase gene (fbpB) occurs early after infection of human monocytes. Mol. Microbiol. 39813-821. [DOI] [PubMed] [Google Scholar]
- 54.Xia, T., J. Song, G. Zhao, H. Aldrich, and R. A. Jensen. 1993. The aroQ-encoded monofunctional chorismate mutase (CM-F) protein is a periplasmic enzyme in Erwinia herbicola. J. Bacteriol. 1754729-4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu, S., Y. Yang, R. Jin, M. Zhang, and H. Wang. 2006. Purification and characterization of a functionally active Mycobacterium tuberculosis prephenate dehydrogenase. Protein Exp. Purif. 49151-158. [DOI] [PubMed] [Google Scholar]
- 56.Zahrt, T. C. 2003. Molecular mechanisms regulating persistent Mycobacterium tuberculosis infection. Microbes Infect. 5159-167. [DOI] [PubMed] [Google Scholar]