Abstract
Clostridium perfringens is an anaerobic, gram-positive, spore-forming bacterium responsible for the production of severe histotoxic and gastrointestinal diseases in humans and animals. In silico analysis of the three available genome-sequenced C. perfringens strains (13, SM101, and ATCC13124) revealed that genes that encode flagellar proteins and genes involved in chemotaxis are absent. However, those strains exhibit type IV pilus (TFP)-dependent gliding motility. Since carbon catabolite regulation has been implicated in the control of different bacterial behaviors, we investigated the effects of glucose and other readily metabolized carbohydrates on C. perfringens gliding motility. Our results demonstrate that carbon catabolite regulation constitutes an important physiological regulatory mechanism that reduces the proficiencies of the gliding motilities of a large number of unrelated human- and animal-derived pathogenic C. perfringens strains. Glucose produces a strong dose-dependent inhibition of gliding development without affecting vegetative growth. Maximum gliding inhibition was observed at a glucose concentration (1%) previously reported to also inhibit other important behaviors in C. perfringens, such as spore development. The inhibition of gliding development in the presence of glucose was due, at least in part, to the repression of the genes pilT and pilD, whose products are essential for TFP-dependent gliding proficiency. The inhibitory effects of glucose on pilT and pilD expression were under the control of the key regulatory protein CcpA (catabolite control protein A). The deficiency in CcpA activity of a ccpA knockout C. perfringens mutant strain restored the expressions of pilT and pilD and gliding proficiency in the presence of 1% glucose. The carbon catabolite repression of the gliding motility of the ccpA mutant strain was restored after the introduction of a complementing plasmid harboring a wild-type copy of ccpA. These results point to a central role for CcpA in orchestrating the negative effect of carbon catabolite regulation on C. perfringens gliding motility. Furthermore, we discovered a novel positive role for CcpA in pilT and pilD expression and gliding proficiency in the absence of catabolite regulation. Carbon catabolite repression of gliding motility and the dual role of CcpA, either as repressor or as activator of gliding, are analyzed in the context of the different social behaviors and diseases produced by C. perfringens.
Motility is an important attribute utilized by many pathogenic and nonpathogenic bacteria to colonize new environments, to search for nutrients, and to allow the formation of complex architectural structures (e.g., biofilms and fruiting bodies) (14, 15, 25). Translocation in liquid medium (swimming) is mediated by flagella, and the swimming model has been traditionally used to describe bacterial motility. However, it is also true that in nature, most bacteria are associated with surfaces and therefore have evolved diverse mechanisms of translocation on solid or semisolid biotic and abiotic surfaces which are important for rapid dissemination and colonization during the course of an infection (21). Among the surface-associated mechanisms of motility that play key roles in cell-cell and cell-surface interactions, it is worth mentioning (i) swarming, a flagellum-dependent social form of translocation where planktonic cells differentiate into giant and hyperflagellated cells that move in groups (“swarms”) to explore and colonize new habitats (10); (ii) twitching, a flagellum-independent but type IV pilus (TFP)-dependent form of intermittent and jerky surface motility (4, 14, 15), which has been demonstrated to occur in the opportunistic pathogen Pseudomonas aeruginosa (5), in the pathogen Neisseria gonorrhoeae (23), and recently in Haemophilus influenzae, which was previously considered a nonmotile bacterium (2); and (iii) gliding, which has been studied in detail in model organisms, such as Myxococcus xanthus and Synechocystis and Anabaena spp. (19). In M. xanthus, two types of mechanisms for gliding motility are utilized: adventurous gliding and social gliding (16). Adventurous gliding motility is observed when cells are isolated or in low numbers without contact with each other. In this sense, gliding motility is defined as a smooth movement of rod-shaped cells in the direction of their long axes on a surface. It has been proposed that the driving force for adventurous gliding motility in M. xanthus is generated by the action of a nozzle-like structure that produces a polysaccharide slime trail. Social gliding motility, similar to twitching motility, is dependent on the active extension and retraction of TFP (16). For other bacteria (e.g., Cytophaga spp., Flavobacterium spp., and Mycoplasma mobile), gliding takes place in the absence of TFP and ATP consumption. Instead, they generate motility with motility proteins anchored to the membrane that in turn use proton motive force and/or polar polysaccharide extrusion to achieve cellular motility over surfaces; all these gliding examples point out the extreme diversity of gliding mechanisms (4, 14, 15, 22, 24). Extracellular TFP appendages are polymers of the small protein PilA, or pilin. In P. aeruginosa, PilA is initially translated as a prepilin with a short leader peptide cleaved by PilD (48); PilB provides energy for assembly. PilT and PilU are nucleoside triphosphate binding proteins that have been implicated in pilus retraction during twitching motility (4). In P. aeruginosa, almost 40 genes are involved in the biosynthesis and function of TFP (23, 47). Besides the key role of TFP in twitching and gliding, these appendages also mediate other important phenomena, such as adherence, fruiting body formation, bacteriophage absorption, DNA uptake, cytotoxicity, activation of host cell responses, and biofilm development (1, 14, 23, 24, 31).
Clostridium perfringens is a gram-positive, anaerobic, spore-forming bacterium that causes severe gastrointestinal and histotoxic infections in humans and animals (28, 32, 33). This pathogen has been traditionally described as a nonmotile bacterium, as no genes that encode flagellar proteins or genes involved in chemotaxis were identified in the complete genomic sequences of the three human-pathogenic C. perfringens strains 13, SM101, and ATCC13124 (26, 36). However, sequence analysis (data not shown) suggests that the three strains carry genes which code for TFP components (such as pilA to pilD and pilT) (26, 36). Recently, Varga et al. demonstrated that C. perfringens strains 13, SM101, and ATCC13124 exhibited social gliding motility on brain heart infusion agar (BHIA) plates, and TFP were detected on the surfaces of the bacteria (41). This study also demonstrated that pilT and pilC mutants failed to produce detectable pili and were nonmotile on BHIA (41). Other putative TFP biosynthesis genes found in C. perfringens remain to be characterized, and the exact mechanism of TFP assembly and its physiological role, importance, and regulation are currently unknown.
In this respect, diverse bacterial behaviors, such as biofilm development, sporulation, fruiting body formation, and surface-associated motility, are regulated by environmental, metabolic, and quorum-sensing signals (9, 10, 16, 28, 46). In particular, carbon catabolite regulation is a widespread phenomenon in bacteria where the expressions of a number of genes are regulated by the presence of a preferred carbon source, such as glucose or fructose (27, 39, 43). In C. perfringens, and many low-G+C-content, gram-positive bacteria, carbon catabolite control is under the regulation of CcpA (catabolite control protein A), a pleiotropic transcriptional regulator belonging to the LacI/GalR family of transcription factors (27, 39, 43). CcpA functions as a DNA binding protein, either activating or repressing genes generally in the presence of a preferred carbon source (35, 43). More precisely, surface-associated motility is an interesting example of social bacterial behavior that could be regulated by nutrient (i.e., carbon) availability in nature. Interestingly, in P. aeruginosa, swarming (but not twitching) motility is carbon source regulated; poor swarming activity is observed in the presence of glucose (37). Therefore, the ability of the pathogen to carry out surface-associated social motility during the course of the infection is of crucial importance (2, 10, 14, 21, 24). In this work, we have investigated the effects of glucose and other readily metabolized carbohydrates on the TFP-dependent social gliding motilities of a collection of pathogenic C. perfringens strains isolated from human and animal sources. Our results clearly demonstrate that carbon catabolite repression is a general and common regulatory mechanism of gliding motility in C. perfringens, independent of the source of isolation (human or animal) or type of infection (diarrhea or myonecrosis). We demonstrate that the repressive effect of glucose on gliding motility is partially due to the CcpA-mediated down-regulation of TFP biosynthesis genes. In addition, we have discovered and analyzed a novel carbon catabolite-independent positive role for CcpA in C. perfringens gliding motility.
MATERIALS AND METHODS
Strains and culture conditions.
The C. perfringens strains used in this study are listed in Table 1. The growth media employed to propagate strains were fluid thioglycolate (FTG) and TGY (3% tryptic soy broth, 2% glucose, 1% yeast extract, and 0.1% cysteine) (8, 32). TY medium results from the omission of glucose from the TGY formula. All cultures were grown under anaerobic conditions in anaerobic jars containing GasPak (BD) at 37°C.
TABLE 1.
C. perfringens strains and plasmids used in this study
| Strain or plasmid | Isolation source or phenotype | Source or reference |
|---|---|---|
| C. perfringens strains | ||
| Human isolates | ||
| Strain 13 | Gas gangrene | 36 |
| KO13 | ccpA mutant derivative of strain 13 | This study |
| NCTC8239 | Food poisoning | 7 |
| NCTC10239 | Food poisoning | 7 |
| SM101 | Food poisoning | 49 |
| 191-10 | Food poisoning | 38 |
| F4406 | Sporadic diarrhea | 7 |
| F4969 | Sporadic diarrhea | 7 |
| F5603 | Sporadic diarrhea | 7 |
| B11 | Antibiotic-associated diarrhea | 7 |
| B41 | Antibiotic-associated diarrhea | 7 |
| NB16 | Antibiotic-associated diarrhea | 7 |
| Animal isolates | ||
| JGS1807 | Diarrheic pig | 45 |
| JGS1818 | Diarrheic pig | 45 |
| 294442 | Diarrheic dog | R. Carman |
| 317206 | Diarrheic dog | R. Carman |
| AHT327 | Diarrheic horse | 44 |
| AHT2911 | Diarrheic horse | 44 |
| Plasmids | ||
| pMRS127 | sigK-gusA in pJIR751 | 30 |
| pEJZ2 | pilT promoter fused with gusA in pMRS127 | This study |
| pPilDgus-127 | pilD promoter fused with gusA in pMRS127 | This study |
| KO2 | This study | |
| pJIR750 | C. perfringens-E. coli shuttle vector | 3 |
| pIH100 | This study |
Motility assays.
C. perfringens strains were grown overnight in FTG at 37°C, and 300 μl of this culture was inoculated and propagated in TGY for 5 h at the same temperature. Next, 1 ml of culture was centrifuged and concentrated twofold. Five microliters of the concentrated cell suspension was spotted onto a predried (1 h at 37°C) BHIA, TGY agar (TGYA), or TY agar (TYA) plate. All plates were supplemented with 0.7% agar. The inoculated plates were then incubated anaerobically for 48 to 96 h at 37°C. Photographs of the plates were taken with a Canon Power Shot SD550 digital camera.
Construction of gusA fusion plasmids and the β-glucuronidase assay.
The PCR-amplified product carrying the upstream region of pilD or pilT was first cloned into the pCR-XL-TOPO vector by using a TOPO-XL cloning kit (Invitrogen). Briefly, the DNA fragment carrying the promoter region of pilT or pilD from SM101 or strain 13 was amplified by PCR using primers CPP53/CPP55 or CPP230/CPP231, respectively (Table 2). The SalI site was incorporated into the forward primer and the PstI site into the reverse primer of each primer pair. These PCR products were then cloned into the pCR-XL-TOPO vector. The recombinant clones carrying the expected DNA fragment were confirmed by restriction enzyme digestion, PCR analysis, and then DNA sequencing. The SalI-PstI fragments carrying the promoter region of pilD or pilT from pCR-XL-TOPO clones were then recloned into the SalI/PstI sites of pMRS127 to create pilD-gusA or pilT-gusA fusion constructs derived from either strain SM101 or strain 13. These reporter plasmids were then introduced by electroporation and Cmr selection (8) into the corresponding wild-type C. perfringens strains 13 and SM101 and the isogenic derivates deficient in CcpA.
TABLE 2.
Primers used in this study
| Primer | Primer sequencea | Positionsb | Gene | Usec |
|---|---|---|---|---|
| CPP53 | 5′ GCGTCGACGAGATATGGTCTTTTAGATGG 3′ | −333 to −312 | pilT promoter | GUS |
| CPP55 | 5′ GCTGCAGCAGATGCTCCTTCTTTAACTG 3′ | +28 to + 48 | pilT promoter | GUS |
| CPP230 | 5′ GCGTCGACCTTGAAGATTTAGATAAGCCTC 3′ | +19 to + 41 | pilD promoter | GUS |
| CPP231 | 5′ GCTGCAGCCTTCCAATTATTAATCCAAATAA 3′ | −361 to −341 | pilD promoter | GUS |
| CCP-F | 5′ AACTAGGATATAGACCTAAT 3′ | +172 to +192 | ccpA | MP |
| CCP-R | 5′ TGATCCCATATCATACATTG 3′ | +876 to +896 | ccpA | MP |
| KO-F | 5′ CTGGAGTGTCAATAGCAAC 3′ | +34 to +53 | ccpA | PROBE |
| KO-R | 5′ TCTCCTAGTGATGAACTCAT 3′ | +366 to +386 | ccpA | PROBE |
| CPP265 | 5′ ATATCCATCAAGTTCATCTATAGAG 3′ | −505 to −481 | ccpA | COMP |
| CPP266 | 5′ TATGTTACCTAATGATTATGCATT 3′ | +1012 to +1036 | ccpA | COMP |
| P1 | 5′ ATGCTGATTACTCAGAAGCT 3′ | −774 to −754 | ccpA upstream | PCR |
| P2 | 5′ CTCATAAGCCCTTGATGAAC 3′ | +1461 to +1484 | ccpA downstream | PCR |
| M13-F | 5′ GTA AAA CGA CGG CCA GT 3′ | pUC18 vector | PCR | |
| M13-R | 5′ CAGGAAACAGCTATGAC 3′ | pUC18 vector | PCR |
Restriction sites that have been added are underlined.
The nucleotide position numbering begins from the first codon and refers to the relevant position within the respective gene sequence (36).
GUS, construction of plasmid for β-glucuronidase assay; MP, construction of mutator plasmid; PROBE, construction of DNA probe for southern blot analysis; COMP, construction of complementing plasmid.
The strains carrying the gusA fusions were grown overnight in FTG at 37°C, and 150 μl of each culture was inoculated into TY and TGY. At various time points, 1 ml of each culture was centrifuged for 2 min at 9,000 rpm, and the pellet was stored frozen at −20°C until used. The cell pellets were resuspended with 1 ml of buffer Z (8.54 g of Na2HPO4, 5.5 g of NaH2PO4·H2O, 0.75 g of KCl, 0.25 g of MgSO4, and 1.4 ml of β-mercaptoethanol·7H2O, pH 7, per liter) (13, 28). Adequate aliquots of the resuspended cells were brought to a final volume of 730 μl with buffer Z. Next, 10 μl of 10 mg/ml lysozyme was added and the mixture was incubated for 30 min at 37°C, followed by the addition of 10 μl 10% Triton X-100 (Sigma). The enzymatic reactions were initiated by adding 100 μl of 6 mM 4-nitrophenyl β-d-glucuronide (Sigma). After 15 min of incubation at 37°C, the reaction was stopped by adding 150 μl of 1 M Na2CO3. The absorbance was measured at 405 nm, and the β-glucuronidase activity was calculated using the following equation: (A405 × 1,000)/(optical density at 600 nm [OD600]pls cross chk inf or sup, edi file not provided × culture volume [in milliliters] × time [in minutes]) (19).
Insertional inactivation of ccpA gene in strain 13.
To disrupt ccpA in strain 13, an internal 760-bp fragment of the ccpA gene was amplified by PCR using primers CCP-F and CCP-R (Table 2) and cloned into the SmaI site of pUC18 to create pKO1. For selection, an erythromycin resistance cassette (ermBP) was ligated into the HincII site of pKO1, creating pKO2. The plasmid pKO2 (which has no origin of replication for C. perfringens) was transformed into strain 13 by electroporation and Emr selection. One Emr clone (KO13) was analyzed (see Fig. S1 in the supplemental material) for correct integration by a single crossover event involving homologous recombination of the suicidal mutator plasmid (pKO2) on ccpA and utilized in the present study.
The insertional disruption of wild-type ccpA in KO13 was first demonstrated by PCR analysis of DNA isolated from the mutant (see Fig. S1 in the supplemental material). As expected, a 1.6-kb product was amplified from KO13 DNA by using primers P1 and M13F and a 2.5-kb product by using primers P2 and M13R. However, no PCR product was obtained with either P1 and M13R or P2 and M13F. These PCR results are consistent with the suicidal mutator plasmid pKO2 having been inserted into the wild-type ccpA gene in KO13. Southern blot analyses showed that a 2.4-kb HindIII DNA fragment from wild-type strain 13 hybridized with our ccpA-specific probe. However, two hybridizing bands, of 2.9 and 4.3 kb, were observed with DNA from mutant strain MO13. This profile is consistent with the expected result since the inserted pKO2 plasmid has a HindIII site.
Southern blot analysis.
A 350-bp internal ccpA DNA fragment was amplified from strain 13 by PCR using primers KO-F and KO-R (Table 2) and labeled with alkaline phosphatase using the Gene Images AlkPhos direct labeling and detection system (Amarsham Bioscience) (28, 44). Isolated C. perfringens DNA samples, prepared as described previously (8, 38), were digested with HindIII, separated by electrophoresis on 1% agarose gels, and transferred by Southern blotting. The blot was hybridized with the AlkPhos-labeled ccpA probe, and the hybridized probe was then detected by CDPstar chemiluminescence (Amersham Bioscience).
Construction of the CcpA-complementing plasmid pIH100.
A 1,539-bp fragment containing the ccpA open reading frame and its 450-bp upstream region was amplified by PCR using primers CPP265 and CPP266 (Table 2) and then cloned into pCR-TOPO-XL (Invitrogen) to generate pCcpA-comp-XL. Next, the ∼1.5-kb KpnI/XbalI fragment was cloned into the KpnI/XbaI sites of shuttle vector pJIR750 to generate the ccpA-complementing plasmid pIH100.
RESULTS
Glucose represses the gliding motilities of wild C. perfringens strains isolated from human and animal infections.
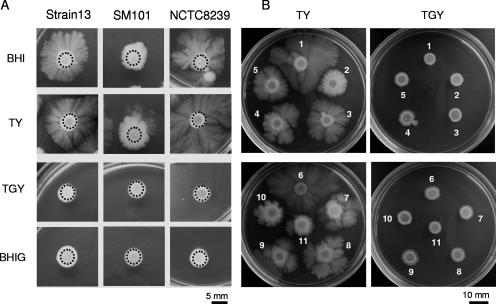
A recent study demonstrated TFP-dependent gliding motility in three human-derived C. perfringens strains for which genome sequences have been previously determined (41). TFP-dependent gliding motility constitutes an important bacterial behavior also known as twitching (14, 15, 24). Taking into account that the referred study (41) was limited to the analysis of only three strains, all of human origin, when there exists a great diversity of C. perfringens isolates causing diverse infections (i.e., gas gangrene, food poisoning, antibiotic-associated diarrhea, etc.) in human beings and animals, we considered it of interest to evaluate whether gliding motility was an intrinsic and general property of wild and undomesticated C. perfringens isolates. To this end, we performed a gliding motility analysis on a collection of 17 different pathogenic human- and animal-derived C. perfringens strains (including those strains whose gliding behavior was previously reported) (Table 1). In our study, motility is defined as the ability of cells to spread away from the edge of inoculation point by at least 0.4 cm in a curved flare pattern after 72 h at 37°C. When culture aliquots (see Materials and Methods for details) of strains 13, SM101, and NCTC8239 were spotted onto BHIA or TYA plates, the pattern of colony translocation was similar to that of the gliding motility previously observed (41), showing a distinctive curved flare pattern (Fig. 1A). Interestingly, both on TGYA and on BHIGA plates (which contained 2% glucose), the spotted cultures of strains 13, SM101, and NCTC8239 remained in the inoculation point, indicating that gliding motility was inhibited. In addition, we noted that the degree of motility was more evident on TYA than on the BHIA that was used in the previous study (41) (Fig. 1A and data not shown). Therefore, all subsequent motility experiments were conducted using TYA plates.
FIG. 1.
Glucose represses the gliding motilities of C. perfringens strains isolated from different mammalian hosts. (A) Gliding phenotypes of the three genome-sequenced human pathogen C. perfringens strains 13, SM101, and NCTC8239. Gliding was developed after inoculation of a 5-μl drop of a concentrated middle-log-phase culture of the corresponding strain on BHIA or TYA medium with or without 2% glucose supplementation. Top-bottom photographs were taken after 72 h of anaerobic incubation at 37°C. BHIA, TYA medium without glucose; BHIG and TGY, media supplemented with 2% glucose. Black dotted circles show the diameters of the initial inoculation spots (see Materials and Methods for details). (B) Gliding phenotypes of a collection of human and animal pathogenic C. perfringens strains: 1, NB16; 2, JGS1818; 3, 294442; 4, NCTC 10239; 5, 317206; 6, AHT327; 7, B11; 8, B41; 9, F5603; 10, F4969; and 11, AHT2911 (see Table 1 for the origin and type of infection produced for each pathogenic strain).
When the gliding motility assay was performed on the entire collection of C. perfringens isolates (Table 1), all the strains exhibited full proficiency in social gliding motility on TYA plates. Gliding motility was, in contrast, completely blocked in the presence of glucose (Fig. 1B and data not shown). Therefore, taking into consideration that glucose is a known inhibitor of other bacterial social behaviors, such as sporulation (28, 30) and biofilm formation (39), we concluded that the glucose added to the BHIA and TYA plates, yielding BHIG and TGYA, respectively, was responsible for the repressive effect on the gliding motilities of all the surveyed C. perfringens isolates.
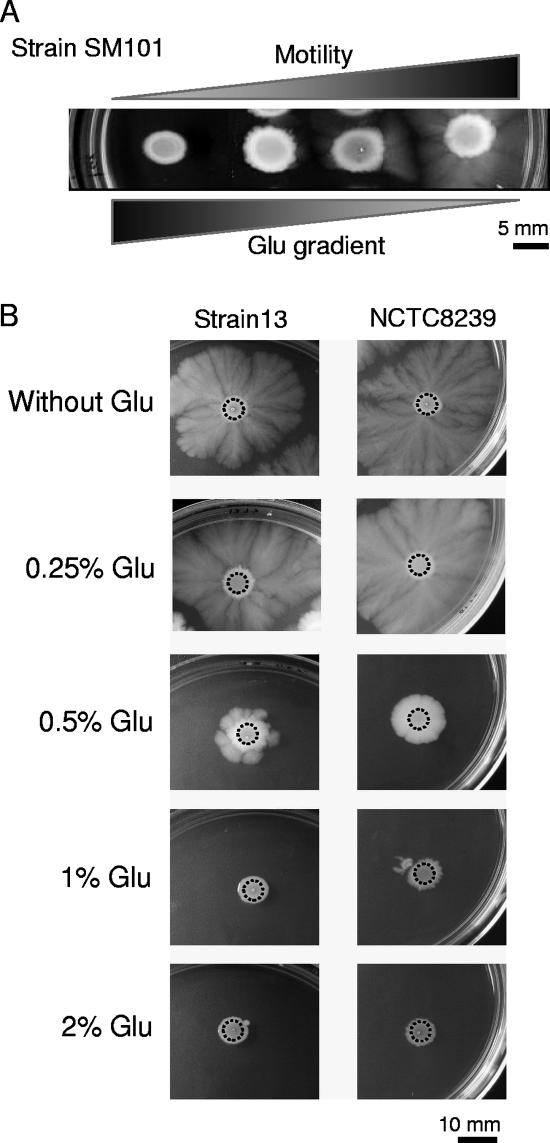
To determine how gliding motility was affected by different glucose levels, a glucose gradient was generated on a TYA plate (see Materials and Methods). Five microliters of a concentrated middle-log phase culture of the food poisoning strain SM101 was inoculated at various positions distributed along the TYA glucose concentration testing plate (see Materials and Methods). As observed in Fig. 2A, the extent of gliding motility exhibited by the pathogenic food poisoning SM101 strain was inversely proportional to the glucose concentration; as the concentration of glucose decreases, the extent of gliding motility increases.
FIG. 2.
Dose-dependent repressive effect of glucose (Glu) on C. perfringens gliding proficiency. (A) Glucose gradient effect on the gliding motility of the enterotoxigenic cpe+ food poisoning C. perfringens strain SM101. One-third of a TYA plate was cut out and replaced with 7 ml of melted TGYA medium, which contained 2% glucose (see Materials and Methods and elsewhere in text for details). After solidification of the added TGYA, a natural glucose gradient was generated (shown from left to right) due to the diffusion of glucose molecules from the TGYA section (which contained 2% added glucose) to the TYA portion (which contained 0% added glucose) of the plate. (B) Evaluation of the minimum glucose concentration required to inhibit gliding proficiency of the food poisoning and gas gangrene producer strains NCTC8239 and 13. For panels A and B, strains were grown on TYA plates as indicated in Fig. 1 and supplemented with glucose as shown in the figure. Top-bottom photographs were taken after 72 h of anaerobic incubation at 37°C.
To determine the minimum concentration of glucose required to inhibit gliding motility, five different concentrations of the sugar were independently assayed: 0.1%, 0.25%, 0.5%, 1%, and 2%. As shown in Fig. 2B, at a glucose concentration of 1%, no gliding motility was observed in the gas gangrene producer strain 13 or in the food poisoning isolate NCTC8239. In contrast, these two strains were able to glide slightly on TYA plates supplemented with 0.5% glucose. No inhibition of gliding motility was observed when these strains were spotted on TYA plates supplemented with 0.25% and 0.1% glucose (Fig. 2B and data not shown). Therefore, glucose repression of social gliding motility in C. perfringens was concentration dependent and apparently triggered at a glucose concentration of 0.5%.
Carbon catabolite repression of C. perfringens gliding motility.
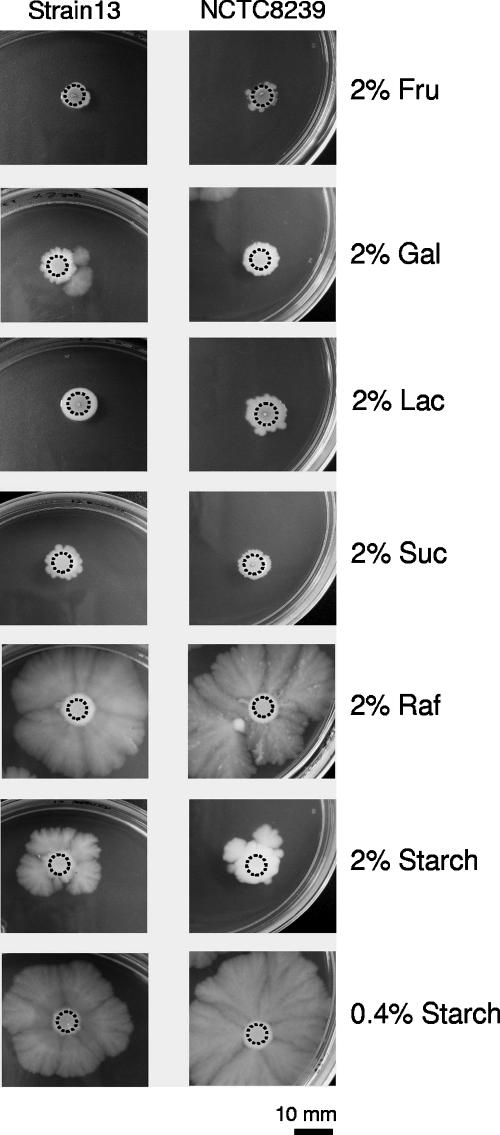
In order to determine if the observed inhibitory effect of glucose on gliding motility was a general phenomenon of carbon catabolite regulation (repression), other readily metabolized carbohydrates, such as galactose, fructose, lactose, and sucrose, were tested. The motilities of strains NCTC8239 (food poisoning) and 13 (gas gangrene) were inhibited when these strains were grown on plates containing 2% of either fructose, galactose, lactose, or sucrose (Fig. 3). Next, we analyzed the effects on gliding of complex carbohydrates (e.g., raffinose and starch), which are slowly metabolized but are required for efficient sporulation of C. perfringens and for C. perfringens enterotoxin (CPE) production (17, 18). When 2% raffinose was added to TYA plates, no inhibition of gliding motility was observed (Fig. 3). With 2% starch, the extents of motility in both strains (13 and NCTC8239) were suppressed to a minor degree, although gliding was not affected in the presence of 0.4% starch (Fig. 3). When similar experiments were performed on the rest of the C. perfringens isolates listed in this work (Table 1), no gliding motility was observed for all tested strains grown on TYA plates supplemented with 2% fructose, galactose, lactose, or sucrose, while gliding motility was observed with 2% raffinose supplementation. However, on TYA plates supplemented with 2% starch, all tested strains were nonmotile on the agar surface, with the exception of the strains NCTC10239 (food poisoning), which exhibited partial gliding motility, and JGS1807 (diarrheic pig), which was highly motile even in the presence of 2% starch (data not shown). The overall results indicated that the complex carbohydrates raffinose and starch did not affect gliding motility at the concentrations routinely used (2% and 0.4%, respectively) for C. perfringens spore formation and CPE production (17, 18, 28). More importantly, we demonstrate for the first time that C. perfringens gliding motility is subject to carbon catabolite repression.
FIG. 3.
Effects of simple and complex carbohydrates on C. perfringens gliding motility. Rapidly metabolized carbohydrates (Fru, fructose; Gal, galactose; Lac, lactose; Suc, sucrose) and complex carbohydrates (Raf, raffinose) commonly used to enhance sporulation and CPE production in Spo0A-proficient C. perfringens strains were assayed for their abilities to affect gliding motility. The gliding phenotypes of the gas gangrene producer and Spo0A-deficient strain 13 and the Spo0A-proficient and food poisoning strain NCTC8239 are shown. Top-bottom photographs were taken after 96 h of anaerobic incubation at 37°C on TYA plates supplemented with the corresponding concentrations of sugar as indicated.
Kinetics of TFP-dependent gliding motility in C. perfringens.
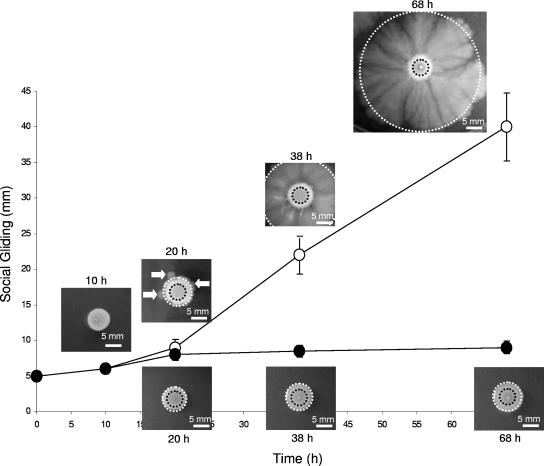
Proficiency in surface-associated motility is an important feature of many human and animal pathogens (2, 10, 14, 21, 24). In the cases of virulent and invasive C. perfringens isolates, an efficient mechanism of translocation on solid or semisolid surfaces could be favorable for escape from host defenses and for rapid dissemination of the infection (i.e., an aggressive gangrene can progress at a rate of 1 cm h−1). Therefore, we consider it of interest to characterize the kinetics and efficiency of the gliding motility of strain 13 (gas gangrene producer) on semisolid TYA plates in the absence and presence of added glucose. As observed in Fig. 4, it is possible to visualize two clear phases of colony growth and expansion. Initially, the growing colony expanded on the TYA surface from the inoculation point at a rate comparable to the speed of colony expansion expected from nonswimming bacteria. During the first 20 h of incubation, the size of the growing colony was evenly increased by a factor of 1.75 as a consequence of cell division, reaching a speed of colony spreading of approximately 125 μm h−1 (Fig. 4 and data not shown). After this initial phase of even colony spreading, a striking change occurred: social gliding was initiated by groups of cells located at the edge of the colony (see marked burgeoning cells in Fig. 4 at a developmental time of 20 h). After the appearance of these first signs of active surface gliding motility (the emergence of up-and-coming cells from the budding sites), a robust and uniform gliding was strongly induced, reaching a maximal speed of 600 to 700 μm h−1 (Fig. 4), a value for gliding speed comparable to the average values (200 μm h−1 to 1,000 μm h−1) for social gliding and twitching speed previously reported for other microorganisms (10, 14, 24).
FIG. 4.
Kinetics of gliding development of the gas gangrene producer C. perfringens strain 13 in the absence and presence of added glucose. Five microliters of a concentrated middle-log-phase culture of strain 13 grown in TY broth was inoculated in the center of each 100-mm petri dish, containing TYA medium with or without supplementation with 2% glucose. Inoculated petri dishes were anaerobically incubated at 37°C, and gliding proficiency was recorded at different times, measuring the distance traveled (in mm) from the initial inoculation point (black dotted circles) to the edge of the expanding colony (white dotted circles). Open and closed symbols represent the experiments performed in the absence and the presence of 2% added glucose, respectively. White arrows indicate the onset (under the influx of unknown signals; see text for details) of gliding development. The onset of gliding was observed only in TYA plates without glucose supplementation, while in the presence of glucose, gliding was never initiated. Photographs are representative of six independent experiments, and plotted values are the averages for those repetitions.
Interestingly, in the presence of 2% glucose, gliding motility was not generated even after long incubation times (Fig. 4 and data not shown). The morphology of the C. perfringens colony developed on TYA plates supplemented with glucose (gliding-deficient phenotype) was quite similar to the morphology of the colony developed on TYA plates without glucose supplementation just before the onset of active gliding; it was also very similar to that of the central part of a mature colony that has glided for more than 40 h on TYA plates (Fig. 4 and data not shown). These observations indicate that glucose and other sugars (data not shown) did not affect the initial phase of vegetative colony spreading. However, they do show that the sugar completely blocked (with carbon catabolite repression) the onset of the second phase of surface translocation, which relied on the development of active gliding.
Effects of glucose on pilT and pilD expression of gas gangrene and food poisoning-producing C. perfringens strains.
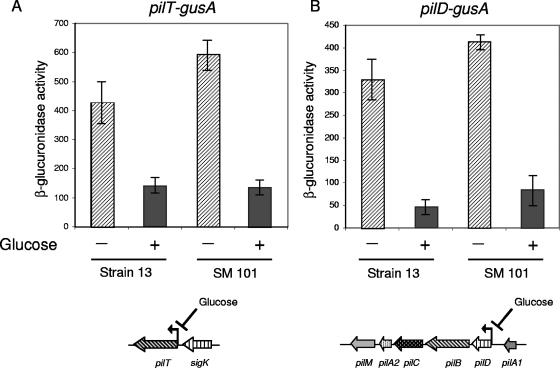
The recent study of Varga et al. demonstrated that the gliding of C. perfringens strain 13 depended on the products of pilT and pilC, which are required for TFP assembly (41). The pilT mutant of strain 13 does not spread out from the inoculation spot as the wild type does, and no pili were detected using field emission-scanning electron microscopy (41). Since glucose and other readily metabolized carbohydrates completely suppressed gliding motility in all the surveyed C. perfringens strains (Table 1) that we analyzed (Fig. 1 to 4 and data not shown), we considered the scenario where the transcription of pil genes might be affected by carbon catabolite repression. Therefore, to test this hypothesis, we measured the expression levels of two other genes required for TFP-dependent motility, pilD and pilT, in TY medium with and without glucose supplementation (Fig. 5). The β-glucuronidase pilD-gusA and pilT-gusA reporter fusions were introduced separately into strains 13 and SM101 by DNA electroporation, and pil-driven β-glucuronidase activity was assayed (see Materials and Methods for details). There were no significant differences in the growth of strains carrying the pil-gusA fusions in medium with or without added glucose (data not shown), although the final cellular yield was slightly higher in TY medium supplemented with glucose than in nonsupplemented TY medium (data not shown). Interestingly, in the presence of 1% glucose, there were dramatic down-regulations of pilD and pilT promoter activities in both C. perfringens strains (Fig. 5). The expression levels of pilT were reduced by 60% in the gas gangrene producer strain 13 and by 75% in the food poisoning isolate SM101 when these strains were grown in the presence of 1% glucose (Fig. 5A). Similarly, significant reductions in pilD-gusA expression were observed in cultures of strains 13 (∼80% reduction) and SM101 (∼75% reduction) when these strains were grown in the presence of 1% glucose in comparison with their expression levels in the absence of glucose (Fig. 5B). These results demonstrated that glucose strongly down-regulates the expression levels of the pilT and pilD genes. They also suggest that the inhibitory effect of carbon catabolite regulation on gliding motility took place, at least partially, at the level of TFP expression.
FIG. 5.
Glucose represses pilT and pilD transcription in gas gangrene and food poisoning C. perfringens strains. Transcription of pilT (A) and pilD (B) promoters measured by a β-glucuronidase assay of C. perfringens strains 13 and SM101, harboring reporter pilT-gusA (A) and pilD-gusA (B) transcriptional fusions. Strains were grown on TY broth with or without the addition of 1% glucose as indicated in the figure (+ or −, respectively). Accumulated β-glucuronidase activity was measured after 30 h of growth. A representative result from three independent assays is shown. Gene arrangements of pilT (A) and pilD (B) chromosomal regions in C. perfringens strains 13 and SM101 are shown at the bottom of the figure, with the repressive effects of glucose on gene transcription indicated (see text for details).
Dual role of the carbon catabolite protein CcpA in C. perfringens gliding motility.
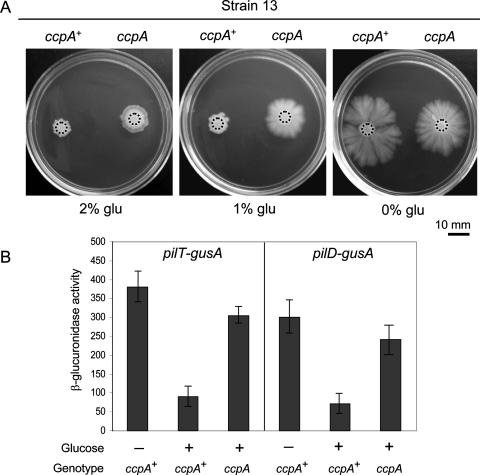
It is known that catabolite control protein A (CcpA) plays a key role in low-G+C-content, gram-positive bacteria, interconnecting carbon metabolism with several cellular responses, such as virulence, spore formation, and biofilm development (39, 43). In C. perfringens, CcpA is required for efficient sporulation and expression of the enterotoxin (CPE) that is responsible for the symptoms of food poisoning and diarrhea in humans and animals (42). Due to the observed repressive effect of glucose on the expression levels of pil genes (Fig. 5) that are essential for the ability of C. perfringens to glide on a solid substrate (41), we were tempted to analyze whether CcpA has a role in the carbon catabolite repression of C. perfringens gliding motility. To explore this possibility, we constructed a C. perfringens ccpA mutant strain (see Materials and Methods and Fig. S1 in the supplemental material) to assay the gliding phenotypes and the activities of TFP genes (pilT and pilD) of isogenic ccpA+ and ccpA C. perfringens strains grown in the presence or absence of added glucose. When gliding proficiency was evaluated in the presence of 1% and 2% glucose, the CcpA-proficient strain 13 was, as expected, unable to move at both sugar concentrations, remaining situated at the point of inoculation (round and smooth colony morphology). In contrast, the isogenic ccpA mutant derivate KO13, deficient in CcpA production and inoculated on the same plates, was able to retain more than 50% of its gliding ability in the presence of 1% glucose and also showed a low but perceptible and reproducible level of gliding proficiency on TYA plates supplemented with 2% glucose (Fig. 6A). Under similar conditions, no gliding motility was observed in the presence of 1% glucose with the ccpA mutant strain harboring the CcpA-complementing plasmid pIH100 (Table 1 and data not shown), suggesting that functional CcpA was required for carbon catabolite repression of C. perfringens social gliding motility.
FIG. 6.
CcpA mediates carbon catabolite repression of C. perfringens gliding motility. (A) Gliding motility phenotypes of the CcpA-proficient (ccpA+) C. perfringens strain 13 and its isogenic CcpA-deficient derivate KO13 (see Materials and Methods for details). A motility assay was performed according to the protocol described in the above figure legends. Top-bottom photographs were taken after 40 h of anaerobic incubation on TYA plates supplemented or not supplemented with glucose (glu) as indicated in the figure. Black dotted circles indicate the initial sizes of the colonies immediately after drop inoculation. (B) Expression of pilT-gusA and pilD-gusA reporter fusions in ccpA+ and ccpA cultures of isogenic C. perfringens strains 13 and KO13 grown for 30 h on TY broth in the absence (−) or presence (+) of 1% glucose. β-Glucuronidase activity was calculated as indicated in Fig. 5. For panels A and B, a representative set of results obtained from five independent experiments is shown.
In order to further confirm the role of CcpA in carbon catabolite repression of C. perfringens gliding motility, we compared the expression levels of pilT and pilD in isogenic CcpA-proficient and CcpA-deficient C. perfringens cultures grown in TY broth plus/minus 1% glucose. As shown in Fig. 6B, the CcpA-deficient cultures, but not the cultures that express CcpA, showed active expression of pilT and pilD in the presence of glucose at almost the same level observed for wild-type cultures grown in the absence of glucose. Collectively, these results demonstrate that CcpA was actively involved in the carbon catabolite repression of C. perfringens gliding motility and the glucose-induced down-regulation of TFP biosynthetic genes.
An intriguing observation was that the gliding ability of the CcpA-deficient C. perfringens strain KO13 grown in the absence of sugar supplementation was delayed in comparison with the gliding proficiency of the wild-type (ccpA+) strain 13 (see right panel, no added glucose, in Fig. 6A). This observation could uncover an additional, and unexpected, carbon-independent positive requirement of CcpA for full proficiency of C. perfringens in gliding motility. It is worth indicating that the slight but reproducible catabolite-independent positive effect of CcpA on social behaviors has been previously documented during biofilm formation in Bacillus subtilis (39) and spore development in C. perfringens (42). In these bacteria, the absence of CcpA activity was reflected in a low level of proficiency in biofilm development and spore formation in comparison with the abilities of the wild-type (CcpA-proficient) strains (39, 42).
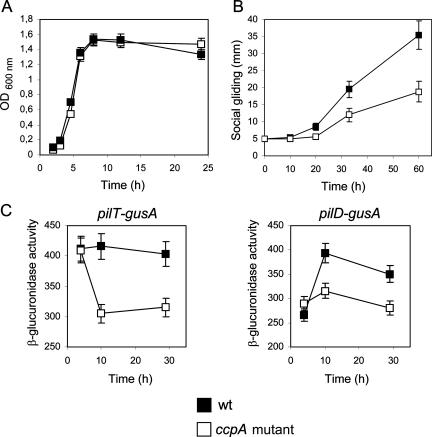
We confirmed our hypothesis by quantifying the gliding developed by the ccpA+ (strain 13) and its isogenic ccpA mutant derivate on TYA plates in the absence of glucose supplementation. Previously, we determined that introduction of the ccpA mutation in strain 13 did not affect its growing ability and the final cellular yield after growth in liquid TY medium (Fig. 7A). In contrast to what occurred in broth, where the growth phenotypes of the CcpA-proficient and CcpA-deficient cultures were the same, we observed clear differences in the speed and kinetics of gliding between the ccpA+ and ccpA strains. The gliding proficiency of the ccpA mutant strain showed a reduced speed compared with the velocity of gliding of the wild-type strain: 250 μm h−1 and 670 μm h−1 for the CcpA-deficient and CcpA-proficient strains, respectively (Fig. 7B). Also, the gliding of the ccpA mutant strain stopped before the gliding of the wild-type strain did (Fig. 6A, right, and 7B). These observations strongly suggest a novel and overlooked positive role for CcpA in the proficiency of gliding motility of C. perfringens in the absence of sugar supplementation. To confirm this conclusion, we measured the expression levels of pilD and pilT (whose products are essential for TFP-dependent gliding motility) in wild-type and CcpA-deficient C. perfringens cultures grown in TY broth without sugar supplementation. As observed in Fig. 7C, there are unambiguous down-regulations of pilT and pilD expression in the cultures deficient in CcpA production. This positive role of CcpA in TFP expression (Fig. 7C) and social gliding proficiency (Fig. 6A and 7B) in the absence of sugar supplementation and its opposite (negative) effect on the same social behavior (gliding motility) under conditions of carbon catabolite regulation (Fig. 6, presence of sugar) suggest a novel, dual (activating and repressing) role for CcpA in regulating C. perfringens gliding motility (Fig. 8).
FIG. 7.
CcpA has a novel catabolite-independent positive role in C. perfringens gliding motility. (A) Growth curves of the ccpA+ C. perfringens strain 13 and its isogenic ccpA derivate KO13. Growth was monitored over time, measuring the OD600s of both cultures developed on TY broth at 37°C. A representative set of results obtained from five independent experiments is shown. Closed symbols, wild type (wt); open symbols, ccpA mutant. (B) Kinetics of gliding motility of strain 13 (wt) and its isogenic CcpA-deficient derivate KO13 (ccpA mutant) developed on TYA plates without sugar supplementation. Gliding was recorded as indicated in Fig. 4. Average values of gliding obtained from five independent experiments are plotted. (C) Requirement of CcpA activity for full expression of pilT and pilD genes of C. perfringens strain 13 grown on TY broth without glucose supplementation. β-Glucuronidase activities driven from CcpA-proficient and CcpA-deficient C. perfringens cultures (strains 13 and KO13, respectively) harboring reporter pilT-gusA (left) and pilD-gusA (right) transcriptional fusions are shown. The four cultures were grown on TY broth without addition of glucose; accumulated β-glucuronidase activity was measured at the times indicated in the figure. Closed and open symbols represent CcpA-proficient and CcpA-deficient isogenic cultures, respectively. A representative set of results obtained from five independent experiments is shown.
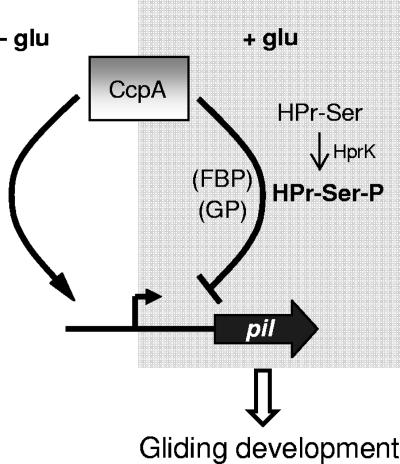
FIG. 8.
A dual (positive and negative) role for CcpA in C. perfringens gliding development. The present cartoon depicts one hypothetical model that might explain our actual knowledge of the gliding phenotypes of CcpA-proficient and CcpA-deficient C. perfringens strains grown in the presence and absence of sugar supplementation. In the absence of glucose (−glu; left) or other catabolite-repressing sugars, CcpA by itself might be able to bind to the positive regulatory regions of genes (pil) involved in TFP expression (i.e., pilT and pilD), producing a positive effect on transcription and hence stimulating gliding proficiency. In support of this view, it has also been reported that in vitro and in vivo, CcpA-DNA mediated interactions do occur in the absence of added sugars (20, 29, 35). In the presence of catabolite-repressing amounts of glucose (right), the phosphotransferase enzyme of the sugar-specific phosphotransferase system Hpr-Ser would be phosphorylated by HprK (35). Hpr-Ser-Pi would bind to CcpA, and the newly formed Hpr-Ser-Pi::CcpA complex would interact with repressor sites located on the regulatory regions of pil (pilT and pilD) and therefore interfere with gliding proficiency. Also shown in the picture is the possibility that the coeffectors fructose 1,6-bisphosphate (FBP) and glucose 6-phosphate (GP) would function as adjunct corepressors to enhance and to fine-tune the response of CcpA to the metabolic needs of the cell (35, 43). Another possibility (an indirect effect of CcpA) that is not illustrated in this model is that CcpA would dually regulate an unidentified factor responsible for switching on and off the expressions of pil genes.
DISCUSSION
Our current study shows several significant contributions toward the understanding of the physiology and regulation of TFP-dependent gliding motility in C. perfringens. First, we extended the analysis to a total of 17 different C. perfringens strains isolated from diverse infections (diarrhea, food poisoning, myonecrosis) produced not only in human beings but also from animal origins (Table 1). Interestingly, all the analyzed strains exhibited active proficiencies in social gliding motility on agar surfaces (Fig. 1 and data not shown). These results significantly consolidate and strengthen the idea that gliding motility is an intrinsic property of pathogenic C. perfringens, regardless of its origin of isolation (41).
Our understanding of the environmental and metabolic factors that control surface-associated translocation in pathogenic bacteria is very limited. Precisely, the main contribution of our work is the demonstration that carbon catabolite repression (20, 39, 43, 50) regulates social gliding motility in C. perfringens. In fact, all the surveyed isolates exhibited social gliding motility on BHIA plates (with no glucose supplementation) but not on TGYA medium which contained 2% glucose, suggesting that glucose is capable of inhibiting social gliding motility. The removal of glucose from TGYA allowed the cells to exhibit social motility, while the addition of glucose in BHIA resulted in the inhibition of gliding motility, confirming that glucose plays a crucial role in inhibiting gliding motility (Fig. 1 and data not shown).
In addition to glucose, gliding motility was also inhibited by other rapidly metabolized sugars, such as fructose, lactose, sucrose, and galactose (Fig. 3). This finding confirmed that the repression of gliding in C. perfringens was due to a general process of carbon catabolite repression (43). Interestingly, two complex carbohydrates, raffinose and starch, behaved differently from the single sugars: raffinose did not inhibit motility at any of the assayed concentrations, and starch inhibited gliding only at concentrations higher than 2% (Fig. 3 and data not shown). These results are consistent with previously reported findings that other social behaviors present in C. perfringens, such as sporulation and enterotoxin (CPE) production, were also repressed by rapidly metabolized single sugars, such as glucose and lactose (28, 42), while the complex carbohydrates raffinose and starch were found to induce both events (17, 18). The correlation between carbon catabolite repression of sporulation and surface-associated motility suggests that the two social processes might share, at least in part, a common regulatory network (Fig. 9).
FIG. 9.
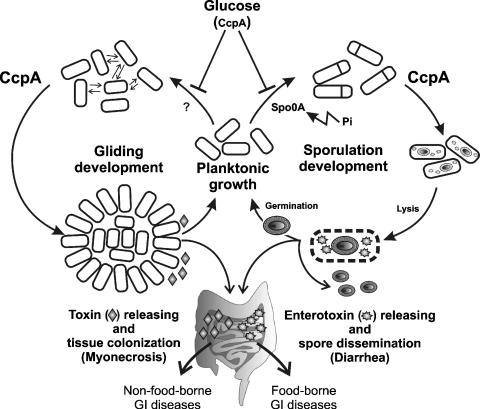
A workable model linking carbon catabolite regulation of social behaviors (gliding, sporulation, and toxin production) with disease progression in C. perfringens. In this hypothetical but realistic scenario, toxigenic, vegetative C. perfringens cells that reach the lumen of a human or animal gastrointestinal (GI) tract, where the basal luminal concentrations of glucose are normally lower than 0.5% (12), have the possibility of undergoing at least two different differentiation pathways: sporulation and/or gliding development. In the first case, the activation of the key transcription factor Spo0A by inorganic phosphate (Pi) present in the intestinal lumen triggers spore morphogenesis and enterotoxin (CPE) production (28). In the case of gliding development (left), unknown signals that might be linked to cell-cell and cell-surface interactions (double arrows) orchestrate the spatial and temporal organization of the cells to the onset of gliding. The progression of either developmental program (sporulation or gliding) would not exclude the occurrence of the other alternative pathway: sporulation and CPE production would take place in the lumen of the GI tract while gliding motility and vegetative toxin synthesis (i.e., collagenase production) would take place in association with the intestinal mucosa. The key role of glucose (representing the occurrence of CcpA-mediated carbon catabolite regulation when the level of the sugar is at least 1%) as a repressor of sporulation (25, 28) and gliding (this study) development is indicated. This regulatory blockage derives from the inhibition of enterotoxin (25, 28) and vegetative-linked toxin (11, 42) production in Clostridium spp. The novel role of CcpA as an activator of sporulation (42) and gliding proficiency (this study) is also shown. The development of inhibitors (e.g., monosaccharide analogs) that block the onsets of gliding and/or sporulation or antagonists that interfere with the positive role of CcpA on toxin production would contribute to combating the outbreak and dissemination of clostridial diseases.
We demonstrate that carbon catabolite repression of gliding motility in C. perfringens occurs through the repression of at least two genes involved in TFP production and functionality, namely, pilD and pilT. As observed in Fig. 5, the addition of 1% glucose to growing cultures of the gliding-proficient reference strains 13 and SM101 resulted in dramatic decreases in pilD-gusA and pilT-gusA expression. The maximum reduction in transcription due to the added glucose occurred approximately after 24 h of growth, which is consistent with the observation that gliding motility on agar plates begins only after 18 to 20 h of growth (Fig. 4 and data not shown).
In low-G+C-content, gram-positive bacteria, carbon catabolite regulation is under the control of the key transcription factor CcpA (carbon catabolite protein A), a member of the LacI-GalR family of transcriptional regulators (43). In the better-known cases of CcpA-mediated carbon catabolite regulation (i.e., in Bacillus subtilis and Lactococcus lactis), a complex and sophisticated signaling network is present (20, 29, 40, 50). Basically, the CcpA-dependent regulatory network utilizes sugar transporters, glycolytic enzymes, and an ATP-dependent, metabolite-activated protein kinase (HprK) and two small HprK target proteins: the phosphotransfer protein Hpr of the phosphotransferase system and the Hpr homologue Crh (35, 43). Moreover, a central role has been reserved for CcpA, which binds to DNA sequences (cis-acting replication element sites) present on the regulatory regions of its target genes (20, 29, 35, 43). For the activation of CcpA binding to the cis-acting replication elements, it is necessary, although not essential (20, 29, 35), for CcpA to bind to the phosphorylated forms of Hpr and/or Crh produced by HprK (35, 43). In C. perfringens, orthologs of ccpA, hpr, and hprK (but not crh) are present on the chromosomes of all the sequenced strains, suggesting that the basic elements for CcpA-mediated carbon catabolite regulation are present in this pathogen (reference 43 and data not shown). In fact, we demonstrated that the repression of C. perfringens gliding motility by glucose was mediated, in large part, by the action of CcpA. As observed in Fig. 6, the inactivation of ccpA significantly restored gliding proficiency (Fig. 6A) and pil expression (Fig. 6B) in the presence of glucose. The reversion to the gliding-deficient phenotype of the ccpA mutant strain in the presence of glucose was obtained after the introduction (by DNA electroporation) of the plasmid pIH100, harboring a wild-type copy of ccpA, which provided direct genetic evidence supporting the strong linkage between CcpA expression and carbon catabolite repression of gliding motility in C. perfringens. These results suggest that CcpA could act as a transcriptional regulator of TFP biosynthesis genes. However, this effect might be indirect since no putative cre sites have been identified in any of the TFP biosynthesis genes analyzed so far (data not shown). It might be possible that other cre-like consensus sequences, different from the ones reported for Bacillus and other low-G+C-content, gram-positive bacteria, exist in clostridia (20, 42, 43). Another possibility is that, apart from CcpA, an unidentified intermediate factor might be involved in regulating TFP gene expression. This suggestion received support based on the observation that the ccpA mutant strain was not able to restore, in the presence of sugar supplementation, full gliding proficiency and pil expression as the levels reached that of the wild-type strain in the absence of added sugars (Fig. 6 and data not shown).
A final and unexpected finding of our study was the observation that, in the absence of added sugar, CcpA has a positive role in gliding motility. As observed in Fig. 6, in the absence of added glucose, the ccpA mutant strain glided on the agar plate to a lesser extent than the isogenic wild-type strain. As observed in Fig. 7B, the wild-type strain (CcpA proficient) reached a maximum speed of gliding of 630 to 670 μm h−1, while its isogenic ccpA derivate (CcpA deficient) reached a maximal speed of gliding of 220 to 250 μm h−1 only. Two observations argue strongly for a positive role for CcpA in gliding development: first, the ccpA mutant strain did not show any growth defect on liquid medium, reaching essentially the same final OD and viable-cell number as the wild-type strain (Fig. 7A); furthermore, the results for the initial phase of colony growth (before the onset of gliding) were very similar for both the ccpA+ and ccpA strains (Fig. 7B and data not shown). This hypothesis was reinforced by the demonstration that CcpA production was required for efficient expression of pilT and pilD in growth media without sugar supplementation (Fig. 7C). These findings indicate that CcpA has a dual role in controlling gliding motility in C. perfringens (Fig. 8). In the presence of rapidly metabolized sugars (e.g., glucose), CcpA has a negative role on the onset of gliding, an effect that is partly mediated through repression of pilT and pilD expression (Fig. 6). In the absence of added sugars, CcpA switches to a positive role on gliding, a novel property that is uncovered by the deficient gliding phenotypes and poor pilT and pilD expression levels of CcpA-deficient cells cultured in the absence of added glucose (Fig. 7). In agreement with our finding, a similar positive role for CcpA in spore formation and cpe expression under conditions without catabolite regulation (in the absence of added sugars) has been reported for C. perfringens (42).
Excess glucose in the environment of C. perfringens not only affects stationary phase phenomena, such as sporulation-linked CPE production (25, 28, 42) and gliding motility (this study), but can also act as a catabolic repressor of collagenase production during vegetative growth (42). Moreover, in the other intestinal pathogenic Clostridium bacterium C. difficile, glucose represses toxin production (11). Importantly, within the context of the development of a clostridial infection, it is plausible to envision that proficiency in gliding associated with toxin production and tissue damage would contribute to the progression of the infectious process (Fig. 9). Luminal glucose concentrations in the small intestines of mammals are in the range of 0.006% to 0.4% (12). Interestingly, in our study the catabolite repression of gliding motility by glucose was concentration dependent; surface motility was observed only when the glucose concentration was less than 0.5% (Fig. 2). This finding opens the possibility that gliding willingly would occur during the course of a clostridial infection (Fig. 9). We are just grasping the regulatory network of surface-associated motility in pathogenic clostridia, and the understanding of how carbon catabolite repression inhibits known and potential virulence processes (sporulation, toxin production, and gliding motility) in C. perfringens (6-8, 21, 26, 36) will contribute to preventing and combating clostridial diseases (Fig. 9).
Supplementary Material
Acknowledgments
This research was supported by grants to M.R.S. from the USDA Food Safety Program (grant 2002-35201-12643) and to R.G. from FONCyT and CONICET (grants PICT2002-0111651 and PIP-8239, respectively). M.M. was a recipient of an international fellowship from the ASM educational board.
We thank Nahid Mahfuz for technical assistance and Denny Weber for editorial comments. R.G. is a former Pew Latin-American Fellow and Fulbright International Scholar.
Footnotes
Published ahead of print on 2 November 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Averhoff, B., and A. Friedrich. 2003. Type IV pili-related natural transformation systes: DNA transport in mesophilic and thermophilic bacteria. Arch. Microbiol. 180385-393. [DOI] [PubMed] [Google Scholar]
- 2.Bakaletz, L. O., B. D. Baker, J. A. Jurcisek, A. Harrison, L. A. Novotny, J. E. Bookwalter, R. Mungur, and R. S. Munson, Jr. 2005. Demonstration of Type IV pilus expression and a twitching phenotype by Haemophilus influenzae. Infect. Immun. 731635-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannam, T. L., and J. I. Rood. 1993. Clostridium perfringens-Escherichia coli shuttle vectors that carry single antibiotic resistance determinants. Plasmid 29233-235. [DOI] [PubMed] [Google Scholar]
- 4.Bardy, S. L., S. Y. Ng, and K. F. Jarrell. 2003. Prokaryotic motility structures. Microbiology 149295-304. [DOI] [PubMed] [Google Scholar]
- 5.Baynham, P. J., D. M. Ramsey, B. V. Gvozdyev, E. M. Cordonnier, and D. J. Wozniak. 2006. The Pseudomonas aeruginosa ribbon-helix-helix DNA-binding protein AlgZ (AmrZ) controls twitching motility and biogenesis of type IV pili. J. Bacteriol. 188132-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergey, D. H., N. R. Krieg, and J. G. Holt. 1984. Bergey's manual of systematic bacteriology. Williams & Wilkins, Baltimore, MD.
- 7.Collie, R. E., and B. A. McClane. 1998. Evidence that the enterotoxin gene can be episomal in Clostridium perfringens isolates associated with non-food-borne human gastrointestinal diseases. J. Clin. Microbiol. 3630-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Czeczulin, J. R., R. E. Collie, and B. A. McClane. 1996. Regulated expression of Clostridium perfringens enterotoxin in naturally cpe-negative type A, B, and C isolates of C. perfringens. Infect. Immun. 643301-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniels, R., J. Vanderleyden, and J. Michiels. 2004. Quorm sensing and swarming migration in bacteria. FEMS Microbiol. Rev. 28261-289. [DOI] [PubMed] [Google Scholar]
- 10.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dupuy, B., and A. L. Sonenshein. 1998. Regulated transcription of Clostridium difficile toxin genes. Mol. Microbiol. 27107-120. [DOI] [PubMed] [Google Scholar]
- 12.Ferraris, R. P., S. Yasharpour, K. C. Lloyd, R. Mirzayan, and J. M. Diamond. 1990. Luminal glucose concentrations in the gut under normal conditions. Am. J. Physiol. 2596822-6837. [DOI] [PubMed] [Google Scholar]
- 13.Griffith, K. L., and R. E. Wolf, Jr. 2002. Measuring beta-galactosidase activity in bacteria: cell growth, permeabilization, and enzyme assays in 96-well arrays. Biochem. Biophys. Res. Commun. 290397-402. [DOI] [PubMed] [Google Scholar]
- 14.Harshey, R. 2003. Bacterial motility on a surface: many ways to a common goal. Annu. Rev. Microbiol. 57249-273. [DOI] [PubMed] [Google Scholar]
- 15.Henrichsen, J. 1972. Bacterial surface translocation: a survey and a classification. Bacteriol. Rev. 36478-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jelsbak, L., and L. Sogaard-Andersen. 2003. Cell behavior and cell-cell communication during fruiting body morphogenesis in Myxococcus xanthus. J. Microbiol. Methods 55829-839. [DOI] [PubMed] [Google Scholar]
- 17.Labbe, R., E. Somers, and C. Duncan. 1976. Influence of starch source on sporulation and enterotoxin production by Clostridium perfringens type A. Appl. Environ. Microbiol. 31455-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Labbe, R. G., and D. K. Rey. 1979. Raffinose increases sporulation and enterotoxin production by Clostridium perfringens type A. Appl. Environ. Microbiol. 371196-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lancero, H., N. B. Caberoy, S. Castaneda, Y. Li, A. Lu, D. Dutton, X. Y. Duan, H. B. Kaplan, W. Shi, and A. G. Garza. 2004. Characterization of a Myxococcus xanthus mutant that is defective for adventurous motility and social motility. Microbiology 1504085-4093. [DOI] [PubMed] [Google Scholar]
- 20.Lorca, G. L., Y. J. Chung, R. Barabote, W. Weyler, C. Schilling, and M. Saier, Jr. 2005. Catabolite repression and activation in Bacillus subtilis: dependency on CcpA, Hpr, and HprK. J. Bacteriol. 187:7826-7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macfarlane, S., M. Hopkins, and G. Macfarlane. 2001. Toxin synthesis and mucin breakdown are related to swarming phenomenon in Clostridium septicum. Infect Immun. 691120-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Margolin, W. 2006. Gliding motility: anticipating the next move with a molecular clock. Curr. Biol. 1685-87. [DOI] [PubMed] [Google Scholar]
- 23.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56289-314. [DOI] [PubMed] [Google Scholar]
- 24.McBride, M. 2001. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu. Rev. Microbiol. 5549-75. [DOI] [PubMed] [Google Scholar]
- 25.Melville, S. B., R. Labbe, and A. L. Sonenshein. 1994. Expression from the Clostridium perfringens cpe promoter in C. perfringens and Bacillus subtilis. Infect. Immun. 625550-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myers, G. S., D. A. Rasko, J. K. Cheung, J. Ravel, R. Seshadri, R. T. DeBoy, Q. Ren, J. Varga, M. M. Awad, L. M. Brinkac, S. C. Daugherty, D. H. Haft, R. J. Dodson, R. Madupu, W. C. Nelson, M. J. Rosovitz, S. A. Sullivan, H. Khouri, G. I. Dimitrov, K. L. Watkins, S. Mulligan, J. Benton, D. Radune, D. J. Fisher, H. S. Atkins, T. Hiscox, B. H. Jost, S. J. Billington, J. G. Songer, B. A. McClane, R. W. Titball, J. I. Rood, S. B. Melville, and I. T. Paulsen. 2006. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 161031-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Toole, G. A., K. A. Gibbs, P. W. Hager, P. V. Phibbs, Jr., and R. Kolter. 2000. The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 182:425-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Philippe, V. A., M. B. Mendez, I. H. Huang, L. M. Orsaria, M. R. Sarker, and R. R. Grau. 2006. Inorganic phosphate induces spore morphogenesis and enterotoxin production in the intestinal pathogen Clostridium perfringens. Infect. Immun. 743651-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puri-Taneja, A., S. Paul, Y. Chen, and M. Hulett. 2006. CcpA causes repression of the phoPR promoter through a novel transcription start site, PA6. J. Bacteriol. 1881266-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raju, D., M. Waters, P. Setlow, and M. R. Sarker. 2006. Investigating the role of small, acid-soluble spore proteins (SASPs) in the resistance of Clostridium perfringens spores to heat. BMC Microbiol. 650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rakotoarivonina, H., G. Jubelin, M. Hebraud, B. Gaillard-Martinie, E. Forano, and P. Mosoni. 2002. Adhesion to cellulose of the Gram-positive bacterium Ruminococcus albus involves type IV pili. Microbiology 1481871-1880. [DOI] [PubMed] [Google Scholar]
- 32.Sarker, M. R., R. P. Shivers, S. G. Sparks, V. K. Juneja, and B. A. McClane. 2000. Comparative experiments to examine the effects of heating on vegetative cells and spores of Clostridium perfringens isolates carrying plasmid genes versus chromosomal enterotoxin genes. Appl. Environ. Microbiol. 663234-3240. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Sauer, U., J. D. Santangelo, A. Treuner, M. Buchholz, and P. Durre. 1995. Sigma factor and sporulation genes in Clostridium. FEMS Microbiol. Rev. 17331-340. [DOI] [PubMed] [Google Scholar]
- 34.Schaeffer, P., J. Millet, and J. P. Aubert. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schumacher, M. A., G. Seidel, W. Hillen, and R. Brennan. 2007. Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors glucose 6-phosphate and fructose 1,6-bisphosphate. J. Mol. Biol. 3681042-1050. [DOI] [PubMed] [Google Scholar]
- 36.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 99996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shrout, J. D., D. L. Chopp, C. L. Just, M. Hentzer, M. Givskov, and M. R. Parsek. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 621264-1277. [DOI] [PubMed] [Google Scholar]
- 38.Sparks, S. G., R. J. Carman, M. R. Sarker, and B. A. McClane. 2001. Genotyping of enterotoxigenic Clostridium perfringens fecal isolates associated with antibiotic-associated diarrhea and food poisoning in North America. J. Clin. Microbiol. 39883-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stanley, N. R., R. A. Britton, A. D. Grossman, and B. A. Lazazzera. 2003. Identification of catabolite repression as a physiological regulator of biofilm formation by Bacillus subtilis by use of DNA microarrays. J. Bacteriol. 1851951-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takahashi, I. 1979. Catabolite repression-resistant mutants of Bacillus subtilis. Can. J. Microbiol. 251283-1287. [DOI] [PubMed] [Google Scholar]
- 41.Varga, J. J., V. Nguyen, D. K. O'Brien, K. Rodgers, R. A. Walker, and S. B. Melville. 2006. Type IV pili-dependent gliding motility in the Gram-positive pathogen Clostridium perfringens and other Clostridia. Mol. Microbiol. 62680-694. [DOI] [PubMed] [Google Scholar]
- 42.Varga, J. J., V. L. Stirewalt, and S. B. Melville. 2004. The CcpA protein is necessary for efficient sporulation and enterotoxin gene (cpe) regulation in Clostridium perfringens. J. Bacteriol. 1865221-5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warner, J. B., and J. Lolkema. 2003. CcpA-dependent carbon catabolite repression in bacteria. Microbiol. Mol. Biol. Rev. 67475-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waters, M., D. Raju, H. S. Garmory, M. R. Popoff, and M. R. Sarker. 2005. Regulated expression of the beta2-toxin gene (cpb2) in Clostridium perfringens type A isolates from horses with gastrointestinal diseases. J. Clin. Microbiol. 434002-4009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Waters, M., A. Savoie, H. S. Garmory, D. Bueschel, M. R. Popoff, J. G. Songer, R. W. Titball, B. A. McClane, and M. R. Sarker. 2003. Genotyping of beta2-toxigenic Clostridium perfringens fecal isolates associated with gastrointestinal diseases in piglets. J. Clin. Microbiol. 413584-3591. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Welsh, K. M., K. A. Trach, C. Folger, and J. A. Hoch. 1994. Biochemical characterization of the essential GTP-binding protein Obg of Bacillus subtilis. J. Bacteriol. 1767161-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitchurch, C. B., S. A. Beatson, J. C. Comolli, T. Jakobsen, J. L. Sargent, J. J. Bertrand, J. West, M. Klausen, L. L. Waite, P. J. Kang, T. Tolker-Nielsen, J. S. Mattick, and J. N. Engel. 2005. Pseudomonas aeruginosa fimL regulates multiple virulence functions by intersecting with Vfr-modulated pathways. Mol. Microbiol. 551357-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whitchurch, C. B., A. J. Leech, M. D. Young, D. Kennedy, J. L. Sargent, J. J. Bertrand, A. B. Semmler, A. S. Mellick, P. R. Martin, R. A. Alm, M. Hobbs, S. A. Beatson, B. Huang, L. Nguyen, J. C. Commolli, J. N. Engel, A. Darzins, and J. S. Mattick. 2004. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 52873-893. [DOI] [PubMed] [Google Scholar]
- 49.Zhao, Y., and S. B. Melville. 1998. Identification and characterization of sporulation-dependent promoters upstream of the enterotoxin gene (cpe) of Clostridium perfringens. J. Bacteriol. 180136-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zomer, A. L., G. Buist, R. Larsen, J. Kok, and O. Kuipers. 2007. Time-resolved determination of the CcpA regulon of Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 1891366-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.