Abstract
The Drosophila heterochromatin protein 1 (HP1) regulates epigenetic gene silencing and heterochromatin formation by promoting and maintaining chromatin condensation. Here we report the identification and characterization of an HP1-interacting protein (Hip). Hip interacts with HP1 in vitro and is associated with HP1 in vivo. This interaction is mediated by at least three independent but similar HP1-binding modules of the Hip protein. Hip and HP1 completely colocalize in the pericentric heterochromatin, and both haplo- and triplo-dosage mutations act as dominant suppressors of position effect variegation. These findings identify a player in heterochromatinization and suggest that Hip cooperates with HP1 in chromatin remodeling and gene silencing.
Keywords: Drosophila, heterochromatin
In eukaryotes, a large portion of the genome is organized as heterochromatin. Heterochromatin replicates late, contains only few genes but a large number of repetitive sequences, and is predominantly found near centromeres and telomeres. Heterochromatin can be distinguished from euchromatin by specific histone tail modifications (reviewed in ref. 1) and by the localization of specific nonhistone chromosomal proteins.
Heterochromatin protein 1 (HP1) is evolutionary conserved from yeast to humans (2). In Drosophila melanogaster it localizes to centric and telomeric heterochromatin, the banded small fourth chromosome, as well as to ≈200 sites throughout the euchromatin (3–5). It is encoded by the Su(var)2-5 gene, which has been identified as a suppressor of position effect variegation (PEV) (6). The PEV system is characterized by a mosaic expression pattern of genes that become positioned close to centric heterochromatin after chromosomal rearrangements (7). Genetic modification of the PEV phenotype is indicative of a function in heterochromatin formation, and this system has been successfully applied to identify novel players in heterochromatinization in the past (8). Heterochromatin-induced silencing coincides with chromatin modifications to form higher-order nucleosomal arrays, which is believed to result in reduced promoter accessibility (9). The essential function of HP1 in this process is demonstrated in Su(var)2-5 mutant flies that suppress PEV and lack reporter gene silencing.
The 206-aa Drosophila HP1 protein contains an N-terminal chromo domain, which mediates the association of HP1 to pericentric heterochromatin by binding to the N-terminal tail of histone H3 methylated at lysine-9 (H3mK9) (10, 11). A flexible hinge region connects the chromo and the C-terminal chromo shadow domains. The hinge is required for nuclear localization and contributes to protein–protein interactions and HP1 association with chromosomes during mitosis (12, 13). The chromo shadow domain promotes HP1 homodimer formation (14) and provides the surface for the interaction with various other chromosomal proteins (15). One of these interacting proteins is SU(VAR)3-9, a histone methyltransferase (HMTase) that sets the H3mK9 epigenetic mark that in turn is recognized by the chromo domain (11, 16). This interaction is suggestive of a self-propagating model to explain chromatin spreading.
Once established, heterochromatic structures need to be maintained, and HP1 contributes to maintenance by stabilizing histone–histone and histone–nonhistone interactions. Biochemical investigations and the identification of interaction partners indicate that the establishment, spreading, and maintenance of heterochromatin are mediated by an HP1 multiprotein complex (reviewed in ref. 17). However, thus far only a few components of this complex, such as SU(VAR)3-9, have been identified. Yet, to understand the mechanism of heterochromatinization and gene silencing, the identification of the remaining players is essential.
Here we describe the identification and characterization of a Drosophila HP1-interacting protein (Hip). Hip and HP1 interact in vitro and in vivo, and we find that both proteins colocalize in heterochromatic regions and that Hip binding depends on HP1 function. The interaction with HP1 is mediated by three binding modules of the Hip protein. The presence of three binding surfaces suggests that Hip functions as a bridging protein to cross-connect multiple HP1 proteins, which would contribute to the stabilization of a higher-order chromatin conformation. Such a function is supported by our observation that hip acts as a haplo- and triplo-suppressor of PEV. We propose a model for the function of Hip in HP1-mediated heterochromatin spreading and the role of Hip in gene silencing.
Results and Discussion
Identification of Hip as an HP1-Interacting Protein.
To identify novel players in chromatin regulation we analyzed global changes in transcription by DNA microarray experiments (our unpublished data) after overexpression of jumeuaux [jumu, also known as Domina (Dom)]. jumu encodes a Drosophila winged-helix domain transcription factor of pleiotropic function (18, 19). Previously, we have shown that jumu modifies chromatin structure and acts as a haplo-suppressor/triplo-enhancer of PEV (ref. 18 and G.K., unpublished data). Accordingly, a number of chromatin binding proteins were highly enriched in our microarray analysis, including the HP1 encoding gene Su(var)2-5. However, the highest magnitude in expression level changes of >20-fold was observed for a gene of unknown function, CG8044, which we named hip for HP1-interacting protein.
Information from the Drosophila protein interaction database (20) suggested that the protein product of CG8044 could interact with three other proteins, including HP1. For our functional analysis we first amplified the hip coding region by RT-PCR according to the predicted sequence of the longer of the two annotated CG8044 transcripts named hip-a (described below). To verify the putative HP1/Hip interaction we performed yeast two-hybrid assays using the Hip isoform A (amino acids 1–106) as bait (21). For the target construct we fused the Gal4 activation domain to the HP1 coding sequence (amino acids 1–206) (22). Yeast cells carrying both constructs showed strong activation of the lacZ reporter, confirming the predicted HP1/Hip interaction, whereas the controls lacked signals (data not shown).
Analysis of the hip Gene Structure.
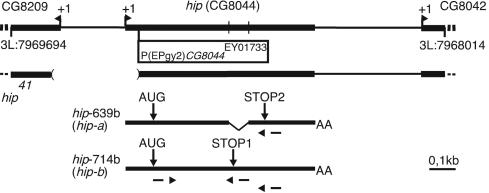
The Berkeley Drosophila Genome Project (BDGP) annotation of the Drosophila genome predicts two CG8044 transcripts of 640 and 714 nt, which we refer to as hip-a and hip-b, respectively (Fig. 1). The conceptual translation results in a 106-aa protein (CG8044-PA, Hip-A) and a 100-aa isoform (CG8044-PB, Hip-B). To confirm the structure of the hip gene, we cloned several cDNAs from third-instar larvae salivary glands by RT-PCR using transcript-specific primers (Fig. 1). The sequences of clones generated with hip-a primers confirmed the predicted spliced variant. In contrast, we found that in hip-b cDNAs the intron is not removed, which results in a stop codon six codons downstream of the predicted alternative splice junction (Fig. 1). Thus, we conclude that the two annotated transcripts hip-a and hip-b exist in vivo. This finding is further supported by two BDGP full insert cDNA sequences for the spliced hip-a (RE44783) and the unspliced hip-b variant (GM01628).
Fig. 1.
The structure of the hip gene. (Upper) The genomic region containing the hip/CG8044 gene and the flanking genes (CG8209 and CG8042) on chromosome 3L are shown. Transcription start site (arrowhead and +1), the insertion site of P(EPgy2)CG8044EY01733, position of splice sites, and the deletion in hip41 are indicated. (Lower) Predicted hip transcripts. The transcript hip-639b (hip-a) contains two exons and encodes a 106-aa Hip isoform A (CG8044-PA) that uses stop codon STOP2. The single exon transcript hip-714b (hip-b) encodes a 100-aa Hip isoform B (CG8044-PB) that uses stop codon STOP1. The primers used in the RT-PCR analysis are indicated.
The two Hip isoforms A and B share the first 94 amino acid residues but differ in the remaining C-terminal sequences, residues 95–106 for isoform A and 95–100 for isoform B. To analyze the expression of these proteins we immunized rabbits with bacterially expressed GST-Hip fusion protein. The obtained two antisera both detect a polypeptide of ≈17 kDa on immunoblots [supporting information (SI) Fig. 6]. This corresponds well to the predicted size of the two Hip isoforms of 12.1 and 11.6 kDa. However, we could not detect distinct bands that correspond to the two Hip isoforms A and B that differ by 6 aa in length. These findings confirm that hip is expressed and translated; however, the lack of two separable products could suggest that in the examined tissue (Drosophila third-instar salivary gland nuclear extracts) only one isoform is translated or could be attributed to the difficulties in resolving a size difference of 6 aa by SDS/PAGE.
Characterization of Hip/HP1 Interaction.
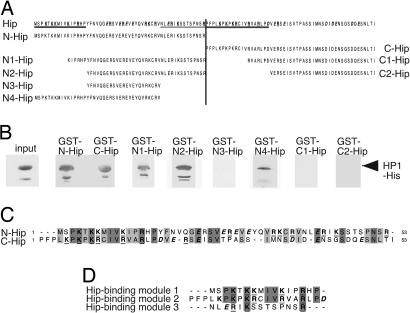
Next we set out to investigate the interaction of Hip with HP1 by in vitro GST pull-down assays. A bacterially expressed GST-Hip-A fusion protein was used to coprecipitate recombinant (his)-tagged HP1 protein from Escherichia coli cell extract followed by Western blot analysis. We find that GST-Hip interacts very efficiently with HP1 in vitro (Fig. 2B). Recombinant GST-Hip was also found to bind specifically to the endogenous Drosophila HP1 from salivary gland nuclear extract (Fig. 2C).
Fig. 2.
Recombinant and endogenous HP1 interact with Hip using its chromo shadow domain. (A) GST pull-down experiments with a GST-Hip fusion protein or GST alone as a control and full-length and truncated His-tagged HP1 proteins were performed. (B) Eluted proteins were probed with anti-His antibody. Recombinant HP1-His protein migrates as two polypeptides of different sizes, which might be due to degradation. Full-length HP1 interacts with Hip and the HP1 chromo shadow domain (HP1-cs) but not the HP1 chromo domain (HP1-cd) or the hinge region (HP1-hinge). (C) Same conditions as in B, but this time nuclear extracts from larval salivary glands were used as a source of endogenous HP1 protein. (D) Anti-HP1 Western blot analysis after immunoprecipitation (IP) from salivary gland nuclear extracts with a combination of both anti-Hip antisera detected HP1 protein. Mock precipitation with preimmune sera did not retain HP1, indicating that Hip and HP1 interact in vivo.
To map the Hip interaction domain in HP1, we applied the same GST pull-down strategy using several his-tagged subdomains of the HP1 protein (Fig. 2A). These experiments revealed that the C-terminal chromo shadow domain of HP1 is sufficient for HP1/Hip interaction (HP1-cs-His) (Fig. 2B). In contrast, the N-terminal 86 aa of HP1 that contain the chromo domain (HP1-cd-His) and the hinge region (HP1-hinge-His) do not show interaction with Hip, indicating that the HP1 chromo shadow domain is necessary and sufficient for binding of Hip.
One possibility to explain this protein–protein interaction is that Hip and HP1 are associated in a protein complex in vivo. To test this, we performed immunoprecipitation assays from salivary gland nuclear extracts. The Hip antibody efficiently coimmunoprecipitates HP1 whereas the preimmune serum did not precipitate HP1 (Fig. 2D). These results establish that Hip and HP1 specifically interact in vitro and in vivo and suggest that both proteins are associated in a protein complex in vivo.
Hip Protein Colocalizes with HP1, and Chromatin Binding Depends on HP1.
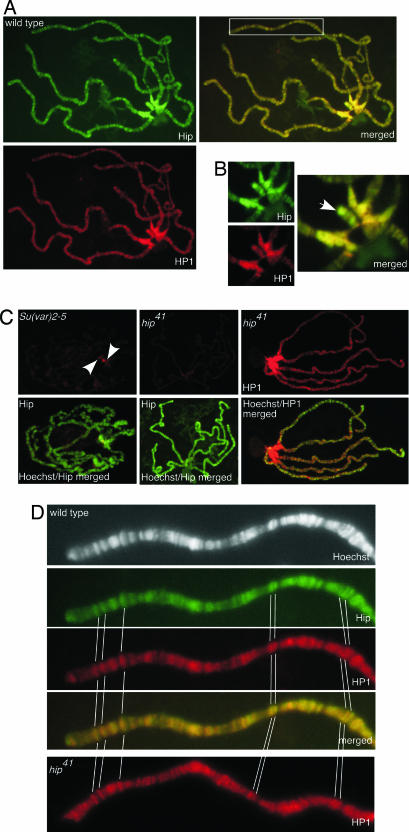
Based on our interaction studies and the detection of Hip in salivary gland nuclear extracts we expected that both proteins colocalize in a chromatin-associated complex. Immunofluorescence microscopy of salivary gland polytene chromosomes revealed that Hip localizes to both the heterochromatic regions and distinct locations within the euchromatin (Fig. 3A). The protein is distributed mainly in pericentric heterochromatin, telomeric heterochromatin, the mostly heterochromatic small fourth chromosome, and in numerous bands scattered throughout the euchromatic chromosomal arms of polytene chromosomes. To identify chromosomal loci that are targets of both Hip and HP1, we double stained with anti-Hip and anti-HP1 antibodies. Both proteins completely colocalize in the heterochromatic chromocenter and the telomeres (Fig. 3 A and B). In contrast, along the arms of all polytene chromosomes the overlap is not as perfect at the >100 binding sites of both proteins (Fig. 3D).
Fig. 3.
Immunolocalization of Hip and HP1 on polytene chromosomes. (A) Both proteins colocalize in the heterochromatic chromocenter (see enlarged detail in B), telomeres, and euchromatic sites (boxed area in merged image is enlarged in D). Third-instar larvae salivary gland chromosomes were immunostained with anti-Hip antibody (green) and anti-HP1 antibody (red). Sites where both proteins colocalize appear yellow in the merged image. (B) Colocalization of Hip and HP1 in the chromocenter and on the fourth chromosome (arrow). (C) Binding of Hip depends on the presence of HP1. Shown are stainings with anti-Hip or anti-HP1 antibody (both in red) on polytene chromosomes counterstained with Hoechst 33258 (green). In Su(var)2-5-deficient larvae almost no HP1 staining could be detected (data not shown). Hip binding to polytene chromosomes is strongly diminished in Su(var)2-5-deficient larvae. A very weak Hip signal remains at the chromocenter (Left, arrowheads). On chromosomes of mutant hip41 homozygous larvae virtually no Hip protein could be detected (Center) whereas the binding of HP1 appears unaffected (Right). (D) An enlarged view of a part of chromosome arm 2L is shown as an example for colocalization of Hip and HP1 at euchromatic sites. In hip41 HP1 binding to euchromatic sites is unchanged. Prominent binding sites are marked.
To gain insight into the mode of Hip/HP1 binding to chromatin we next addressed the question of whether Hip and HP1 are recruited to chromatin in an independent fashion or whether their binding is interdependent. In HP1-deficient third-instar larvae (23, 24) Hip binding to chromatin is dramatically reduced as compared with control stainings of chromosomes from wild-type larvae. Although the overall signal on the mutant chromosomes was reproducibly weak some residual signal could be detected in the central part of the chromocenter (Fig. 3C). Confirming the HP1 mutant condition, almost no staining could be detected with anti-HP1 antibody. However, a closer inspection revealed that very weak HP1 staining signal remained in the central part of the chromocenter (data not shown) as described above for the residual Hip staining. We speculate that some binding of Hip to residual maternal HP1 protein persists in the central part of the chromocenter in HP1-deficient third-instar larvae. Surprisingly, Hip localization to the numerous sites scattered throughout the euchromatic arms of polytene chromosomes was completely abolished in HP1-deficient larvae, even at those positions where no colocalization with HP1 could be detected by immunohistochemistry (compare Fig. 3A). To explain this behavior we speculate that levels of HP1 protein that are not detected by our immunofluorescent stainings are sufficient to recruit Hip protein. To rule out that the lack of Hip signal on HP1-deficient polytene chromosomes is caused by a general reduction in protein level, we quantified Hip protein levels by Western blot analysis. Compared with wild type, Hip levels are not significantly reduced in the HP1 mutant condition (SI Fig. 6). Together, these results suggest that Hip binds to heterochromatin and to specific sites within the euchromatin and that this binding depends on HP1 function.
Hip Has Three HP1-Binding Modules.
Database searches for sequences similar to D. melanogaster Hip uncovered similar proteins in other Drosophila species. Despite the lack of recognizable protein motifs or sequences that show conservation beyond closely related species, Hip reveals some exceptional biochemical properties such as an unusual amino acid composition. Both isoforms are serine-rich with 13.2% and 12.0% serine content for Hip-A and Hip-B, respectively. Furthermore, both proteins contain a high proportion of the charged amino acids K, R, E, and D (Hip-A, 31.2%; Hip-B, 32%), which calculates to an isoelectric point (pI) at 9.79 (Hip-A) and 10.09 (Hip-B). Despite the lack of significant sequence conservation we speculate that functional analogies could exist in nonhomologous sequences that share these biochemical properties.
The direct comparison of the Hip sequence with characterized HP1 binding proteins did not point to putative HP1 interaction sequences within Hip. Therefore, we experimentally dissected the Hip protein to identify HP1 binding domains. In a first attempt, we separated the 106 aa of Hip-A into an N-terminal half (amino acids 1–53; N-Hip) and a C-terminal half (amino acids 54–106; C-Hip) and fused each to GST (Fig. 4A). These constructs were subjected to GST pull-down assays as described above. Surprisingly, we find that both the N-terminal and the C-terminal halves of Hip are sufficient to bind HP1 (Fig. 4B). Prompted by this finding we reanalyzed the sequences of N-Hip and C-Hip and found similar repeats in both fragments (Fig. 4C) suggesting that Hip could contain at least two similar HP1-binding interfaces, one in the N-terminal half and another in its C-terminal half. To further test this hypothesis we successively truncated N-Hip and C-Hip constructs (Fig. 4A) to fine-map the binding sites by GST pull-down experiments (Fig. 4B). Three constructs derived from the N-terminal half, N1-, N2-, and N4-Hip, showed binding to HP1-His. A fourth N-terminal construct, N3-Hip, and both C-terminal fragments, C1-Hip and C2-Hip, lack this ability (Fig. 4). Together these results identify three HP1-binding modules within the 106-aa sequence of Hip-A. Each of these sequences appears to be sufficient for HP1 binding. An alignment of the stretches that are necessary for HP1 binding revealed some sequence similarities. Each binding module is composed of a central positively charged K residue flanked by other positively charged K and R residues (Fig. 4 A and D). Taken together, we have identified three HP1-binding modules that share some recognizable sequence similarity with each other. The presence of three binding interfaces in Hip implies a mode of cooperative binding suited to cross-link multiple HP1 molecules. Such a mechanism could contribute to the stabilization and maintenance of heterochromatin.
Fig. 4.
Hip contains three HP1-binding interfaces. (A) The Hip protein sequence (at the top) is unusually rich in charged amino acid residues, such as K, R, E, and D (bold type; positively and negatively charged amino acids are underlined and in italics, respectively). The N- and C-terminal fragments used for GST pull-down assays are indicated. (B) His-tagged full-length HP1 was used in GST pull-down assays with different Hip N-terminal (GST-N-Hip, GST-N1-Hip, GST-N2-Hip, GST-N3-Hip, and GST-N4-Hip) or C-terminal (GST-C-Hip, GST-C1-Hip, and GST-C2-Hip) fragments. Comparable amounts of protein were used in each experiment. Western blot analysis with anti-His tag antibody revealed that the Hip N- and C-terminal fragments are sufficient for HP1 binding. Fragments N1-Hip, N2-Hip, N4-Hip, but not N3-Hip, C1-Hip, or C2-Hip, interact with HP1. Three sequences that are necessary for HP1 binding are underlined in A. (C) Sequence comparison of the N- and C-terminal parts of Hip shows striking similarity. Identical residues are shaded in dark gray, chemically similar residues are indicated in light gray, and charged amino acids are in bold type. (D) Comparison of the three HP1-binding modules (underlined in A) reveals some similarity, especially in the position of charged residues, such as K and R.
Changes in hip Expression Levels Suppress PEV.
To analyze the hip function we generated a mutation in the hip gene by P-element mobilization. From >200 excision lines of the P(EPgy2)CG8044EY01733 P-Element (Fig. 1) we identified hip41. This mutant line carries a deletion of 333 bp caused by imprecise excision, which we verified by sequencing and Southern blot experiments (data not shown). The small deletion removes the predicted hip promoter and additional flanking regions (see Fig. 1). hip41 homozygous animals are viable and fertile, but compared with wild-type flies their individual development is delayed for 1 day. RT-PCR experiments revealed that transcript levels are significantly reduced in the homozygous hip41 condition (SI Fig. 6). Consistent with this observation, no Hip protein could be detected on polytene chromosomes by immunohistochemistry (Fig. 3C) and in immunoblots with salivary gland protein extracts (SI Fig. 6). Interestingly, the chromosomal distribution of HP1 appears unaffected in hip41 homozygous larvae (Fig. 3 C and D). We conclude that Hip is not an essential factor for the chromosomal association of HP1, whereas chromatin binding of Hip depends on HP1.
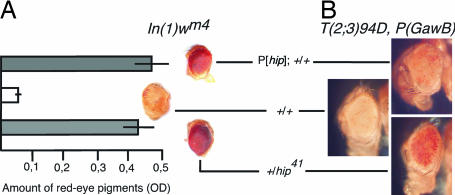
We tested hip41 mutant flies for their ability to modify variegation of In(1)wm4 and found that hip41 causes a dominant suppression of PEV in a heterozygous condition (Fig. 5A). It therefore belongs to the group of haplo-suppressors of PEV that also includes Su(var)2-5 (25, 26). The similar behavior of hip and Su(var)2-5 mutations further supports a role of hip in heterochromatin formation and gene silencing. Surprisingly, the increase of hip dosage with a transgenic hip P-element (P[hip]; for details see Materials and Methods) also shows suppression of PEV (Fig. 5A). Thus, hip acts as a haplo-suppressor and triplo-suppressor of the In(1)wm4 PEV phenotype. To verify this result we used a different PEV system, a P(GawB) translocation adjacent to chromocentric heterochromatin (G.K., unpublished data). Again, the haplo condition as well as hyperexpression of hip suppress heterochromatin-mediated inactivation of miniw expression (Fig. 5B). At a first glance the concurrent function as a haplo-suppressor and triplo-suppressor appears contradictory; however, we envisage that extra Hip protein might titrate HP1 protein. An increase in the cellular concentration of Hip could alter the stoichiometric ratio of Hip:HP1 to withdraw HP1 from heterochromatic regions. A decrease of chromatin-bound HP1 would result in the decondensation of heterochromatin to suppress PEV. Such a dominant negative squelching effect is in good agreement with the presence of three HP1 binding interfaces in each Hip molecule. Thus, the in vitro and in vivo binding properties of Hip presented in this study are sufficient to explain the triplo-abnormal suppression PEV phenotype.
Fig. 5.
hip is a dominant suppressor of PEV. (A) Variegating eye pigmentation in In(1)wm4 flies heterozygous for the hip41 allele (haplo condition) and the P insertion P[hip] (triplo condition). Both decreasing and increasing the dose of hip causes an increase in pigmentation when compared with control eyes. Eye pigmentation was quantified to determine wm4 expression. Representative eye phenotypes of hemizygous wm4 males are shown. (B) In a different system, a translocation of the P(GawB) transposon juxtaposed to the chromocentric heterochromatin results in mottled eye pigmentation. As in A, hyperexpression as well as reduction of hip suppresses heterochromatin-mediated inactivation of miniwhite expression. +, wild-type hip allele.
Summary and Model for Hip Function in Heterochromatin Formation and Maintenance.
HP1 is a key player in maintaining a heterochromatic state; however, the mechanisms of HP1-mediated heterochromatin formation, spreading, and maintenance are only partially understood to date. HP1 recognizes modified histone H3 (H3mK9) and interacts with the HMTase SU(VAR)3-9 that catalyzes the histone H3 K9 methyl modification. HP1 is believed to interact with a wide variety of other nuclear proteins, of which only a subset have been identified to date (reviewed in ref. 17). Our study identifies Hip, an HP1-interacting protein. Both proteins interact in vitro and in vivo and colocalize on polytene chromosomes, and Hip chromatin binding depends on HP1. We show that HP1 and Hip association is mediated by the HP1 chromo shadow domain that has been previously identified to interact with numerous other proteins and also mediates self-association of HP1 (14). By functionally dissecting the Hip sequence we identified three HP1-binding interfaces in this protein. The previously described pentapeptide motif (PxVxL), which is commonly associated with HP1 binding, is found in a number of HP1-interacting proteins and functions in HP1 homodimerization (27–29). This PxVxL site is very degenerate and poorly conserved, making it hard to recognize (30). Future biochemical experiments will be necessary to assess the relationship of the three Hip-binding interfaces with the previously described HP1-interacting sequences. Although other regulators of heterochromatin formation, such as Su(var)2-5, act as haplo-suppressors and triplo-enhancers of PEV, we were surprised that hip functions as a haplo-suppressor as well as a triplo-suppressor. However, such a behavior is consistent with the proposed molecular function of Hip. The function as a haplo-suppressor is intuitively understandable. A decrease in the concentration of HP1 cross-linking modules is expected to weaken the stability of the heterochromatin forming HP1–protein complexes. On the other hand, an increased dose of hip could lead to suppression of PEV via a dominant negative effect. Each additional Hip molecule adds three HP1-binding valencies. This overload in cross-linking capacity can be expected to deprive HP1 from binding to chromatin, thus causing a weakening of heterochromatic structures.
However, the observation that hip mutant flies show a delay in development but are viable and fertile suggests either that the hip function is dispensable or that redundant functions compensate for the loss of Hip. We favor the second explanation, because our observation that hip functions as a modulator of PEV demonstrates that it contributes to heterochromatin formation and gene silencing in a dose-dependent manner. Functional redundancy might also account for the fact that hip has not been uncovered by forward genetic screens so far. The identification of this HP1-interacting protein adds a player to the investigation of heterochromatin formation and stabilization. Recently, four Drosophila heterochromatin proteins were identified in an independent study of which three [HP3, HP4 (identical to Hip), and HP5] were characterized to directly bind HP1 (31). This study is in excellent agreement with our findings. In particular, the HP1-dependent chromatin localization that was analyzed in tissue culture cells and a function of a P-element insertion in HP4/hip in PEV provide compelling support.
Whereas Greil et al. (31) used the P(EPgy2)CG8044EY01733 insertion in the hip genomic region for their PEV analysis we have generated a small deletion in this region by imprecise excision of this transgene. The molecular characterization of the resulting hip41 mutation revealed that the small deletion in the hip upstream genomic region includes the putative promoter and results in a strong reduction in transcript levels that we could quantify by RT-PCR and Western analysis (SI Fig. 6). Because of the close proximity of hip and CG8209 on the opposite strand this deletion also removes the upstream region of CG8209 and parts of the coding sequence of this gene, which results in a strong decrease in CG8209 transcript levels (SI Fig. 6). Although no functional information is available for CG8209 it is predicted to encode a protein with an N-terminal UBA (ubiquitin-associated) domain and a C-terminal UBX (ubiquitin-like) domain. We cannot rule out that this gene is involved in chromatin regulation; however, we believe that the observed PEV effects in the hip41 mutant are due to hip rather than CG8209 function based on the characterized interaction of Hip with HP1 and the fact that our P[hip] transgene does not include any coding sequences of CG8209 but functions as a suppressor of PEV.
In summary, we have characterized the interaction of HP1 with the Hip protein in vitro and in vivo and demonstrated that this factor contributes to heterochromatin formation and/or maintenance. The concurrent identification of other HP1 binding proteins together with the identification of three binding modules in Hip and the future dissection of functional residues within these modules will add to our understanding of the HP1-dependent multiprotein complex in Drosophila heterochromatinization. It is reasonable to expect that these findings will also aid the discovery of similar mechanisms in other species even if they are not conserved at the sequence level.
Materials and Methods
Cloning and RT-PCR.
RT-PCR on third-instar larvae salivary glands was performed to amplify hip-a and hip-b with the primers hipfwdBamHI (gccgcggatccATGTCACCGAAGACTAAAA) and hipGSTrevBamHI (cggccggatcCTAGATTGTCAGATTAGACT) for hip-a or hipfwdBamHI and hipGSTrevHindIII (cggcggaagcttCTAGAAGACCTGGCACC) for hip-b (introduced BamHI or HindIII sites are indicated by lowercase letters). PCR products were cloned into pGEM-T vector (Promega) and sequenced. To generate a GST fusion construct hip-a was subcloned in pGEX2T (Amersham Pharmacia) by using BamHI.
Two-Hybrid Assays.
Yeast two-hybrid assays were performed in strain Y187 (22) using plasmids pAS2 (22) and pACT2 (21) according to standard protocols provided by Clontech.
Antibodies and Chromosome Immunostaining.
Bacterially expressed GST-Hip-A was purified and used to immunize two rabbits (Pineda Antikörper Service). The specificity of the resulting anti-Hip I and anti-Hip II antisera was tested by immunoblots using Drosophila salivary gland nuclear extract (data not shown). HP1 antibody was a gift of S. C. R. Elgin (Washington University, St. Louis). Polytene chromosomes were stained as described in ref. 32 by using Cy3-conjugated goat anti-mouse (Jackson ImmunoResearch) and Alexa Fluor 488 goat anti-rabbit (Molecular Probes) secondary antibodies. DNA was counterstained with Hoechst 33258. Because of the difficulties in quantifying protein levels by immunostainings we analyzed a large number (>20) of independent preparations for each condition under comparable excitation and detection conditions.
Immunoprecipitation and Direct Protein Interaction Assays.
Coimmunoprecipitation reactions containing 90 μl of salivary gland nuclear extract (described in ref. 33) and 50 μl each of anti-Hip antisera I and II were essentially performed as described in ref. 34. As a negative control, a combination of preimmune sera I and II was used.
GST-Hip constructs (GST-N-Hip, GST-N1-Hip, GST-N2-Hip, GST-N3-Hip, GST-N4-Hip, GST-C-Hip, GST-C1-Hip, and GST-C2-Hip; amino acid sequences shown in Fig. 4A) were generated by PCR and cloned in pGEX2T vector (Amersham Pharmacia). HP1 constructs were made by cloning fragments that correspond to amino acids 1–216 (for HP1-His), 1–86 (for HP1-cd-His), 137–206 (for HP1-cs-His), and 74–146 (for HP1-hinge-His) into pET21c (Novagen). Primer sequences for GST-Hip and HP1 constructs can be obtained upon request. HP1 His6-tagged recombinant proteins and GST-Hip constructs were expressed in BL21(DE3) cells. GST fusion proteins were immobilized on glutathione Sepharose-4B beads (Amersham Pharmacia). The beads were incubated with HP1 fusion proteins from crude bacterial cell extracts or Drosophila salivary gland nuclear extract as a source of endogenous HP1. Immunoblots were probed with anti-His tag antibody (Novagen) or HP1 antibody. Further details of the GST pull-down experiments can be obtained upon request.
Fly Strains and PEV Experiments.
Transheterozygous HP1 mutants were generated by crossing Df(2L)TE128-22/CyOGFP and Su(var)2-504/CyOGFP (gift from G. Reuter, Universität Halle, Halle, Germany). The mutant hip41 allele was generated by imprecise excision of P(EPgy2)CG8044EY01733. PCR on hip41 flies identified a 333-bp deletion upstream of the P-element insertion site including the predicted hip promoter region and flanking regions (see Fig. 1). Inversion In(1)wm4 and the P(GawB) translocation (G.K., unpublished data) were used for examination of PEV. Eye pigments were measured to determine wm4 expression as described in ref. 18. To express transgenic hip (P[hip]) a 1,295-bp genomic fragment flanked by CG8209 and CG8042 was amplified with the primers triplo-hipfwdXhoI (gcgtcgctcgagAGATCTGTCGATAAGTGG) and triplo-hiprevXhoI (cggcgctcgagATAAATTAAGACAAGGAGC) (XhoI sites are in lowercase letters). The amplicon contains the hip coding sequences and putative regulatory regions. This genomic fragment was inserted in the SalI site of pCarnegie 20 (pC20) (35), and transgenic lines were generated by germ-line transformation of the ry506 strain.
Supplementary Material
ACKNOWLEDGMENTS.
We thank S. C. R. Elgin for providing anti-HP1 antibody; G. Reuter for fly stocks; and K. Dünnbier, M. Brünner, and R. Lintermann for technical assistance. This work was financially supported by pilot project grants of the Freie Universität Berlin (to A.S. and G.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705595105/DC1.
References
- 1.Lachner M, O'Sullivan RJ, Jenuwein T. J Cell Sci. 2003;116:2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 2.Singh PB, Georgatos SD. J Struct Biol. 2002;140:10–16. doi: 10.1016/s1047-8477(02)00536-1. [DOI] [PubMed] [Google Scholar]
- 3.James TC, Eissenberg JC, Craig C, Dietrich V, Hobson A, Elgin SC. Eur J Cell Biol. 1989;50:170–180. [PubMed] [Google Scholar]
- 4.Fanti L, Berloco M, Piacentini L, Pimpinelli S. Genetica. 2003;117:135–147. doi: 10.1023/a:1022971407290. [DOI] [PubMed] [Google Scholar]
- 5.Sun FL, Cuaycong MH, Craig CA, Wallrath LL, Locke J, Elgin SC. Proc Natl Acad Sci USA. 2000;97:5340–5345. doi: 10.1073/pnas.090530797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eissenberg JC, James TC, Foster-Hartnett DM, Hartnett T, Ngan V, Elgin SC. Proc Natl Acad Sci USA. 1990;87:9923–9927. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiler KS, Wakimoto BT. Annu Rev Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- 8.Wakimoto BT. Cell. 1998;93:321–324. doi: 10.1016/s0092-8674(00)81159-9. [DOI] [PubMed] [Google Scholar]
- 9.Wallrath LL, Elgin SC. Genes Dev. 1995;9:1263–1277. doi: 10.1101/gad.9.10.1263. [DOI] [PubMed] [Google Scholar]
- 10.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 11.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 12.Smothers JF, Henikoff S. Mol Cell Biol. 2001;21:2555–2569. doi: 10.1128/MCB.21.7.2555-2569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Badugu R, Yoo Y, Singh PB, Kellum R. Chromosoma. 2005;113:370–384. doi: 10.1007/s00412-004-0324-2. [DOI] [PubMed] [Google Scholar]
- 14.Brasher SV, Smith BO, Fogh RH, Nietlispach D, Thiru A, Nielsen PR, Broadhurst RW, Ball LJ, Murzina NV, Laue ED. EMBO J. 2000;19:1587–1597. doi: 10.1093/emboj/19.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eissenberg JC, Elgin SC. Curr Opin Genet Dev. 2000;10:204–210. doi: 10.1016/s0959-437x(00)00058-7. [DOI] [PubMed] [Google Scholar]
- 16.Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Dorn R, Reuter G. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Kirschmann DA, Wallrath LL. Proc Natl Acad Sci USA. 2002;99:16462–16469. doi: 10.1073/pnas.162371699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stroedicke M, Karberg S, Korge G. Mech Dev. 2000;96:67–78. doi: 10.1016/s0925-4773(00)00371-3. [DOI] [PubMed] [Google Scholar]
- 19.Cheah PY, Chia W, Yang X. Development (Cambridge, UK) 2000;127:3325–3335. doi: 10.1242/dev.127.15.3325. [DOI] [PubMed] [Google Scholar]
- 20.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 21.Durfee T, Becherer K, Chen PL, Yeh SH, Yang Y, Kilburn AE, Lee WH, Elledge SJ. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 22.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 23.Lu BY, Emtage PC, Duyf BJ, Hilliker AJ, Eissenberg JC. Genetics. 2000;155:699–708. doi: 10.1093/genetics/155.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fanti L, Giovinazzo G, Berloco M, Pimpinelli S. Mol Cell. 1998;2:527–538. doi: 10.1016/s1097-2765(00)80152-5. [DOI] [PubMed] [Google Scholar]
- 25.Henikoff S. BioEssays. 1996;18:401–409. doi: 10.1002/bies.950180510. [DOI] [PubMed] [Google Scholar]
- 26.Reuter G, Spierer P. BioEssays. 1992;14:605–612. doi: 10.1002/bies.950140907. [DOI] [PubMed] [Google Scholar]
- 27.Thiru A, Nietlispach D, Mott HR, Okuwaki M, Lyon D, Nielsen PR, Hirshberg M, Verreault A, Murzina NV, Laue ED. EMBO J. 2004;23:489–499. doi: 10.1038/sj.emboj.7600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smothers JF, Henikoff S. Curr Biol. 2000;10:27–30. doi: 10.1016/s0960-9822(99)00260-2. [DOI] [PubMed] [Google Scholar]
- 29.Cowieson NP, Partridge JF, Allshire RC, McLaughlin PJ. Curr Biol. 2000;10:517–525. doi: 10.1016/s0960-9822(00)00467-x. [DOI] [PubMed] [Google Scholar]
- 30.Eskeland R, Eberharter A, Imhof A. Mol Cell Biol. 2007;27:453–465. doi: 10.1128/MCB.01576-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greil F, de Wit E, Bussemaker HJ, van Steensel B. EMBO J. 2007;26:741–751. doi: 10.1038/sj.emboj.7601527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehmann M, Korge G. EMBO J. 1996;15:4825–4834. [PMC free article] [PubMed] [Google Scholar]
- 33.Lehmann M, Korge G. EMBO J. 1995;14:716–726. doi: 10.1002/j.1460-2075.1995.tb07050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bardwell VJ, Treisman R. Genes Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 35.Rubin GM, Spradling AC. Nucleic Acids Res. 1983;11:6341–6351. doi: 10.1093/nar/11.18.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.