Abstract
Lamins are nuclear intermediate filament proteins and the major building blocks of the nuclear lamina. Besides providing nuclear shape and mechanical stability, lamins are required for chromatin organization, transcription regulation, DNA replication, nuclear assembly, nuclear positioning, and apoptosis. Mutations in human lamins cause many different heritable diseases, affecting various tissues and causing early aging. Although many of these mutations result in nuclear deformation, their effects on lamin filament assembly are unknown. Caenorhabditis elegans has a single evolutionarily conserved lamin protein, which can form stable 10-nm-thick filaments in vitro. To gain insight into the molecular basis of lamin filament assembly and the effects of laminopathic mutations on this process, we investigated mutations in conserved residues of the rod and tail domains that are known to cause various laminopathies in human. We show that 8 of 14 mutant lamins present WT-like assembly into filaments or paracrystals, whereas 6 mutants show assembly defects. Correspondingly, expressing these mutants in transgenic animals shows abnormal distribution of Ce-lamin, abnormal nuclear shape or change in lamin mobility. These findings help in understanding the role of individual residues and domains in laminopathy pathology and, eventually, promote the development of therapeutic interventions.
Keywords: filament assembly, laminopathic diseases, nuclear envelope
The nuclear lamins are evolutionarily conserved nuclear-specific intermediate filaments (IFs) (1, 2). They are divided into A- and B-types. B-type lamins are encoded by two genes in mammals and by a single gene in Caenorhabditis elegans. A-type lamins are found only in more complex metazoans and are expressed mainly in differentiated cells (3). Lamins support many nuclear functions, including maintaining nuclear shape, DNA replication, regulation of gene expression, transcription elongation by Pol II, nuclear positioning, segregation of chromosomes, meiosis, and apoptosis (4–7).
Mutations in the human LMNA gene cause at least 11 different heritable diseases, collectively termed laminopathies (8–10). These include the muscle diseases Emery–Dreifuss muscular dystrophy (EDMD), limb–girdle muscular dystrophy (LGMD), and dilated cardiomyopathy (DCM), all of which include cardiac conduction defects, the metabolic diseases familial partial lipodystrophy, Dunnigan Type (FPLD), Seip syndrome, and diabetes; the axonal neuronal disease Charcot–Marie–Tooth disease type 2; the premature aging diseases Hutchinson–Gilford progeria syndrome (HGPS); atypical Werner's syndrome and Mandibuloacral dysplasia; and the prenatal disease restrictive dermopathy. The disease causing mutations are distributed throughout the entire LMNA gene (7).
Lamins, like all IFs, have a short N-terminal “head” domain, an α-helical coiled-coil “rod” domain, and a globular carboxyl “tail” domain. In the Xenopus germinal vesicle, lamins are organized as a 10-nm-thick filamentous meshwork of short and branching filaments (11). A similar organization of lamin filaments has not been observed in other cell types (12). The smallest soluble units generated during mitosis are dimeric coiled-coils (13). One lamin assembly model suggests that the dimers assemble into thin “head to tail” fibrils. These fibrils are then laterally anneal into 10-nm-thick filaments and further into paracrystalline structures (11, 14, 15). Recently, it has been shown that the B-type C. elegans lamin (Ce-lamin) is able to form stable 10-nm-thick filaments in vitro (16, 17). The in vitro association reactions occur via an intermediate step of ≈20-nm-thick filaments, they are extremely fast and significant filamentous assemblies are formed within seconds, depending on the used ionic conditions (17).
Most biological roles of mammalian lamins are evolutionarily conserved in C. elegans. In addition, the single C. elegans lamin probably functions both as A- and B-type lamin, including its presence in the nucleoplasm (12). To date, the effects of specific mutations on the structural properties of lamins are largely unknown. Therefore, we investigated how mutations in specific residues of Ce-lamin corresponding to laminopathic disease-causing mutations in the human LMNA gene affect filament and paracrystal assembly in vitro and lamin organization and dynamics in vivo. Our results show that, although some mutants assemble into seemingly normal lamin filaments or paracrystals, other mutants interfere with the assembly process, presumably during filament assembly and maturation. Correspondingly, the mutants with in vitro assembly defects yield abnormal distribution and mobility of Ce-lamin in transgenic animals.
Results
Point Mutations in Disease-Linked Conserved Residues Affect Ce-Lamin Assembly in Vitro.
We inserted missense mutations in the rod and tail regions of Ce-lamin in conserved residues that cause EDMD, FPLD, DCM, or HGPS when mutated in human lamin A [Fig. 1, Table 1, and supporting information (SI) Fig. 5B]. Each of the 14 generated mutants contained a different point mutation was bacterially expressed, purified to near homogeneity (SI Fig. 5A) (18), and used to prepare either filaments or paracrystals as we described in ref. 17. The filaments and paracrystals were studied by negative staining electron microscopy (Figs. 2 and 3).
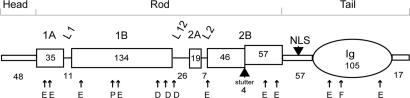
Fig. 1.
Schematic diagram of C. elegans lamin. The numbers represent the number of amino acids in each subdomain, including the amino (head) and carboxyl (tail) domains and the different coils (1A, 1B, 2A, and 2B) and linker regions (L1, L12, and L2) in the rod domain. Also marked are the positions of the stutter in coil 2B, the NLS, and the Ig fold. Arrows indicate the location of mutated amino acid residues. E, EDMD; P, HGPS; D, DCM; F, FPLD.
Table 1.
Summary of the effects of different Ce-lamin mutations on filament and paracrystal assembly and on GFP::Ce-lamin cellular localization
| Position in C. elegans | Position in human lamin A | Disease |
In vitro assembly |
In vivo cellular localization | |
|---|---|---|---|---|---|
| Filament | Paracrystal | ||||
| Y59C | Y45C | EDMD | X | V | X |
| R64P | R50P | EDMD | X | X | X |
| R103C | R89C | EDMD | V | V | nd |
| Q159K | E145K | HGPS | X | X | X |
| T164P | T150P | EDMD | V | V | X |
| N209K | N195K | DCM | V | V | X |
| D217K | E203K | DCM | X | V | X |
| L229P | L215P | DCM | V | V | V |
| Y281C | Y267C | EDMD | V | V | nd |
| E358K | E358K | EDMD | V | V | nd |
| E381A | E381A | EDMD | V | V | nd |
| R460W | R453W | EDMD | V | X | X |
| G472D | G465D | FPLD | V | V | X |
| L535P | L530P | EDMD | V | X | X |
V, normal; X, abnormal; nd, not determined.
Fig. 2.
The R64P lamin protein makes only short filaments. (A) (a–d) WT Ce-lamin. (e–h) R64P mutant protein. The assembly reaction was stopped after 5 sec (a and e), 15 sec (b and f), 5 min (c and g), or overnight (d and h) by adding an equal amount of 0.2% (vol/vol) glutaraldehyde (17). Samples were negatively stained and visualized by electron microscopy. (Scale bars: 100 nm.) (B) R64P Ce-lamin was assembled overnight under standard conditions and assembly products were centrifuged at 11,500 × g. The supernatant was transferred into a new tube and centrifuged at 26,000 × g, and the procedure was repeated at 104,000 × g. The precipitants of each step and the supernatant that remained after the last centrifugation step (SN) were analyzed in PAGE/12% SDS.
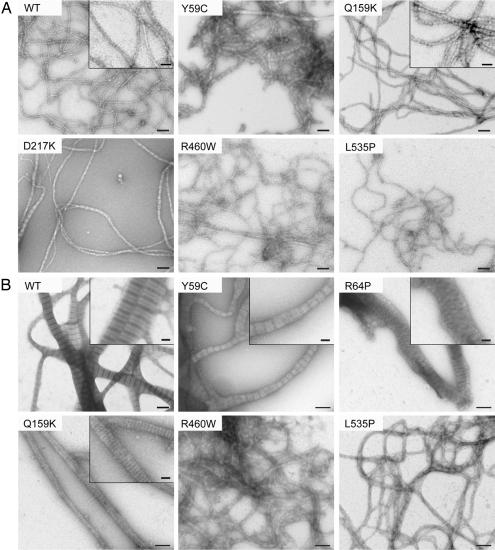
Fig. 3.
In vitro assembly of WT and mutant Ce-lamin filaments (A) and paracrystals (B). Filaments and paracrystals were negatively stained and visualized by electron microscopy. The inlets give enlarged details of the filaments and paracrystals. (Scale bars: A and B, 100 nm; Insets, 40 nm.)
Wildtype Ce-lamin formed filaments when diluted in the assembly buffer (17). These filaments were 10 nm thick, branched, and irregular as described in ref. 17 (Figs. 2A and 3A). Under similar conditions, the 14 Ce-lamin mutants assembled into various types of filaments (Figs. 2 and 3 and Table 1). A dramatic effect on filament assembly was observed in the R64P mutant (Fig. 2). After 5 sec of assembly, WT Ce-lamin was organized as both short and long branched filaments (Fig. 2Aa). In contrast, the R64P mutant formed 50- to 300-nm-long head to tail fibrils, corresponding to the length of only 1–6 lamin dimers (SI Table 2), with an average thickness of 15.5 nm ± 4 nm (n > 40) (Fig. 2Ae). After 15 sec, at which time the WT Ce-lamin protein was assembled into long and branched filaments (Fig. 2Ab), the R64P mutant remained assembled as short filaments (Fig. 2Af). Even after a 5-min incubation, the R64P mutant formed short filaments only (Fig. 2Ag). After an overnight assembly, most R64P filaments were still up to few hundred nanometers long and 20 ± 4 nm thick with few longer filaments that were up to 800 nm long (arrow in Fig. 2Ah and SI Table 2). Next, we centrifuged the R64P protein after overnight assembly at different gravitation values, transferring the supernatant of each centrifugation to a new tube and analyzing the precipitants and supernatants by SDS/12% PAGE (Fig. 2B). After centrifugation at 11,500 × g, ≈75% of the WT Ce-lamin was in the pellet fraction, whereas only ≈25% of the R64P mutant appeared in that fraction. At 26,000 × g and 104,000 × g, <4% of WT Ce-lamin was in the pellet, whereas 35% and 19% of the R64P mutant molecules, respectively, were in the pellet. Approximately 20% of R64P and 18% of WT Ce-lamin still remained in the supernatant. These data again suggest that most R64P molecules did not assemble into long filaments.
Four of the 14 examined missense mutations caused filament assembly phenotypes (Fig. 3 and Table 1). These include Y59C and R64P in coil 1A and Q159K and D217K in coil 1B (Fig. 1). All other tested lamin mutants gave apparently normal filaments (Table 1). There were two other kinds of abnormal filaments, beside the R64P assembly phenotype (Fig. 2A). Ce-lamin with the Q159K mutation assembled into “striped” filaments (Fig. 3A, see inlet), suggesting that the organization of the head to tail fibrils with respect to each other is different from in WT Ce-lamin filaments. The Y59C and D217K mutants both gave filaments that were between 10 and 40 nm thick with an average thickness of ≈20 ± 6 nm (n = 40) (Fig. 3A), which could represent either a previously observed intermediate step of 20-nm-thick filaments in the maturation process of 10-nm Ce-lamin filaments (17) or an aberrant filament assembly.
Point Mutations in Disease-Linked Residues Affect Paracrystal Assembly.
Ce-lamin formed paracrystals when dialyzed from urea into a Tris·HCl buffer containing CaCl2 (16, 17). The paracrystals exhibited a distinct transverse banding pattern with a 24- to 25-nm axial repeat (Fig. 3B) as described in ref. 17. Four of the 14 examined missense mutations (R64P, Q159K, R460W, and L535P) caused paracrystal assembly defects. All other tested mutants showed apparently normal paracrystals (Table 1). The R64P mutant resulted in unorganized structures (Fig. 3B), the Q159K showed paracrystals with stripes of equal width and distribution (Fig. 3B Inset), and the tail domain mutants R460W and L535P formed only thick fibers with an average width of 20–22 ± 4 nm (minimum 10.5 nm, maximum ≈30 nm; n > 40 for each mutant). Interestingly, the Y59C and D217K mutants that caused abnormal filament assembly did not show paracrystal assembly phenotypes (Fig. 3B and data not shown), whereas R460W and L535P mutants that affect paracrystal assembly (Fig. 3B) had no apparent effect on filament assembly (Fig. 3A). We conclude that point mutations in conserved disease-linked residues can be divided into four groups, based on their effects on lamin assembly in vitro: (i) no apparent effect on the lamin filaments or paracrystals; (ii) affecting only filaments; (iii) affecting only paracrystals; or (iv) affecting both filaments and paracrystals.
Point Mutations in Conserved Disease-Linked Residues Affect Lamin Localization in Vivo.
Ten of the 14 Ce-lamin disease-linked missense mutations, including the 6 mutations that gave in vitro assembly phenotypes, were also tested for their effects on lamin localization and mobility in vivo (Table 1). The gfp gene was fused in frame to the 5′ end of the lmn-1 gene (WT or containing a specific mutation). Each construct was used to generate 2–5 individual transgenic strains, using the microparticle bombardment technique (19). To avoid high overexpression of the transgene, in many cases we used the baf-1 promoter to drive gfp::lmn-1 expression, because its expression pattern overlaps that of the lmn-1 promoter (20) (SI Tables 3 and 4). Western blot analysis revealed that the levels of overexpression of the WT or mutant GFP::Ce-lamin proteins in the transgenic lines were between 5% and 40% of the endogenous Ce-lamin with no apparent degradation of the GFP::Ce-lamin (data not shown). When driven by the baf-1 promoter, the constructs containing the R64P or R460W mutations did not produce viable C. elegans transgenic strains, and, therefore, we used the heat shock promoter hsp16-2 (21).
When driven by the lmn-1 promoter (22), the baf-1 promoter (Fig. 4 a and b) or the hsp16-1 promoter (data not shown) WT GFP::Ce-lamin was localized at the nuclear periphery with a weaker signal in the nucleoplasm suggesting that all these promoters are suitable for this study. A dramatic phenotype was observed in Y59C or R64P mutant transgenic lines where the mutant proteins were localized mainly in the nucleoplasm without affecting nuclear shape (Fig. 4 c–g). In most cells, GFP::Ce-lamin containing a Q159K progeria mutation was localized similarly to WT Ce-lamin. However, in late embryos and adult hypodermis and muscle, ≈75% of the cells showed GFP::Ce-lamin aggregation and/or nuclear lobulation, which is one of the hallmarks of laminopathic mutations in human cells (Fig. 4 h–j). The most affected tissue was the hypodermis surrounding the vulva. Ce-lamin containing the T164P mutation, which assembled into apparently normal filaments and paracrystals in vitro (Table 1), showed WT localization in embryos before the 100-cell stage. However, in older embryos and in adult stages, the T164P protein aggregated at the nuclear periphery, and both nuclear aggregation and lobulation were observed in adult pharynx and tail nuclei (Fig. 4 k and l). Interestingly, these aggregates contained Ce-emerin (compare the nucleus in Fig. 4k with that in SI Fig. 6).
Fig. 4.
Selected pictures of single nuclei derived from embryos, hypodermis, intestine, or the tail region of WT or mutant strains expressing GFP::Ce-lamin. (a and b) Wildtype embryo (a) and intestine (b). (c and d) Y59C intestine (c) and embryo (d). (e–g) R64P intestine (e), embryo (f), and hypodermis (g). (h–j) Q159K embryo (h) and hypodermis (i and j). (k and l) T164P embryo (k) and hypodermis (l). (m and n) D217K embryo (m) and adult tail (n). (o and p) N209K young (o) and old (p) embryos. (q–s) R460W intestine (q), hypodermis (r) and embryo (s). (t and u) R460C intestine (t) and embryo (u). (v and w) G472D intestine (v) and embryo (w). (x and y) L535P embryo (x) and intestine (y). (Scale bars: 4 μm.) (z) Cells from worms expressing GFP::Ce-lamin (WT or containing a specific missense mutation) were photobleached in either the nuclear periphery (Left) or nucleoplasm (Right), and GFP::Ce-lamin fluorescence recovery was measured. The normalized fluorescence intensity in the bleached area is plotted as a function of time.
Although most cells expressing the N209K mutation showed WT localization, after the 1-fold stage embryos expressing this mutant protein had few nuclei with nonhomogeneous GFP localization and GFP::Ce-lamin aggregation (Fig. 4 o and p). There was also very low cytoplasmic background in some of the late embryos expressing the N209K mutant. This background could be due to low levels of degraded protein, which were below the level of detection in the Western blots. Surprisingly, the GFP::Ce-lamin containing a D217K mutation, which affected filament assembly in vitro, was localized mostly at the nuclear periphery, albeit with a much lower intensity in embryos (Fig. 4 m and n). Interestingly, although localization seemed normal, expression of the G472D mutant caused lobulation of embryonic and intestine nuclei (Fig. 4 v and w). Other nuclei were apparently normal (data not shown).
GFP::Ce-lamin with a R460W mutation showed many small aggregates in all embryonic, larva and adult cells (Fig. 4 q–s). The GFP::Ce-lamin containing a R460C mutation, which is not known to be involved in a laminopathic disease, showed WT localization (Fig. 4t, u). In late embryos and intestinal adult cells, GFP::Ce-lamin containing a L535P mutation showed intra nuclear aggregates, which were bigger and fewer than in R460W GFP::Ce-lamin (Fig. 4x, y).
Staining with anti-Ce-lamin antibodies, which label both GFP::Ce-lamin and endogenous Ce-lamin, revealed that, in all tested strains, the endogenous Ce-lamin was still localized at the nuclear periphery, even when the nuclear shape was affected (data not shown). Staining with anti-emerin antibodies showed labeling at the nuclear periphery (SI Fig. 6) (23).
We conclude that mutations that affect the in vitro assembly of Ce-lamin also affected its localization in all or some cells in vivo. The other mutations either did not affect Ce-lamin localization or caused a change in localization only in specific cells.
Point Mutations in Disease-Linked Residues Can Affect Lamin Mobility in Vivo.
Fluorescence recovery after photobleaching (FRAP) in living worms expressing GFP::Ce-lamin showed no measurable recovery of WT GFP::Ce-lamin after >60 min (n > 10) (Fig. 4z shows data until 25 min). The lack of recovery was observed for both peripheral and nucleoplasmic Ce-lamin. This indicates that Ce-lamin is apparently more stable than human lamin B1 (t½ of ≈2 h; see ref. 24). GFP:Ce-lamin containing T164P, G472D, or L535P mutations gave FRAP values that were similar to WT GFP::Ce-lamin (Fig. 4z). In contrast, R64P caused a dramatic increase in GFP::Ce-lamin mobility (t½ of ≈20 sec) (Fig. 4z). This change in mobility is probably due to the impaired ability of the R64P protein to form long filaments (Fig. 2). Surprisingly, the Y59C mutant, which formed abnormal filaments and apparently normal paracrystals, also showed a significant increase in mobility (t½ of ≈20 sec) (Fig. 4z). We conclude that Ce-lamin is a very stable protein and that specific mutations that interfere with filament assembly can also cause an increase in its dynamics.
Discussion
Ce-lamin Mutations Affect Filament and Paracrystal Assembly.
There is an unprecedented number of disease-causing heritable mutations in the human LMNA gene (8). However, although assumed, it was unknown whether and how these mutations affect lamin assembly. The C. elegans lamin is unique, so far being the only known lamin that forms both stable 10-nm filaments and paracrystals in vitro (16, 17). In this study, we took advantage of the assembly properties of Ce-lamin in vitro and tested 14 missense mutations in the Ce-lamin rod and tail domains in conserved residues, which cause laminopathic diseases when present in human lamin A, for their effects on assembly in vitro and localization and mobility in vivo. Six of the 14 tested missense mutations interfered with the assembly of Ce-lamin filaments or paracrystals. The observation that eight missense mutations had no visible effects on the assembly suggests that defects in lamin filament assembly can not be the only mechanism leading to laminopathic diseases. Interestingly, two of the rod domain mutations that affected filament assembly (Y59C and D217K) had no apparent effect on paracrystal assembly. Likewise, whereas none of the tested Ce-lamin tail mutations caused abnormal filament assembly, under the standard assembly conditions, two of these mutations (R460W and L535P) severely affected the assembly of paracrystals. It should be noted, however, that under very different assembly conditions [25 mM Tris·HCl (pH 7.45) and 30 mM NaCl], these lamin mutants formed paracrystals (data not shown). These data suggest that beyond the assembly of dimers and head to tail fibrils, there are differences in higher order structural organization between lamin filaments and lamin paracrystals that depend on the lamin rod domain, and on its tail domain (25). Two of the tested mutations (R64P and Q159K) affected assembly of both filaments and paracrystals indicating that other features of assembly are common to both filaments and paracrystals.
The R64P mutant assembled into very short filaments between 50 nm and several hundred nanometers long, suggesting that R64P does not support efficient head to tail association. Interestingly, these filaments were up to 20 nm thick. Because the expected thickness of a single head to tail fibril is ≈2 nm (3), these data suggest that even short head to tail fibrils have the potential to assemble laterally, similarly to the assembly of unit length filaments of cytoplasmic IFs (25). Short and thick filaments are also observed when the assembly reaction of WT Ce-lamin filaments is stopped at 5 sec (17), suggesting that lateral assembly of filaments in WT Ce-lamin can also occur in parallel to the head to tail association. The Y59C and D217K mutants both assembled into filaments that were ≈20 nm wide, which is also an intermediate stage of compaction seen during the assembly of WT Ce-lamin filaments (17). However, determining whether these Ce-lamin mutations affect specific intermediate steps in the assembly requires higher resolution analyses of the WT and mutant filaments.
Both filaments and paracrystals had a “striped” appearance in mutant Q159K Ce-lamin. One possible explanation for this different appearance is that although the head to tail fibrils are assembled normally, the lateral association of these fibrils with each other is abnormal in the mutant protein.
Abnormal Filament Assembly in Vitro Can Help Explain Several Mutations That Affect Ce-lamin Localization and Dynamics in Vivo.
To try to correlate between the effects of the disease-linked mutations on Ce-lamin assembly in vitro and Ce-lamin organization in vivo, we generated transgenic C. elegans strains, each expressing GFP::Ce-lamin with a specific missense mutation. The integration sites of the transgenes probably did not affect the results obtained, because each mutant construct was used to generate two to five independent strains, and only identical results obtained in two or more strains were considered. In addition, our results were probably not affected by the expression levels of the transgenes, because they were only between 5% and 40% of endogenous Ce-lamin levels, and GFP::Ce-lamin localization in strains overexpressing WT Ce-lamin or the nondisease R460C mutant were similar to those of endogenous Ce-lamin (Fig. 4 and ref. 22).
Mutations that affected filament and/or paracrystal assembly showed abnormal nuclear localization of Ce-lamin in some or all cells. There were two major types of localization defects: loss of nuclear peripheral localization caused by mutations in coil 1A in the rod domain and aggregation of the protein caused by mutations in the rod and tail domains. We also found that at least one disease-linked mutation (T164P) can affect Ce-lamin nuclear localization without affecting filament or paracrystal assembly in vitro. This is not surprising, because Ce-lamin binds many partners in vivo, which in turn can potentially affect its localization. If a mutation impairs the ability of Ce-lamin to bind such partners in some or all cells, it can affect its nuclear organization in vivo (5). Indeed, emerin localization was abnormal in the T164P mutant strain. In addition, the transgenic strains allowed us to analyze the localization phenotypes of lamin mutations in the context of a whole organism and in many different cell types. Although some of the mutations caused localization phenotypes in all cells, others had developmental and/or tissue-specific localization phenotypes. For example, the Q159K mutant was mislocalized in a fraction of late embryonic cells and in the adult hypodermis but not in other cells, and the L535P mutation caused aggregation only in all late embryonic cells and adult intestine cells. Likewise, the T164P mutation caused aggregation in late embryos with >100 cells and in adult pharynx and tail cells.
Nuclear lobulation in primary fibroblasts is associated with several human lamin A disease mutations (6) and is found in C. elegans embryos down-regulated for Ce-lamin (26). Therefore, it was interesting that nuclear lobulation was observed for some mutations in a stage- and tissue-specific manner. In human fibroblasts, the mutant protein is normally expressed with the background of one WT allele, and in the transgenic animals it was expressed with the background of two WT alleles. It would therefore be interesting to repeat these analyses in animals deleted for one or both lmn-1 alleles.
Materials and Methods
Antibodies, Constructs, and Bacterial Expression of Polypeptides Derived from Ce-lamin.
Ce-lamin 3931 and 3932 antibodies and Ce-emerin antibodies are described in refs. 23 and 27. GFP polyclonal antibodies were from Santa Cruz Biotechnology, and mouse-anti-GFP 3E6 monoclonal antibody was from Qbiogene. All secondary antibodies were from The Jackson Laboratory. C. elegans were fixed and stained by indirect immunofluorescence as described in ref. 28.
Ce-lamin cDNA in pET24d (17) was mutagenized by site directed mutagenesis, using the Pfu Turbo DNA polymerase (Stratagene) and specific primers. The list of primers used for mutagenesis appears in SI Materials. All constructs were verified by DNA-sequencing. Escherichia coli BL21DE3 (codon plus-RIL) cells were used to express the different Ce-lamin constructs. All constructs were purified with a His-bind resin (Qiagen). The genomic WT lmn-1 was PCR amplified from the initiation codon to the stop codon and inserted in the pGEM T-vector (Promega). Mutagenesis was with similar primers used to mutagenize the cDNA. All constructs were verified by DNA sequencing. WT lmn-1 was cloned into the pSMuncGFP vector that includes the lmn-1 promoter, the unc-119 gene, and the gfp gene containing C. elegans introns. WT and all mutants lmn-1 constructs were cloned into the pAD010 vector, which is similar to the pSMuncGFP vector, but contains the baf-1 promoter (20) instead of the lmn-1 promoter. All constructs except for G472D and L535P were also cloned into the pADHSp vector containing the hsp16-2 promoter (29) instead of the lmn-1 promoter. The final constructs were introduced in worms by microparticle bombardment as described in ref. 19. All strains used in this study are listed in SI Table 3.
Electron Microscopy.
Proteins in urea (0.1–0.2 mg/ml) were dialyzed at room temperature and assembled into filaments or paracrystals as described in ref. 17, the only difference being that grids with paracrystals were not washed with double-distilled water. The samples were visualized with a transmission electron microscope (Technai 12; Philips) equipped with a MegaView II CCD camera.
Sedimentation Experiments.
Sedimentation experiments were carried out by using a Beckman ultracentrifuge (model L8–70) in a LP42TI rotor at 23°C for 25 min. Pellet and supernatant fractions were separated, and the pellet was resuspended in protein elution buffer containing 8 M urea. The samples were loaded on SDS/12% PAGE. Band-intensity quantification was performed by Image Gauge (Fuji Film).
Microscopy and FRAP.
Images were taken either with an Axiocam CCD camera mounted on a Zeiss Axioplan II microscope equipped for fluorescence and differential interference contrast or with an an FV-1000 confocal microscope (Olympus) equipped with an IX81 inverted microscope and a ×60/1.4 oil-immersion objective. For the FRAP experiments, worms expressing GFP::Ce-lamin WT, T164P, G472D or L535P were photobleached in defined regions of specific nuclei with 100% power of a 488-nm laser and imaged with a 488-nm laser line for excitation and a 505- to 525-nm filter for emission. Five frames every 30 sec were taken before the photobleach, and 10 frames were taken every 30 sec after the photobleach, and 30 frames were taken every 2.5 min. Y59C and R64P GFP::Ce-lamin mutants were photobleached by a 405-nm laser and imaged as above. Fifteen frames were taken before the photobleach, 2 frames were taken during the photobleach, and 200 frames were taken after the photobleach. Frames were taken every 1.107 sec. FRAP analysis, fluorescence intensity in the bleached area (ROI), the background area (BG), and the total nucleus area (Tot) were measured as a function of time before and after bleaching. The measurements were corrected and normalized according to the formula ((ROIt − BGt)/(Tott − BGt)) × ((Tott0 − BGt0)/(ROIt0 − BGt0)) essentially as described in ref. 30.
Supplementary Material
ACKNOWLEDGMENTS.
This work was supported by EURO-Laminopathies research project of the European Commission Contract LSHM-CT-2005–018690 (to Y.G., H.H., and U.A.), the Israel Ministry of Health, the USA-Israel Binational Science Foundation, the Israel Science Foundation (to Y.G.), and a Minerva short-term research grant (to N.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708974105/DC1.
References
- 1.Schirmer EC, Florens L, Guan T, Yates JR, Gerace L. Science. 2003;531:1380–1382. doi: 10.1126/science.1088176. [DOI] [PubMed] [Google Scholar]
- 2.Herrmann H, Aebi U. Annu Rev Biochem. 2004;74:749–789. doi: 10.1146/annurev.biochem.73.011303.073823. [DOI] [PubMed] [Google Scholar]
- 3.Stuurman N, Heins S, Aebi U. J Struct Biol. 1998;122:42–66. doi: 10.1006/jsbi.1998.3987. [DOI] [PubMed] [Google Scholar]
- 4.Goldman RD, Gruenbaum Y, Moir RD, Shumaker DK, Spann TP. Genes Dev. 2002;16:533–547. doi: 10.1101/gad.960502. [DOI] [PubMed] [Google Scholar]
- 5.Gruenbaum Y, Margalit A, Shumaker DK, Wilson KL. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- 6.Worman HJ, Courvalin JC. Int Rev Cytol. 2005;246:231–279. doi: 10.1016/S0074-7696(05)46006-4. [DOI] [PubMed] [Google Scholar]
- 7.Broers JL, Ramaekers FC, Bonne G, Yaou RB, Hutchison CJ. Physiol Rev. 2006;86:967–1008. doi: 10.1152/physrev.00047.2005. [DOI] [PubMed] [Google Scholar]
- 8.Worman H, Bonne G. Exp Cell Res. 2007;313:2121–2133. doi: 10.1016/j.yexcr.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vlcek S, Foisner R. Curr Opin Cell Biol. 2007;19:298–304. doi: 10.1016/j.ceb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Mattout A, Dechat T, Adam SA, Goldman RD, Gruenbaum Y. Curr Opin Cell Biol. 2006;18:1–7. doi: 10.1016/j.ceb.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 11.Aebi U, Cohn J, Buhle L, Gerace L. Nature. 1986;323:560–564. doi: 10.1038/323560a0. [DOI] [PubMed] [Google Scholar]
- 12.Melcer S, Gruenbaum Y, Krohne G. Exp Cell Res. 2007;313:2157–2166. doi: 10.1016/j.yexcr.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Gerace L, Blobel G. Cell. 1980;19:277–287. doi: 10.1016/0092-8674(80)90409-2. [DOI] [PubMed] [Google Scholar]
- 14.Heitlinger E, Peter M, Lustig A, Villiger W, Nigg EA, Aebi U. J Struct Biol. 1992;108:74–89. doi: 10.1016/1047-8477(92)90009-y. [DOI] [PubMed] [Google Scholar]
- 15.Klapper M, Exner K, Kempf A, Gehrig C, Stuurman N, Fisher PA, Krohne G. J Cell Sci. 1997;110:2519–2532. doi: 10.1242/jcs.110.20.2519. [DOI] [PubMed] [Google Scholar]
- 16.Karabinos A, Schunemann J, Meyer M, Aebi U, Weber K. J Mol Biol. 2003;325:241–247. doi: 10.1016/s0022-2836(02)01240-8. [DOI] [PubMed] [Google Scholar]
- 17.Foeger N, Wiesel N, Lotsch D, Mucke N, Kreplak L, Aebi U, Gruenbaum Y, Herrmann H. J Struc Biol. 2006;155:340–350. doi: 10.1016/j.jsb.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 18.Mattout A, Goldberg M, Tzur Y, Margalit A, Gruenbaum Y. J Cell Sci. 2007;120:77–85. doi: 10.1242/jcs.03325. [DOI] [PubMed] [Google Scholar]
- 19.Praitis V, Casey E, Collar D, Austin J. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Margalit A, Neufeld E, Feinstein N, Wilson KL, Podbilewicz B, Gruenbaum Y. J Cell Biol. 2007;178:661–673. doi: 10.1083/jcb.200704049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hajdu-Cronin YM, Chen WJ, Sternberg PW. Genetics. 2004;168:1937–1949. doi: 10.1534/genetics.104.028423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haithcock E, Dayani Y, Neufeld E, Zahand AJ, Feinstein N, Mattout A, Gruenbaum Y, Liu J. Proc Natl Acad Sci USA. 2005;102:16690–16695. doi: 10.1073/pnas.0506955102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee KK, Gruenbaum Y, Spann P, Liu J, Wilson KL. Mol Biol Cell. 2000;11:3089–3099. doi: 10.1091/mbc.11.9.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moir RD, Spann TP, Lopez-Soler RI, Yoon M, Goldman AE, Khuon S, Goldman RD. J Struct Biol. 2000;129:324–334. doi: 10.1006/jsbi.2000.4251. [DOI] [PubMed] [Google Scholar]
- 25.Herrmann H, Foisner R. Cell Mol Life Sci. 2003;60:1607–1612. doi: 10.1007/s00018-003-3004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Rolef-Ben Shahar T, Riemer D, Spann P, Treinin M, Weber K, Fire A, Gruenbaum Y. Mol Biol Cell. 2000;11:3937–3947. doi: 10.1091/mbc.11.11.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tzur YB, Hersh BM, Horvitz HR, Gruenbaum Y. J Struct Biol. 2002;137:146–153. doi: 10.1006/jsbi.2002.4452. [DOI] [PubMed] [Google Scholar]
- 28.Fridkin A, Mills E, Margalit A, Neufeld E, Lee KK, Feinstein N, Cohen M, Wilson KL, Gruenbaum Y. Proc Natl Acad Sci USA. 2004;101:6987–6992. doi: 10.1073/pnas.0307880101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stringham GE, Dixon DK, Jones D, Candido EPM. Mol Biol Cell. 1992;3:221–233. doi: 10.1091/mbc.3.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rabut G, Ellenberg J. Photobleaching Techniques to Study Mobility and Molecular Dynamics of Proteins in Live Cells: FRAP, iFRAP and FLIP. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.