Abstract
Large conductance voltage and Ca2+-dependent K+ channels (BKCa) are activated by both membrane depolarization and intracellular Ca2+. Recent studies on bacterial channels have proposed that a Ca2+-induced conformational change within specialized regulators of K+ conductance (RCK) domains is responsible for channel gating. Each pore-forming α subunit of the homotetrameric BKCa channel is expected to contain two intracellular RCK domains. The first RCK domain in BKCa channels (RCK1) has been shown to contain residues critical for Ca2+ sensitivity, possibly participating in the formation of a Ca2+-binding site. The location and structure of the second RCK domain in the BKCa channel (RCK2) is still being examined, and the presence of a high-affinity Ca2+-binding site within this region is not yet established. Here, we present a structure-based alignment of the C terminus of BKCa and prokaryotic RCK domains that reveal the location of a second RCK domain in human BKCa channels (hSloRCK2). hSloRCK2 includes a high-affinity Ca2+-binding site (Ca bowl) and contains similar secondary structural elements as the bacterial RCK domains. Using CD spectroscopy, we provide evidence that hSloRCK2 undergoes a Ca2+-induced change in conformation, associated with an α-to-β structural transition. We also show that the Ca bowl is an essential element for the Ca2+-induced rearrangement of hSloRCK2. We speculate that the molecular rearrangements of RCK2 likely underlie the Ca2+-dependent gating mechanism of BKCa channels. A structural model of the heterodimeric complex of hSloRCK1 and hSloRCK2 domains is discussed.
Keywords: BK channel, circular dichroism, MaxiK, RCK, structural modeling
BKCa channels are formed by the assembly of four identical pore-forming α subunits. They can couple the membrane potential to the intracellular Ca2+ level (1–4), playing critical roles in cell excitability, for example, by controlling smooth muscle tone and neurotransmitter release (1, 5–7). Each BKCa α subunit possesses a transmembrane voltage sensor (8–10) and two distinct high-affinity Ca2+ sensors (11–15) located within the large intracellular carboxyl terminus. A well studied Ca2+-binding site corresponds to a C-terminal region that includes five consecutive negatively charged aspartates (D894–D898), christened the “Ca bowl” by the Salkoff laboratory (16, 17). The Ca bowl binds Ca2+ with high affinity (18–21) and strongly contributes to the channel's Ca2+ sensitivity (18–20) [supporting information (SI) Fig. 6]. A second high-affinity Ca2+-sensing region that is impaired by neutralization of two aspartates (D362/D367) (11, 15) or methionine 513 (22) has been identified ≈400 aa upstream the Ca bowl.
Most likely, these two high-affinity Ca2+-binding sites form parts of a complex functional domain that converts the free energy of Ca2+ binding into mechanical work to open the channel. Indeed, specialized intracellular motifs regulating the conductance of K+ channels (RCK domains) have been recently described in prokaryotic cells and identified also in the BKCa channel (23–25). In the bacterial Ca2+-activated K+ channel MthK and the KtrAB K+ transporter, RCK domains are thought to assemble in octameric structures responsible for channel opening by formation of a gating ring (26–28) reviewed in refs. 25 and 29. Based on these findings, two RCK domains were proposed to exist also in the BKCa channel α subunit, thus accounting for a total of eight RCK domains in the functional channel (two RCK domains per subunit). The first RCK domain described in BKCa channel (RCK1) encompasses the high-affinity Ca2+ sensor characterized by residues D362/D367 and M513 (11, 15, 22). A second RCK domain (RCK2) has been described recently within the BKCa C terminus (30–32). However, the location and boundaries of the second RCK domain (RCK2) are uncertain because of the poor sequence homology with known RCK domains (25, 31, 33, 34). In addition, the presence or the position of the Ca bowl (high-affinity Ca2+-binding site) within BKCa RCK2 remains unclear (25, 34). In his Ph.D. thesis, Pico (31) proposed an alignment for the putative BK channel RCK2 domain that included the high-affinity Ca bowl. On the other hand, recent literature has suggested that a second RCK domain is located downstream of RCK1. However, focusing on the regions of high sequence homology, the proposed alignments terminated before the Ca bowl region (30, 32).
An elegantly designed functional study from the Magleby laboratory suggested that the interaction between RCK1 and a downstream region that includes the Ca bowl is critical for the Ca2+-dependent activation of the channel (34), implying that the Ca bowl might constitute the high-affinity Ca2+-binding site of the second BKCa channel RCK domain (RCK2).
Because previous alignments (except for ref. 31) did not include a full RCK domain, we propose an evaluation of the positioning of α helices and β sheets within the hypothesized RCK domain.
Using a structure-based multiple-sequence alignment of the C terminus of BKCa channel and several bacterial K+ channel RCK domains, we have identified a region in the human BKCa channel (hSlo) C terminus that folds into an α/β structure that contains similar secondary structure content as the MthK RCK domain. We propose that this region encodes a second RCK domain (hSloRCK2). hSloRCK2 possesses a high-affinity Ca2+ sensor corresponding to the Ca bowl, suggesting functional homology with the hSloRCK1 domain.
We demonstrate that the hSloRCK2 domain undergoes Ca2+-dependent conformational changes in physiological condition and in a range of [Ca2+] concentration relevant to BKCa channel activation.
Results
A Second RCK Domain (hSloRCK2) that Contains the Ca Bowl.
In BKCa and prokaryotic RCK domains, the conservation of RCK-specific sequence motifs occurs mainly within ordered secondary structure elements (23, 24), although they share a low overall amino acid sequence homology (<20%). Therefore, to identify the putative RCK2 domain of the hSlo channel, we performed a structure-based multiple sequence alignment between the hSlo C terminus and several prokaryotic K+ channel RCK domains, focusing primarily on conserved ordered structural elements (α-helix–β-strands). To achieve a functionally relevant structure-based alignment, we based our approach on the following criteria: (i) the pattern of conserved residues should occur within ordered secondary structures and (ii) the length of hSloRCK2 should be similar to hSloRCK1.
The results of this analysis are shown in Fig. 1, which illustrates the position and the predicted structural composition of the hSloRCK2 domain. The hSloRCK2 sequence starts ≈100 aa downstream of hSloRCK1, as previously suggested (30–32), and shares conserved amino acids with the MthK RCK and hSloRCK1 domains within the α/β elements. The proposed hSloRCK2 extends beyond two previously proposed BKCa RCK2 structures (30, 32) and includes a high-affinity Ca2+-binding site (16–18), corresponding to the Ca bowl located between αG and βG. Interestingly, in our proposed alignment, other RCK domains include several negatively charged residues (2–5) within this region (Fig. 1). In one proposed BKCa RCK2 alignment (31), the Ca bowl locates in a nonhomologous region between βE and αE, generating significant differences with the RCK2 structure proposed here. The inclusion of the Ca bowl within hSloRCK2 strengthens the structural and possibly functional homology among hSloRCK1, hSloRCK2, and bacterial RCK domains. Moreover, the Ca bowl region shares significant homology with other RCK domains within the same loop (αG-βG) (Fig. 1).
Fig. 1.
Structure-based multiple sequence alignment of the C terminus of the human BKCa channel and RCK domains of prokaryotic K+ channels. The secondary structure assignment is based on the crystal structure of the MthK RCK domain (Protein Data Bank accession code 2AEF). Bars and arrows show helices and extended strands, respectively; yellow highlighted residues are the semiconserved sequences. Residues in red are critical for high-affinity Ca2+ sensitivity [BKCaD362/D367/M513 and D894–898 (Ca bowl)]. Residues in cyan are the low-affinity Ca2+/Mg2+-binding site (BKCa E374/E399 and MthK D184/E210/E212). hSloRCK1, hSloRCK2 from hSlo (GI: 507922); MthKRCK, Methanobacterium thermautotrophicum (GI: 2622639); A. aeoRCK, Aquifex aeolicus (GI: 2983007); S. spRCK, S. sp (GI: 7447543); EcoliRCK, E. coli (GI: 400124).
Solution-Based Analysis of the Structural Organization of hSloRCK2.
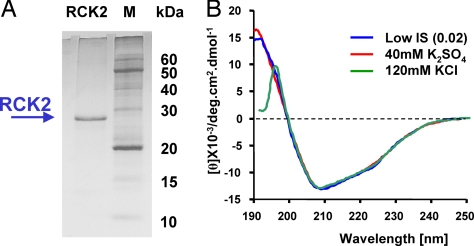
The hSlo C terminus sequence (695CAPK … ILTL936) corresponding to ≈90% of the proposed hsloRCK2 domain was cloned into a bacterial vector for protein expression and obtained with high yield and degree of purity as described in Materials and Methods. After purification, hSloRCK2 migrated as a single band corresponding to the expected molecular mass (≈28 kDa), as shown in Fig. 2A. To determine the secondary structure composition of the hsloRCK2 domain in solution, we used circular dichroism (CD) spectroscopy. Importantly, this powerful technique allowed us to assess the structure of proteins under physiological conditions of ionic strength (IS) 0.12 and pH 7.2 (35). Typical far-UV CD spectra of the purified hsloRCK2 domain are shown in Fig. 2B, displaying a strong signal at 208 and 192 nm, characteristic of the α-helical fraction (35, 36) and also an inflection at 223 nm. The secondary structure of the hSloRCK2 domain was not affected by changes in the ionic strength of the protein solution, as shown by the identical optical shape and ellipticity of the two CD spectra at low and high IS (Fig. 2B). Replacing 40 mM K2SO4 with 120 mM KCl (to mimic intracellular ionic condition) left the CD spectrum profile unmodified, except for wavelengths shorter than 200 nm, which are effected by high Cl− absorbance in this concentration range (35).
Fig. 2.
Purification and structural analysis of the hSloRCK2 domain. (A) 12.5% SDS/PAGE analysis of hSloRCK2 domain. The hSloRCK2 domain was obtained at high purity, suitable for spectroscopy analysis. Left lane, 10 μg of purified protein. Right lane, protein markers. (B) The far-UV spectra of hSloRCK2 domain recorded in solutions with low IS [24 mM Mops, 2 mM EGTA (pH 7.3)] or high IS (120 mM KCl or 40 mM K2SO4) are shown superimposed. No significant differences are observed. The strong absorption of Cl− <200 nm causes a distortion of the CD spectrum in 120 mM KCl.
The far UV CD spectra from five different experiments were analyzed for secondary structure content by using three algorithms, SELCON3, CONTIN/LL, and CDSStr, of the CDpro suite and the SMP56 protein reference set, which includes 13 transmembrane and 43 soluble proteins (37). The program classifies different types of protein secondary structures, providing the best correlation between CD spectra and known crystallographic structures (38). CD spectra analysis shows that the hsloRCK2 domain packs in α/β folds in the proportion of ≈30% α-helix and ≈20% β-strand, which tightly correlates to the predicted secondary structure (26.6% α-helix and ≈22.3% β-strand). From the CD data, we have estimated that the total number of α-helices and β-strands is nine and ten, respectively (see Materials and Methods), in excellent agreement with the theoretical predictions of nine (α) and eight (β) (Table 1). Thus, these results are within the range predicted from the hSloRCK2 alignment proposed (at least in the case of Ca2+-unbound state) supporting the view that the C terminus of the BKCa channel contains the predicted α/β structure of an RCK domain. Importantly, these data also suggest that hSloRCK2 maintains its folding after our purification procedures, which include unfolding and refolding steps.
Table 1.
The secondary structure composition of the hSloRCK2 domain
| Program | α-helix, % |
β-strand, % |
Turn, % | Unord., % | nrmsd | Nh | Ns | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| H(r) | H(d) | ΣH | S(r) | S(d) | ΣS | ||||||
| CONTIN/LL | 16.6 ± 0.8 | 13.8 ± 0.5 | 30.4 ± 1.1 | 10.7 ± 0.4 | 8.5 ± 0.7 | 19.2 ± 0.9 | 21.5 ± 0.5 | 29.4 ± 1.4 | 0.03 ± 0.01 | 8.8 ± 0.34 | 10.6 ± 0.97 |
| SELCON3 | 16.4 ± 0.4 | 13.6 ± 0.6 | 30.0 ± 0.3 | 11.5 ± 0.2 | 8.6 ± 0.4 | 20.1 ± 0.5 | 21.6 ± 0.1 | 28.8 ± 0.4 | 0.21 ± 0.08 | 8.9 ± 0.31 | 10.4 ± 0.24 |
| CDSStr | 18.1 ± 1.2 | 13.8 ± 0.3 | 31.9 ± 1.5 | 11.5 ± 0.7 | 8.1 ± 0.2 | 19.6 ± 0.9 | 19.5 ± 0.4 | 29.2 ± 0.6 | 0.10 ± 0.02 | 8.9 ± 0.09 | 10.1 ± 0.47 |
| Predicted | 26.6 | 22.3 | 9 | 8 | |||||||
CD spectra of hSloRCK2 domain obtained from five recordings were analyzed for the content of secondary structure fractions by using three algorithms of the CDPro software package. The average value is presented STDEV. H(r) and S(r) are for regular α-helix and regular β-strand, H(d) and S(d) is for distorted α-helix and distorted β-strand, respectively. ΣH = H(r) + H (d), ΣS = S(r) + S (d). The total number of helices (Nh) and β-strand (Ns) estimated as described in Materials and Methods. unord., unordered structure; nrmsd, normalized root mean square deviation.
Ca2+ Induces an α-to-β Conformational Transition in hSloRCK2.
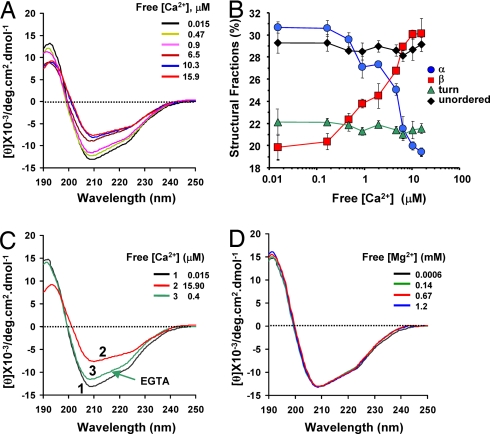
To obtain information on the Ca2+ sensitivity of the hSloRCK2 domain and details on possible structural changes occurring in the hsloRCK2 domain upon its interaction with Ca2+, we applied CD spectroscopy. The far-UV CD spectra of the hSloRCK2 domain were recorded for increasing free [Ca2+] ranging from 0.015 to 15.9 μM, as shown in Fig. 3A. The CD spectra of the hsloRCK2 domain showed a marked Ca2+-induced decrease of the negative ellipticity at 208 nm, accompanied by a change in the ratio between the negative ellipticity at 208 and 223 nm (from 1.24 to 1.12) and a shift of the minimum from 208 to 210 nm. These changes are a manifestation of Ca2+-induced changes in the secondary structure of hSloRCK2 domain occurring in a dose-dependent manner. The Ca2+-induced changes in the secondary structure [calculated by using separate samples from three protein preparations (n = 3)] were estimated by using the CONTIN/LL algorithm, which gave the most reliable analysis, as shown by the lowest normalized root mean square deviation between theoretical and experimental spectra (39) (Table 1).
Fig. 3.
Properties of Ca2+-dependent conformational transitions in hSloRCK2 domain. (A) Superimposed far-UV CD spectra of WT RCK2 domain obtained for increasing free [Ca2+] (from 0.015 μM to 15.9 μM). (B) The corresponding secondary structure fractions, estimated by using the CONTIN/LL algorithm, are plotted as a function of the free [Ca2+]. Each point is an average of three independent experiment (mean ± SEM). (C) Decreasing the free [Ca2+] from 15.9 to 0.4 μM restored the initial conformation; Ca2+-induced conformational transition is reversible. (D) Superimposed far-UV CD spectra of hSloRCK2 domain in the presence of increasing [Mg2+] (as shown). Free [Mg2+] (up to 1.2 mM) did not produce CD spectral changes in hSloRCK2.
The percentage of secondary structure composition is presented as a function of free [Ca2+] in Fig. 3B. We found that the β-strand content increased from ≈20% to ≈30% as the free [Ca2+] was increased from 0.015 to 15.9 μM. This Ca2+-induced increase in the β-strand content was paralleled by a similar decrease in α-helix content from ≈30% to ≈19%, while the turns and unordered fraction remained practically unchanged. The increase of β-strand content at the expense of an equal decrease in the α-helix fraction clearly indicates a Ca2+-induced α-to-β switch in the hsloRCK2 domain.
The α-to-β Conformational Switch Is Reversible and Ca2+-Specific.
BKCa channels are reversibly modulated by intracellular micromolar Ca2+ and display a rather low sensitivity to Mg2+ in the millimolar range (14, 40). We have assessed the reversibility of the Ca2+-induced α-to-β structural transition by lowering the free [Ca2+] from 15.9 to 0.4 μM by addition of EGTA. As shown by the CD spectra in Fig. 3C, this maneuver restored the initial conformation, confirming the reversibility of the Ca2+ effect. On the other hand, free Mg2+ up to 1.2 mM was unable to produce significant conformational transitions, as suggested by the CD spectra shown in Fig. 3D.
Role of the Ca Bowl in the hsloRCK2 Domain.
The hSloRCK2 domain proposed in this study includes a high-affinity Ca2+-binding site (Ca bowl). The neutralization of the five consecutive aspartates within the Ca bowl region has been shown to significantly reduce the Ca2+ sensitivity of the BKCa channel in mouse (16, 41) and Drosophila (19), decreasing Ca2+-binding affinity by >50% in an in vitro assay (19). We have confirmed this finding in the human clone hSlo (41). SI Fig. 6 recapitulates the main Ca2+ and voltage-dependent features of the hSlo BKCa channel expressed in Xenopus oocytes. As expected, increasing [Ca2+]i facilitated channel opening, progressively shifting the activation curve (GV) toward hyperpolarized potentials. The neutralization of the five aspartates within the Ca bowl region (D894–898N) drastically increased the half-activation potential for [Ca2+]i ranging between ≈0.01 and ≈100 μM, so larger depolarizations were required to reach 50% of the conductance in the hSlo D894–898N mutant.
How is this change in the Ca2+ dependence of channel activation related to the conformational changes observed in hSloRCK2? We neutralized the same aspartates (D894–898N) in the hSloRCK2 domain (hSloRCK2-5N) and recorded CD spectra at increasing [Ca2+] as shown in Fig. 4A. The Ca2+-dependence of the various structural components of hSloRCK2-5N estimated by the CONTIN/LL algorithm is plotted in Fig. 4B. As for WT hSloRCK2, Ca2+ increased the β content of hSloRCK2-5N at the expense of a similar decrease in α-helix fraction.
Fig. 4.
Ca2+-dependent conformational transition in the hSloRCK2 domain after neutralization of the Ca bowl (5N). (A) Far-UV CD spectra of (5N) mutant hSloRCK2 were obtained at progressively increasing free [Ca2+] (0.015–15.9 μM). (B) The corresponding estimated secondary structure fractions as a function of free [Ca2+]. (C) The change in β-strand fraction as a function of free [Ca2+] is shown for WT hsloRCK2 (circles) and for the neutralized Ca bowl mutant (squares). Data points are fitted to the sum of two Hill functions suggesting multiple Ca2+-binding sites with different affinity. The neutralization of the five aspartates in the Ca bowl practically abolished the first component, decreasing the overall Ca2+ affinity of the mutated Ca bowl. (D) Parameters used for best fit of the averaged data.
To assess the effect of the Ca bowl neutralization, we have plotted together the dependence of the β fraction vs. [Ca2+] for WT and Ca bowl-neutralized hSloRCK2-5N domains (Fig. 4C). The averaged data points obtained from three separate experiments and protein samples were fit to a linear combination of two Hill functions characterizing the hSloRCK2 biphasic response to Ca2+. In the WT hSloRCK2, the two sequential transitions occur with K0.5_1 = 0.4 μM and K0.5_2 = 5.1 μM, with an increase in β structure content of 4.8% and 5.5%, respectively. The Hill coefficient for the two components was n_1 = 2.2 and n_2 = 5, suggesting multiple binding sites and cooperativity (Fig. 4D, WT).
The neutralization of the five aspartates practically eliminated the most negative component observed in the WT hSloRCK, reducing the first transition from 4.8% to 0.5% and suggesting the loss of a high-affinity binding site (Fig. 4D). These results offer strong evidence that the hSloRCK2 domain undergoes Ca2+-induced conformational transition and that the Ca bowl plays a significant role in the Ca2+-dependence of RCK2. The neutralization of the Ca bowl eliminated the transition occurring at the lowest [Ca2+] (K0.5 = 0.4 μM), reducing the overall extent of the Ca2+-dependent α-to-β transition by >50% (Fig. 6).
Discussion
Two RCK Domains and Two High-Affinity Ca2+-Binding Sites in the BKCa Channel.
Despite the significant progress in understanding the Ca2+-dependent activation properties of BKCa channels, the nature of the molecular events initiated by Ca2+ binding and leading to channel opening are still unresolved. Intracellular regions directly involved in the Ca2+-dependent gating of BKCa channels must meet three indispensable criteria: (i) they must be accessible to intracellular Ca2+, (ii) they must bind Ca2+ in the same range of channel activation, and (iii) they must transduce Ca2+ binding into conformational changes that open the pore. We have searched for an intracellular region of hSlo that satisfied these requirements. Tremendous help came from structural studies of bacterial K+ channels and RCK domains (23, 24, 26, 27). Our solution-based functional characterization of hSloRCK2, together with the structure-based multiple sequence alignment of the C terminus of the human (hSlo) BKCa channel and several prokaryotic K+ channel RCK domains, supports the view that the each α subunit of the BKCa channel encodes two RCK domains. Both domains are characterized by a high-affinity Ca2+-binding site. In our view, the Ca bowl in the hSloRCK2 domain has the same functional Ca2+-binding role as D362/D367/M513 in hSloRCK1 and D185/E210/E212 in MthK RCK (15, 22, 23, 34).
According to the proposed alignment, hsloRCK2 has a length practically identical to hSloRCK1 (268 and 270 aa, respectively). It includes a C-Lobe, a high-affinity Ca site, and maintains all of the conserved motifs and residues present in RCK domains (23, 24) all of the way through to the terminal αJ helix. Interestingly, the hSloRCK2 Ca bowl region shares some homology with other RCK domains within the same loop (αG-βG) region, although functional roles for the negatively charged residues in this region have not been reported.
hSloRCK2 Is a Ca2+ Sensor: Functional Relevance of Ca2+-Induced α-to-β Switch in the RCK2 Region.
This work represents an attempt to resolve the functional properties of the hSloRCK2 domain under physiological condition. We have used CD spectroscopy, a powerful technique for characterizing ligand-dependent structural changes of protein in solution (27, 37). The results presented in Fig. 3B reveal that Ca2+ in the 0.2–10 μM range induces a characteristic dose-dependent conversion of α into β structure. Similar ligand-induced structural transitions have been reported for endoplasmic reticulum chaperones (42) and for a zinc-binding protein (43). Typically, conformational transitions resulting in an increase in β structure have been interpreted as a prelude to protein–protein interactions (42, 44, 45), and the α-to-β conformational transitions may occur at the interfaces between interacting domains of oligomeric structures (46). According to this view, the conversion of the protein to an increased β structure would represent a transition favoring interaction between RCK domains. Although direct interactions between BKCa channel RCK1 and RCK2 have not yet been demonstrated [but have been proposed recently (34)], the Ca2+-induced structural transition in hsloRCK domains could favor the formation or rearrangement of a gating ring structure that in turn controls channel opening (26, 27, 47).
The Ca Bowl Is an Integral Part of hSloRCK2.
The most significant Ca2+-induced event taking place within the hsloRCK2 domain is the conversion of α into β structure. Our CD data showed that this conversion occurs in two Ca2+-dependent structural changes. The transition, with an apparent Ca2+ affinity of K0.5 = 0.4 μM, accounts for ≈50% of the total α-to-β conversion (Fig. 4). The neutralization of the five consecutive aspartates within the “Ca bowl” practically abolishes the first transition, reducing the overall level of α-to-β transition by half. This mutant also drastically reduces the Ca2+ sensitivity of the hSlo channel (SI Fig. 6). Interestingly, these data are in agreement with published results of Ca2+-binding activity in vitro of isolated polypeptides from the C terminus of BKCa channels, showing that neutralization of the five aspartate residues within the Ca bowl significantly decreased, but did not completely eliminate Ca2+ binding in this region (18). Similarly, Bian et al. (19) reported that mutations in the Ca bowl region reduced its Ca2+-binding activity by 56%, suggesting that other residues might be involved in Ca2+ binding. Potential candidates that can contribute to Ca2+ coordination are oxygen-containing residues located close to the five aspartate residues within the Ca bowl (20). This point certainly deserves further investigation.
Ca2+ Specificity of the α-to-β Switch.
The Ca2+ specificity of the conformational changes observed in hsloRCK2 is consistent with electrophysiological data. BKCa channels are sensitive to intracellular Mg2+ (in the mM range). When Ca2+ was replaced by Mg2+ (up to >1 mM), no significant changes in the CD spectra of the hsloRCK2 domain were observed, indicating that Mg2+ is not capable of inducing detectable structural modification in this region. However, low-affinity Ca2+/Mg2+-binding sites have been identified within the RCK1 domain (15, 40).
Molecular Interaction Between hSloRCK1 and hSloRCK2 Domains Proposed by Structural Modeling.
Despite the low primary sequence homology, the strong similarity in the secondary structure between hSlo and bacterial RCK domains (Fig. 2) allowed us to create 3D models of hSloRCK1 and hSloRCK2 domains based on the crystal structure of MthK RCK (Protein Data Bank accession codes 2AEF and 1LNQ).
As shown by the hSloRCK1 and hSloRCK2 models reported in Fig. 5 A and B and SI Fig. 7, the negatively charged residues critical for Ca2+ sensitivity (hsloRCK1) or Ca2+ binding (hsloRCK2) are not spatially correlated with the MthK RCK Ca2+-binding site. Differences in the positioning of Ca2+-binding sites may underlie the different Ca2+ sensitivity of BKCa vs. MthK channels (μM vs. mM). The structural model of hSloRCK1 places M513 (22) in proximity of D362, supporting the idea that mutations in D362/D367 and M513 possibly affect the same binding site (22, 48) (Fig. 5A). Interestingly, the Ca bowl locates at the C-terminal end of the helix-turn-helix motif in hSloRCK2 (after αG; Figs. 1 and 5B), the same region where M513 maps within the hSloRCK1 domain (Figs. 1 and 5A).
Fig. 5.
Structural homology modeling of hSloRCK1, hSloRCK2, and the S5–S6 pore domains of the human BKCa channel. (A and B) Ribbon representations of hsloRCK1 (A) and hSloRCK2 (B) models. The position of the residues known to be involved in Ca2+ sensing [D362/D367 and M513 in hSloRCK1, and the Ca bowl (D894–8) in hSloRCK2] are shown. (C) Ribbon representation of a single BKCa channel pore and two RCK domains. The transmembrane pore region (S5–S6) and the heterodimeric hSloRCK1 and hSloRCK2 structure are shown. The pore and RCK domains were modeled as independent chains and may not be part of the same α subunit. The S5–S6 pore domain and the intracellular hSloRCK1/RCK2 complex were modeled by using the MthK channel structure (Protein Data Bank accession code 1LNQ) based on the sequence alignment of the BKCa pore domain (S5–S6) with TM1-TM2 of MthK and C terminus of the human BKCa channel and RCK domains of prokaryotic K+ channel as presented in Fig. 1. The Ca2+-binding region known as Ca bowl is part of the second BKCa RCK domain (hsloRCK2). We propose that hSloRCK1 and hSloRCK2 interlock through their αG helix-turn-αF motifs, forming a flexible interface.
Possible Structural Organization of the BKCa Channel Ca2+-Sensing Region.
In MthK, the helix-turn-helix motif connecting the two lobes (αG-turn-αF) forms an interface between RCK domains, and two Ca2+ ions bind at the base of the cleft within this interface (23, 26, 27). The placement of the high-affinity Ca2+-binding sites in proximity to the αG helix-turn-αF helix regions suggests a mechanism for Ca2+ gating. We predict that the hSloRCK1 and hSloRCK2 domains form a flexible interface (mediated through the αG helix-turn-αF) as observed in MthK, and that Ca2+ binding at D362/D367, M513 (RCK1), and the Ca bowl (RCK2) induces a structural rearrangement that favors the open state of the channel. This interaction may occur within the same subunit (intrasubunit interaction), as suggested by a recent functional study (34). Fig. 5C shows a structural model of the heterodimeric structure hSloRCK1 and hSloRCK2 domains. The configuration of this dimeric structure was constructed based on the organization of the proposed octameric gating ring structure observed in MthK (Protein Data Bank accession code 1LNQ) (23). A second interface known as “assembly” (fixed) interface may connect hSloRCK1 and hSloRCK2 from adjacent α subunits (intersubunit interface; ref. 34) through the helices αD and αE via hydrophobic interactions (25, 30). Thus, four hSloRCK1/RCK2 dimers may form an octameric ring in the homotetrameric structure of hSlo.
Thus, the proposed configuration of the hsloRCK1/RCK2 complex places the high-affinity Ca2+-binding sites near the interface between two RCK subunits and provides new insights into the structural organization of Ca2+-sensing in the BK channel.
In conclusion, we have proposed, isolated, and characterized a putative second RCK domain within the C terminus region of the human hSlo BKCa channel. The hsloRCK2 domain contains a high-affinity Ca2+-binding site (Ca bowl) that confers to this region the functional role of Ca2+ sensor. The Ca2+ specificity and the sensitivity of the Ca2+-induced conformational transition observed for hsloRCK2 are consistent with the main features of the Ca2+ regulation of the native BKCa channels. It is likely that the structural rearrangements (α-to-β switch) reported in this study underlie BKCa channel Ca2+-dependent gating. The possible organization and interaction between hSlo RCK1 and hSloRCK2 have been investigated by constructing 3D models of the hSlo C terminus based on the crystal structure of MthK RCK domains.
Materials and Methods
Structure-Based Sequence Alignments and Homology Modeling.
Sequences of bacterial RCK domains of K+ channels MthK2M (GI:2622639), A. aeo2TM (GI:2983007), S. sp2TM (GI:7447543), E. coli6TM (GI:400142), and human BKCa channel hSlo (GI:507922) C terminus were aligned by using the program Clustal W (49) followed by visual inspection and correction to account for conserved amino acid regions as reported in previous works (23, 24).
Structural homology modeling of the RCK1, RCK2, and S5–S6 pore domains of human BKCa channel was performed by using Modeller9v2 (49) and DS Modeller 1v7 (Accelrys). The alignment in Fig. 1 was used to model RCK1 and RCK2 by using the isolated MthK RCK (Protein Data Bank accession code 2AEF) as the structural template. The pore domains (S5–S6 of BKCa and TM1–TM2 of MthK) were modeled by using the Clustal W alignment shown in Table 2.
Table 2.
Pore-region sequence alignment of BKCa and MthK channels
| >BKCa (S5–S6) | ISTWLTAAGFIHLVENSGDPWENFQNNQALTYWECVYLLMVTMSTVGYGDVYAKTTLGRLFMVFFILGGLAMFASYV |
| >MthK (TM1–TM2) | PATRILLLVLAVIIYGTAG.…FHFIEGESWTVSLYWTFVTIATVGYGDYSPSTPLGMYFTVTLIVLGIGTFAVAV |
The MthK channel structure (Protein Data Bank accession code 1LNQ) was used as a structural template for the pore–RCK1–RCK2 model. For all three models, no additional restraints were specified. A first round of 11 RCK and pore–RCK1–RCK2 models were constructed and sorted by probability density function (PDF) total energy scores to create three lowest energy initial models. The RCK-only models were then visually inspected to identify poorly modeled loop regions. The loops of these models underwent a further round of optimization by using Modeller's DOPE-based loop modeling protocol. In total, 20 RCK1 and RCK2 loop models were created and evaluated by PDF total energy and DOPE scores. The lowest scoring loop-refined RCK1 and RCK2 structures were then structurally superimposed onto the initial pore–RCK1–RCK2 model to create the model shown in Fig. 5.
Plasmid Constructions and Mutagenesis.
DNA sequences from human BKCa channel RCK2 domain (695CAPK … ILTL936) were amplified by PCR and subcloned into vector plasmid pQE-30 (Qiagen), for Escherichia coli cells. In the plasmid pQE-30, the DNA fragment was fused to an N-terminal His6 sequence by using restriction enzymes BamHI and HindIII. Plasmid constructs with mutant genes were obtained by using QuickChange site-directed mutagenesis (Stratagene).
Electrophysiology.
cRNAs encoding for the hSlo wild type and 5N mutant were injected in Xenopus laevis oocytes (0.05–0.1 mg/ml), and K+ currents were recorded with the patch-clamp technique (inside-out configuration) 2–4 days after injection. Solutions contain 115 mM KMES, 5 mM KCl, 10 mM Hepes, and 5 mM HEDTA. The free [Ca2+] was measured with a Ca2+ electrode (World Precision Instruments).
Expression and Purification of the hSloRCK2 Domain.
The RCK2 domain was expressed and purified from M15 [pREP4] cells (Qiagen). Addition of isopropyl β-d-thiogalactoside to a final concentration of 1 mM induced the hSloRCK2 expression. The protein fractions were solubilized in 100 mM NaH2PO4, 10 mM Tris·HCl, 8M Urea (pH 8.0) containing 1 mg/ml lysozyme and incubated 2 h at room temperature. The supernatant obtained after centrifugation was applied to Ni-NTA resin column, and the protein fractions were eluted at pH 5.9 and dialyzed against 50 mM Tris·HCl, 5 mM EDTA, 1 mM BME (pH 7.8). A final purification step was performed by gel filtration by using Sephacryl S-300 High Resolution (Amersham). The purity of expressed proteins was analyzed by using 12.5% SDS/PAGE. The protein concentration was determined by using the Biuret method and Lowry protein assay.
CD Spectroscopy.
Four to six micromolar purified hSloRCK2 domain was used for CD studies. Spectra were obtained by using a Jasco-715 spectropolarimeter. The protein samples for CD measurement were prepared by dialysis against 24 mM MOPS, 0.5 mM BME, and 2 mM EGTA (pH 7.2) in presence of 40 mM K2SO4 or 120 mM KCl. Far UV spectra were recorded between 190 and 260 nm by using a quartz cell of 0.1-cm path length. Spectra were collected as an average of nine scans. CD data were presented in units of molar ellipticity per residue. The secondary structure fractions of the hSloRCK2 domain were calculated by using the algorithms SELCON, CONTIN/LL, and CDSSRT of CDPro software package. NRMSD (normalized root mean-square deviation) was used as the measure of the goodness of fit between the experimental spectrum and the curve calculated from crystallographic data in SMP56 (protein reference set). NRMSD is defined as [Σ (θexp − θcal)2/Σ (θexp)2]1/2, where θexp and θcal are the experimental and calculated molar ellipticity per amino acid residue, respectively. The total number of helices (Nh) and β-strand (Ns) were estimated by dividing the number of residues included in the distorted helical and β structure by a factor of 4 and 2, respectively (38). Free concentrations were measured by using Ca2+-selective electrodes (World Precision Instruments) or estimated by using the software WEBMAXC Standard (51).
Supplementary Material
ACKNOWLEDGMENTS.
We thank Ligia Toro (University of California, Los Angeles) for the hSlo clone and the members of the R.O. and D. Rees laboratories for constructive discussions. This work was supported by National Institutes of Health/National Institute of Neurological Disorders and Stroke Research Grant RO1NS043240 (to R.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705261105/DC1.
References
- 1.Salkoff L, Butler A, Ferreira G, Santi C, Wei A. Nat Rev Neurosci. 2006;7:921–931. doi: 10.1038/nrn1992. [DOI] [PubMed] [Google Scholar]
- 2.Pallotta BS, Magleby KL, Barrett JN. Nature. 1981;293:471–474. doi: 10.1038/293471a0. [DOI] [PubMed] [Google Scholar]
- 3.Latorre R, Vergara C, Hidalgo C. Proc Natl Acad Sci USA. 1982;79:805–809. doi: 10.1073/pnas.79.3.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen KZ, Lagrutta A, Davies NW, Standen NB, Adelman JP, North RA. Pflügers Archiv-Eur J Physiol. 1994;426:440–445. doi: 10.1007/BF00388308. [DOI] [PubMed] [Google Scholar]
- 5.Lu R, Alioua A, Kumar Y, Eghbali M, Stefani E, Toro L. J Physiol (London) 2006;570:65–72. doi: 10.1113/jphysiol.2005.098913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gribkoff VK, Starrett JE, Jr, Dworetzky SI. Neuroscientist. 2001;7:166–177. doi: 10.1177/107385840100700211. [DOI] [PubMed] [Google Scholar]
- 7.Tang XD, Garcia ML, Heinemann SH, Hoshi T. Nat Struct Mol Biol. 2004;11:171–178. doi: 10.1038/nsmb725. [DOI] [PubMed] [Google Scholar]
- 8.Stefani E, Ottolia M, Noceti F, Olcese R, Wallner M, Latorre R, Toro L. Proc Natl Acad Sci USA. 1997;94:5427–5431. doi: 10.1073/pnas.94.10.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diaz L, Meera P, Amigo J, Stefani E, Alvarez O, Toro L, Latorre R. J Biol Chem. 1998;273:32430–32436. doi: 10.1074/jbc.273.49.32430. [DOI] [PubMed] [Google Scholar]
- 10.Savalli N, Kondratiev A, Toro L, Olcese R. Proc Natl Acad Sci USA. 2006;103:12619–12624. doi: 10.1073/pnas.0601176103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia XM, Zeng X, Lingle CJ. Nature. 2002;418:880–884. doi: 10.1038/nature00956. [DOI] [PubMed] [Google Scholar]
- 12.Niu XW, Magleby KL. Proc Natl Acad Sci USA. 2002;99:11441–11446. doi: 10.1073/pnas.172254699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magleby RL. J Gen Physiol. 2003;121:81–96. doi: 10.1085/jgp.20028721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng XH, Xia XM, Lingle CJ. Biophys J. 2004;86:128A. [Google Scholar]
- 15.Zeng XH, Xia XM, Lingle CJ. J Gen Physiol. 2005;125:273–286. doi: 10.1085/jgp.200409239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schreiber M, Salkoff L. Biophys J. 1997;73:1355–1363. doi: 10.1016/S0006-3495(97)78168-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schreiber M, Yuan A, Salkoff L. Nat Neurosci. 1999;2:416–421. doi: 10.1038/8077. [DOI] [PubMed] [Google Scholar]
- 18.Bao L, Kaldany C, Holmstrand EC, Cox DH. J Gen Physiol. 2004;123:475–489. doi: 10.1085/jgp.200409052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bian S, Favre I, Moczydlowski E. Proc Natl Acad Sci USA. 2001;98:4776–4781. doi: 10.1073/pnas.081072398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheng JZ, Weljie A, Sy L, Ling SZ, Vogel HJ, Braun AP. Biophys J. 2005;89:3079–3092. doi: 10.1529/biophysj.105.063610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Braun AP, Sy L. J Physiol (London) 2001;533:681–695. doi: 10.1111/j.1469-7793.2001.00681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bao L, Rapin AM, Holmstrand EC, Cox DH. J Gen Physiol. 2002;120:173–189. doi: 10.1085/jgp.20028627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang YX, Lee A, Chen JY, Cadene M, Chait BT, MacKinnon R. Nature. 2002;417:515–522. doi: 10.1038/417515a. [DOI] [PubMed] [Google Scholar]
- 24.Jiang YX, Pico A, Cadene M, Chait BT, MacKinnon R. Neuron. 2001;29:593–601. doi: 10.1016/s0896-6273(01)00236-7. [DOI] [PubMed] [Google Scholar]
- 25.Lingle CJ. J Gen Physiol. 2007;129:101–107. doi: 10.1085/jgp.200709739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye S, Li Y, Chen LP, Jiang YX. Cell. 2006;126:1161–1173. doi: 10.1016/j.cell.2006.08.029. [DOI] [PubMed] [Google Scholar]
- 27.Dong JB, Shi N, Berke I, Chen LP, Jiang YX. J Biol Chem. 2005;280:41716–41724. doi: 10.1074/jbc.M508144200. [DOI] [PubMed] [Google Scholar]
- 28.Albright RA, Ibar JL, Kim CU, Gruner SM, Morais-Cabral JH. Cell. 2006;126:1147–1159. doi: 10.1016/j.cell.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 29.Chakrapani S, Perozo E. Nat Struct Mol Biol. 2007;14:180–182. doi: 10.1038/nsmb0307-180. [DOI] [PubMed] [Google Scholar]
- 30.Kim HJ, Lim HH, Rho SH, Eom SH, Park CS. J Biol Chem. 2006;281:38573–38581. doi: 10.1074/jbc.M604769200. [DOI] [PubMed] [Google Scholar]
- 31.Pico A. New York: Rockefeller University; 2003. PhD thesis. [Google Scholar]
- 32.Roosild TP, Le KT, Choe S. Trends Biochem Sci. 2004;29:39–45. doi: 10.1016/j.tibs.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 33.Fodor AA, Aldrich RW. J Gen Physiol. 2006;127:755–766. doi: 10.1085/jgp.200509419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qian X, Niu XW, Magleby KL. J Gen Physiol. 2006;128:389–404. doi: 10.1085/jgp.200609486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelly SM, Jess TJ, Price NC. Biochim Biophys Acta. 2005;1751:119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 36.Turk E, Gasymov OK, Lanza S, Horwitz J, Wright EM. Biochemistry. 2006;45:1470–1479. doi: 10.1021/bi052160z. [DOI] [PubMed] [Google Scholar]
- 37.Sreerama N, Woody RW. In: Methods in Enzymology. Brand L, Johnson ML, editors. Vol 383. San Diego: Elsevier; 2004. pp. 318–351. [Google Scholar]
- 38.Venyaminov SY, Klimtchuk ES, Bajzer Z, Craig TA. Anal Biochem. 2004;334:97–105. doi: 10.1016/j.ab.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 39.Sreerama N, Woody RW. Anal Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 40.Shi JY, Krishnmoorthy G, Yang YW, Hu L, Chaturvedi N, Harilal D, Qin J, Cui JM. Nature. 2002;418:876–880. doi: 10.1038/nature00941. [DOI] [PubMed] [Google Scholar]
- 41.Wallner M, Meera P, Ottolia M, Kaczorowski GL, Latorre R, Garcia ML, Stefani E, Toro L. Recept Channels. 1995;3:185–199. [PubMed] [Google Scholar]
- 42.Corbett EF, Oikawa K, Francois P, Tessier DC, Kay C, Bergeron JJ, Thomas DY, Krause KH, Michalak M. J Biol Chem. 1999;274:6203–6211. doi: 10.1074/jbc.274.10.6203. [DOI] [PubMed] [Google Scholar]
- 43.Paul I, Cui J, Maynard EL. Proc Natl Acad Sci USA. 2006;103:18475–18480. doi: 10.1073/pnas.0604150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomaselli S, Esposito V, Vangone P, van Nuland NAJ, Bonvin AM, Guerrini R, Tancredi T, Temussi PA, Picone D. ChemBioChem. 2006;7:257–267. doi: 10.1002/cbic.200500223. [DOI] [PubMed] [Google Scholar]
- 45.Gales L, Cortes L, Almeida C, Melo CV, Costa MD, Maciel P, Clark DT, Damas AM, Macedo-Ribeiro S. J Mol Biol. 2005;353:642–654. doi: 10.1016/j.jmb.2005.08.061. [DOI] [PubMed] [Google Scholar]
- 46.Zimmer J, Li W, Rapoport TA. J Mol Biol. 2006;364:259–265. doi: 10.1016/j.jmb.2006.08.044. [DOI] [PubMed] [Google Scholar]
- 47.Kuo MM, Baker KA, Wong L, Choe S. Proc Natl Acad Sci USA. 2007;104:2151–2156. doi: 10.1073/pnas.0609085104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang X, Solaro CR, Lingle CJ. J Gen Physiol. 2001;118:607–635. doi: 10.1085/jgp.118.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sali A, Blundell TL. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 51.Patton C, Thompson S, Epel D. Cell Calcium. 2004;35:427–431. doi: 10.1016/j.ceca.2003.10.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.