Abstract
We have developed a miniaturized 3D cell-culture array (the Data Analysis Toxicology Assay Chip or DataChip) for high-throughput toxicity screening of drug candidates and their cytochrome P450-generated metabolites. The DataChip consists of human cells encapsulated in collagen or alginate gels (as small as 20 nl) arrayed on a functionalized glass slide for spatially addressable screening against multiple compounds. A single DataChip containing 1,080 individual cell cultures, used in conjunction with the complementary human P450-containing microarray (the Metabolizing Enzyme Toxicology Assay Chip or MetaChip), simultaneously provided IC50 values for nine compounds and their metabolites from CYP1A2, CYP2D6, and CYP3A4 and a mixture of the three P450s designed to emulate the human liver. Similar responses were obtained with the DataChip and conventional 96-well plate assays, demonstrating that the near 2,000-fold miniaturization does not influence the cytotoxicity response. The DataChip may therefore enable toxicity analyses of drug candidates and their metabolites at throughputs compatible with the availability of compounds at early-stage drug discovery.
Keywords: cytochromes P450, in situ drug metabolism, in vitro cytotoxicity, on-chip cell encapsulation
Recent advances in genomics and proteomics coupled with sequencing of the human genome have led to a dramatic increase in the number of screenable drug targets (1). Combinatorial (2) and diversity-oriented synthesis (3) programs along with increased access to natural products and their structural scaffolds (4) have provided vast numbers of compounds to screen against these targets. However, there has not been a commensurate increase in the number of approved drugs (5). A major reason for this is the high failure rate of drug candidates because of factors that are not typically considered in the early stages of drug discovery, including poor ADME/tox (absorption, distribution, metabolism, excretion, and toxicology) profiles (6, 7). Thus, pharmaceutical companies are beginning to evaluate toxicity of drug candidates early in the discovery process to reduce the chances of late-stage failure (8).
Such early-stage toxicity information requires the development of accurate, reproducible, and predictive in vitro assays. In some cases, validation of in vitro assays has been achieved, such as in percutaneous absorption, skin corrosivity, and phototoxicity (9); however, these tests are limited and are not effective for acute organ-specific toxicity or metabolite toxicity. Moreover, in some European industries (e.g., cosmetics and chemicals), animal testing is being phased out entirely, thereby forcing companies to adopt new in vitro screens that effectively predict human toxicity (10–12). Thus, the need for concordance between in vitro assays and in vivo responses is becoming greater and more pressing, particularly in high throughput that would enable prioritization of compounds for further development involving animal testing of pharmaceutical candidates (13) or direct human testing of cosmetic ingredients.
High-throughput screening (HTS) assays for toxicity routinely use 96- or 384-well plates with 2D cell monolayer cultures (14, 15). The multiwell plate format, however, suffers from several limitations, including inefficient removal of reagents from the wells and the difficulty of subsequent washing of cell monolayers (16). These limitations are further compounded when high-throughput screening of cellular targets is coupled with metabolite synthesis, which requires addition of multiple reagents. More recently, to emulate native microenvironments, 3D cell cultures have been used extensively, particularly in tissue engineering applications, e.g., cell-seeded scaffolds (17) and patterned cocultures (18) as well as in directing cell fate and differentiation (19). Although miniaturization of 3D platforms has been performed for high-throughput applications (20, 21), relatively little effort has been directed toward using 3D cell cultures as screening tools for microscale toxicology assays (12, 22, 23). Herein, we address this technology gap by developing a miniaturized 3D cell-culture array (the Data Analysis Toxicology Assay Chip or DataChip) for high-throughput toxicity screening of drug candidates and their cytochrome P450-generated metabolites. Cell cultures have been performed in volumes as low as 20 nl, which can also be coupled to human P450 metabolite generation (24). As a result, toxicity analysis of drug candidates and their metabolites can be performed at speeds consistent with the large number of compounds present at early stages of drug discovery.
Results and Discussion
The DataChip is a microarray consisting of a spatially addressable pattern of cells encapsulated in a 3D hydrogel matrix, such as collagen or alginate, which supports cell growth at the microscale. Cells are seeded within the matrix material and are spotted onto a functionalized glass microscope slide by using a standard microarrayer. The DataChip can be incubated in culture medium to support cell growth over time scales relevant for toxicity analysis (up to 5 days).
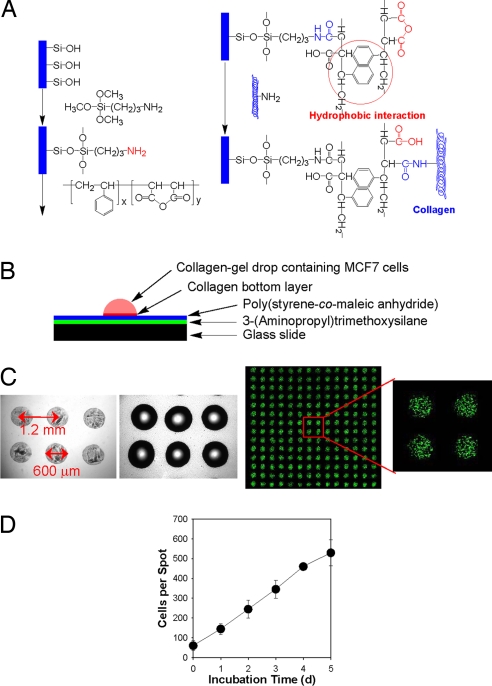
We initially prepared a DataChip by generating 560-spot microarrays of collagen bottom spots (30 nl each, 14 × 40 spots per slide) onto standard glass microscope slides (25 × 75 mm). The slides were first pretreated with 3-aminopropyltrimethoxysilane (APTMS), followed by poly(styrene-co-maleic anhydride) (PS-MA), which increases the hydrophobicity of the surface while providing a reactive functionality to covalently attach to collagen (Fig. 1A). The initial collagen spots were ≈50-μm thick, highly uniform in shape, and distributed uniformly on the slide. Collagen aliquots (60 nl) containing MCF7 cells (initial seeding density of 106 cells per milliliter, representing ≈60 cells per spot) were then spotted on top of the collagen bottom spots (Fig. 1B). The resulting spot diameters were 0.6 mm (close to that expected for hemispherical spots), and the center-to-center distance was 1.2 mm (Fig. 1C). The DataChip was incubated in cell culture medium for up to 5 d (Fig. 1D) and supported nearly linear growth, as has been observed previously in 3D mammalian cell cultures (23). The apparent doubling time ranged from 14 to 19 h, similar to that observed in cell monolayer culture (data not shown). By comparison, MCF7 cells grown in 60-μl 3D cultures in 96-well plates had a doubling time of ≈37 h. The observed difference in growth rates between the DataChip and 96-well plate cell cultures may be due to oxygen transport limitations that are more pronounced in the 1,000-fold-larger volumes of the well-plate cultures. Indeed, Brown et al. (25) found that oxygen transport limitations can be significant for cells cultured in 3D collagen matrices. For the smaller spots of the DataChip, oxygen transport is expected to be less of a limitation than in the well plate, and, as a result, the growth rate should be closer to that observed in monolayer cultures.
Fig. 1.
Preparation and characterization of the DataChip. (A) Chemical modification of a glass slide with 3-(aminopropyl) trimethoxysilane (APTMS) and poly(styrene-co-maleic anhydride) (PS-MA). (B) Construction of the collagen-gel spot containing MCF7 cells on the PS-MA-treated slide. (C) Presented left to right, microscopic photographs of collagen bottom layer (30 nl, 560 spots) on the PS-MA-treated slide, the collagen-gel spots containing MCF7 cells (60 nl, 560 spots) applied on top of the bottom layer, and live-cell stain of the array. (D) Growth of MCF7 cells in collagen-gel spots (60 nl, initial seeding of 1 × 106 cells per milliliter) based on live-cell staining at indicated incubation times.
The DataChip platform was evaluated by measuring the response of MCF7 cells to varying doses of model compounds (doxorubicin, DOX; 5-fluorouracil, 5-FU; and tamoxifen, TAM), all of which are known to be cytotoxic to MCF7 cells. To that end, 60 nl of each compound was dispensed at concentrations up to 1 mM onto a slide containing only the dried collagen islands in a 14 × 40 array (i.e., without the additional 3D collagen spots). This array was then stamped on top of the DataChip (seeded at 106 cells per milliliter), and the dual slide system was incubated for 6 h at 37°C. At the end of the stamping period, any excess compound remaining on the DataChip was washed out, and the cells were grown for an additional 3 d before cell viability was measured.
Dose–response cytotoxicity profiles of all three compounds for MCF7 cells are shown in supporting information (SI) Fig. 5. IC50 values calculated from the DataChip results are summarized in Table 1 along with values for MCF7 cells grown in both 2D monolayers and 3D collagen-gel cultures in conventional 96-well plates. The good agreement between IC50 values from the DataChip and 2D/3D cell cultures in 96-well plates indicates that the DataChip yields accurate cytotoxicity information. Furthermore, despite the ≈2,000-fold scale-down for the DataChip compared with more conventional plate assays, the accuracy of cytotoxicity data is not adversely affected. Finally, the stamping procedure is performed for 6 h; however, for more conventional growth-inhibition studies, the drug or drug candidate remains in contact with the cells for anywhere from 1 to 7 days (23, 26–29). Thus, the DataChip enables rapid cellular uptake that results in sufficient exposure to deliver a representative cytotoxic dose of drug.
Table 1.
Calculated IC50 values (μM) for the DataChip platform compared with 2D and 3D microtiter plates
| Drug | 96-well plate (2D) | 96-well plate (3D) | Collagen DataChip | Alginate DataChip | Alginate DataChip (Hep3B) | Literature values (2D) |
|---|---|---|---|---|---|---|
| Doxorubicin | 0.05 ± 0.00 | 0.09 ± 0.01 | 0.22 ± 0.04 | 0.19 ± 0.08 | 0.29 ± 0.09 | 0.01–55 (27, 28, 36) |
| 5-Fluorouracil | 51.5 ± 3.63 | 56.8 ± 5.09 | 62.2 ± 11.7 | 84.6 ± 18.7 | 66.1 ± 8.8 | 4.1–30.0 (27, 36) |
| Tamoxifen | 7.09 ± 0.24 | 18.9 ± 0.94 | 13.7 ± 2.32 | 13.3 ± 4.80 | 19.9 ± 7.70 | 1.0–8.0 (29, 37) |
Values are for MCF7 cells except where indicated.
Despite the ability of collagen to support human cell culture on the microscale, the material rapidly gels, thereby limiting the time that cell-seeded collagen remains in the liquid state. Moreover, at longer incubation times, the collagen matrix degraded, perhaps because of the presence of matrix metalloproteinases produced by cells in the culture, which resulted in the leaching of cells out from the 3D matrix (SI Fig. 6). To overcome these problems, we switched to an alginate gel matrix, which remained liquid in the absence of a bivalent metal ion and, therefore, allowed more control over DataChip preparation including the generation of spot volumes as small as 20 nl. Alginate is also inert to proteinase-catalyzed degradation.
A 10-nl mixture of poly-l-lysine (PLL) and BaCl2 was spotted onto the PS-MA-coated glass slides. Barium was used instead of the more common calcium because it is stable in the presence of phosphate (30), which is a common constituent of cell culture media. The positively charged PLL promoted attachment of the negatively charged polysaccharide constituent of alginate upon gelation. Formation of the 3D cell culture spots onto the DataChip was then achieved by arraying 20 nl of alginate suspension containing MCF7 or Hep3B cells (cell density of 6 × 106 cells per milliliter or 120 cells per spot). Gelation on the DataChip slide was immediate because of interaction of the alginate with Ba2+ ions. Growth of both MCF7 and Hep3B cells in alginate was similar to that in collagen (SI Fig. 6), but with no spot breakage. Moreover, dose responses to DOX, 5-FU, and TAM in alginate were similar to those obtained with collagen (Table 1). In addition, there was no difference in the growth rate as a function of seeding density. For example, MCF7 cells in 20-nl alginate spots on the DataChip showed little difference in growth rate when seeded at 3 × 106 cells per milliliter or 6 × 106 cells per milliliter (60 or 120 cells per spot, respectively). The doubling times were 26 h and 29 h, respectively. Thus, alginate provides a suitable alternative to collagen and allows for further scale down of the DataChip spots without problems of spot degradation.
Further miniaturization, and enhanced throughput, of the DataChip was achieved by arraying 20 × 54 (1,080) spots by using 20-nl alginate spot volumes. In addition, P450-catalyzed drug metabolism was added to the platform, thereby expanding the DataChip into a more general cytotoxicity screening system for drugs and drug metabolites. P450s catalyze the first-pass metabolism of xenobiotics, thereby generating one or more metabolites (31), some of which may be more or less toxic than the parent compound. The action of specific P450 isoforms can thus alter the cytotoxic dose responses of the DataChip.
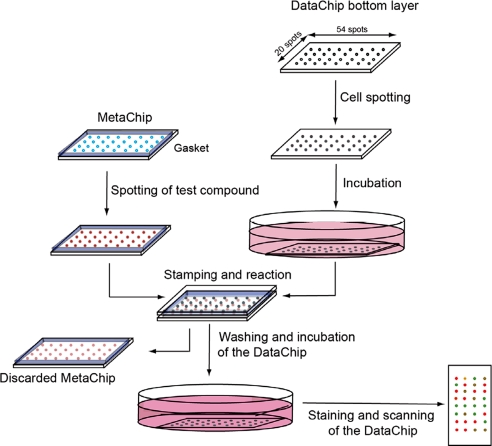
To introduce P450 catalysis into the DataChip platform, we modified the MetaChip (metabolizing enzyme toxicology assay chip), which we used previously to assess the influence of P450 metabolism on prodrug and protoxicant activation (24), to act as the complementary chip ofFig. 2. Specifically, a MetaChip consisting of 20 × 54 alginate spots was prepared; each spot contained a single human P450 isoform (CYP1A2, CYP2D6, or CYP3A4), a mixture of the three isoforms, or no P450 as a test-compound control. Addition of test compounds was carried out by overlay spotting onto the MetaChip, which was then stamped on top of a Hep3B DataChip. The three P450 isoforms were highly active on the alginate MetaChip (see SI Table 2); in all cases, the values of kcat/Km toward a fluorogenic substrate were within a factor of two of the solution-phase reactions.
Fig. 2.
Schematic of the DataChip platform for direct testing of compound toxicity or coupling with the MetaChip for evaluating toxicity of P450-generated metabolites.
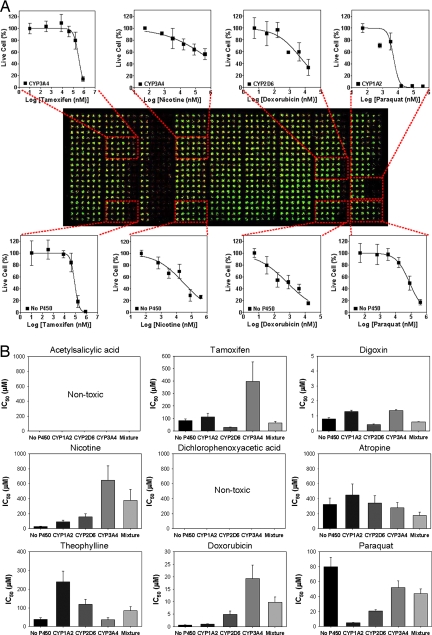
IC50 values for 27 compounds and their P450-generated metabolites were determined against human Hep3B cells by using three DataChips in combination with three MetaChips. The cells were noninduced and were expected to have essentially no intrinsic P450 activity; therefore, they provided a useful model to assess hepatotoxicity with the DataChip platform. Indeed, this was confirmed by growing Hep3B in the absence of inducers, disrupting the cell membranes by using sonication (32), and measuring the activity of CYP3A4, CYP1A2, and CYP2D6 by using standard fluorogenic substrates. No P450 activity was observed (SI Fig. 7). The test compounds were added to the MetaChip, which was then stamped onto the DataChip (Fig. 2) and incubated for 6 h. The DataChip was then removed, rinsed with growth medium, incubated in medium for 3 d, and stained for cell viability. As shown in Fig. 3A, a single DataChip yielded information on the dose–response of nine test compounds, each performed in one of 45 distinct regions of the DataChip. Each region consisted of a 4 × 6 miniarray that accommodated six different doses of a single test compound with four replicates. Representative dose-responses of four compounds are shown graphically in Fig. 3A, whereas the differential effect of P450 metabolism on nine compounds spotted on a single DataChip is presented in Fig. 3B.
Fig. 3.
Results from operation of the MetaChip/DataChip platform with Hep3B cells. (A) The 45 regions are divided into five rows of miniarrays. From top to bottom: equimolar mixture of CYP3A4, CYP1A2, and CYP2D6; CYP3A4, CYP2D6, CYP1A2, and no-P450. Dose responses corresponding to the fluorescent live-cell staining are shown for four of the nine compounds tested on the chip. (B) Comparison of IC50 values determined on the chip.
The DataChip platform is able to rapidly identify metabolic activation or deactivation of xenobiotics through the action of P450 isoforms (Fig. 3 and SI Fig. 8 and SI Table 3). Of the 27 compounds, 19 produced IC50 values <1 mM and two (digoxin and doxorubicin) produced IC50 values <1 μM. In both cases, the values determined on the DataChip were similar (within a factor of two) to literature values for related cell types (27, 33). Moreover, 13 compounds were reactive toward one or more of the P450 isoforms, as evidenced by activation or deactivation of the toxic response by the Hep3B cells (at least 5-fold vs. the parent compound). For example, as expected (34), CYP1A2 strongly activated acetaminophen, converting the compound into a cytotoxic metabolite (presumably the N-acetyl-p-benzoquinone imine). Similar activation was achieved for paraquat (methyl viologen) and isoniazide with CYP1A2 (Fig. 3 and SI Table 3).
Examination of the full DataChip results (SI Table 3) reveals that CYP1A2 deactivated lindane, orphenadrine, ketoconazole, theophylline, and cytarabine. CYP2D6 deactivated lindane, orphenadrine, nicotine, doxorubicin, methotrexate, and cytarabine. CYP3A4 activated acetaminophen and cyclophosphamide while deactivating orphenadrine, nicotine, tamoxifen, and doxorubicin. Thus, the DataChip–MetaChip combination was able to accurately predict the influence of P450-catalyzed first-pass metabolism on a diverse range of xenobiotics.
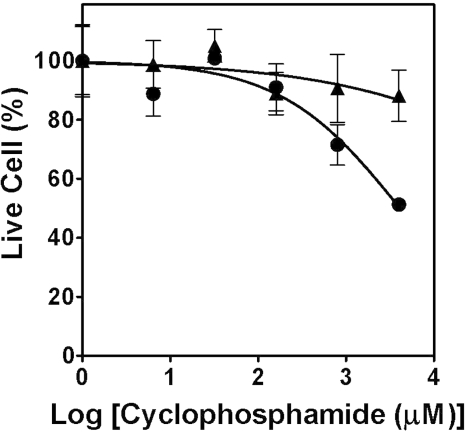
The MetaChip–DataChip platform can also provide information on potential drug interactions. For example, ketoconazole is a well known inhibitor of CYP3A4 (35). To test whether the presence of ketoconazole would suppress the action of CYP3A4 on the microarray platform, we examined the dose–response behavior of cyclophosphamide for Hep3B. As shown in Fig. 4, the addition of 3.2 μM ketoconazole effectively shut down the conversion of cyclophosphamide into the cytotoxic 4-hydroxycyclophosphamide metabolite. In conclusion, these results illustrate that the MetaChip–DataChip platform can evaluate compounds and their P450-generated metabolites individually or in combination at throughputs commensurate with the expanding influx of early-stage drug candidates.
Fig. 4.
Dose–response curves for Hep3B DataChip after stamping with CYP3A4 MetaChip exposed to cyclophosphamide: cyclophosphamide alone (●) and cyclophosphamide supplemented with 3.2 μM ketoconazole (▴).
Methods
Preparation of the DataChips.
Type I collagen from rat tail (3.9 mg/ml from BD Biosciences) was diluted with sterile phosphate-buffer saline (PBS from GIBCO) on ice to a final concentration of 2 mg/ml. The diluted collagen solution was spotted onto the PS-MA-treated slides (30-nl spot each, 14 × 40 spot array) by using a MicroSys 5100-4SQ microarray spotter equipped with an extended head (Cartesian Technologies). A suspension of cells in ice-cold collagen was prepared by mixing the cell suspension in 5% FBS- (GIBCO) supplemented DMEM from Sigma so that the final concentration of cells and collagen were 106 cells per milliliter and 1.3 mg/ml, respectively (see SI Methods for cell culturing techniques). After drying of the collagen bottom-layer spots on the slides for 10 min in a sterile Petri dish, 30 nl of the cell suspension in collagen was spotted atop each collagen spot within 10 min. The humidity in the microarrayer chamber was maintained at 90% to retard evaporation of water during spotting. The DataChip was quickly covered with a sterile glass slide separated by a 1-mm thick gasket (McMaster–Carr) to prevent drying of the gel spots. After 30 min of gelation, the DataChip was placed in a 100-mm-diameter Petri dish containing 16 ml of 5% FBS-supplemented DMEM and incubated in a 5% CO2 incubator (ThermoForma Electron) at 37°C for 18 h before exposing the cells to drug compounds through the stamping process.
The alginate-containing DataChip was prepared as follows. A PLL–barium mixture was prepared by mixing an equal volume of sterile PLL 0.01% (wt/vol) from Sigma) and barium chloride solution in distilled water (0.1 M; Sigma). A suspension of cells in low-viscosity alginate (Sigma) was prepared by mixing a cell suspension in 5% FBS-supplemented DMEM with alginate solution in distilled water so that the final concentration of cells and alginate were 6 × 106 cells per milliliter and 1% (wt/vol), respectively. After spotting and drying of 10 nl of PLL–barium mixture on the PS-MA-treated slide, 20 nl of the cell suspension in alginate was spotted atop each PLL-barium spot. After nearly instantaneous gelation, the alginate-based DataChip was placed in 5% FBS-supplemented DMEM and incubated in the CO2 incubator at 37°C for 18 h before in vitro toxicology assays.
Growth Inhibition Assay with Anticancer Drugs on the DataChip.
Three major P450 isoforms including CYP1A2, CYP2D6, and CYP3A4 and a mixture of the three P450s encapsulated in alginate-gel spots were prepared on the methyltrimethoxysilane (MTMOS)-coated slide (24). The resulting MetaChip was stored at −80°C for 1 day before use. The activities of three P450 isoforms encapsulated in alginate-gel matrix were tested by using blue fluorogenic substrates (see SI Methods). Cytotoxicity studies were performed by spotting 20 nl of test compound in distilled water (0–2 mM) onto the MetaChip spots, followed by partial drying. To these spots, 20-nl solutions of DMEM, containing 2 mM NADP+ and its regeneration system [67 mM glucose-6-phosphate and 8 units/ml glucose-6-phosphate dehydrogenase in 20 mM potassium phosphate buffer (pH 8)], were spotted. This was followed by stamping of the DataChip containing either MCF7 or Hep3B cells onto the MetaChip. After incubating the stamped MetaChip–DataChip combination for 6 h at 37°C, the DataChip was removed, rinsed for 2 h, incubated for 3 d, stained, and scanned by using the microarray scanner (see SI Methods for cell staining, scanning, and data analysis).
Supplementary Material
ACKNOWLEDGMENTS.
This work was supported by the National Institutes of Health (ES-012619) and the New York State Office of Science and Technology (NYSTAR).
Footnotes
Conflict of interest statement: D.S.C. and J.S.D. are cofounders of Solidus Biosciences.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708756105/DC1.
References
- 1.Dobson CM. Nature. 2004;432:824–826. doi: 10.1038/nature03192. [DOI] [PubMed] [Google Scholar]
- 2.Geysen HM, Schoenen F, Wagner D, Wagner R. Nat Rev Drug Discov. 2003;2:222–230. doi: 10.1038/nrd1035. [DOI] [PubMed] [Google Scholar]
- 3.Tan DS. Nat Chem Biol. 2005;1:74–84. doi: 10.1038/nchembio0705-74. [DOI] [PubMed] [Google Scholar]
- 4.Butler MS, Buss AD. Biochem Pharmacol. 2006;71:919–929. doi: 10.1016/j.bcp.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 5.Service RF. Science. 2004;303:1796–1799. doi: 10.1126/science.303.5665.1796. [DOI] [PubMed] [Google Scholar]
- 6.Lin J, Sahakian DC, de Morais SM, Xu JJ, Polzer RJ, Winter SM. Curr Top Med Chem. 2003;3:1125–1154. doi: 10.2174/1568026033452096. [DOI] [PubMed] [Google Scholar]
- 7.van der Waterbeemd H, Gifford E. Nat Rev Drug Discov. 2003;2:192–204. doi: 10.1038/nrd1032. [DOI] [PubMed] [Google Scholar]
- 8.Baillie TA. Chem Res Toxicol. 2006;19:889–893. doi: 10.1021/tx060062o. [DOI] [PubMed] [Google Scholar]
- 9.Bhogal N, Grindon C, Combes R, Balls M. Trends Biotechnol. 2005;23:299–307. doi: 10.1016/j.tibtech.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Piersma AH. Basic Clin Pharmacol Toxicol. 2006;98:427–431. doi: 10.1111/j.1742-7843.2006.pto_373.x. [DOI] [PubMed] [Google Scholar]
- 11.Elmore E, Sun C, Li HR, Wyatt GP, Buckmeier JA, Steele VE, Kelloff GJ, Redpath JL. Anticancer Res. 1999;19:909–918. [PubMed] [Google Scholar]
- 12.Bang H, Lim SH, Lee YK, Chung S, Chung C, Han D-C, Chang JK. J Micromech Microeng. 2004;14:1165–1170. [Google Scholar]
- 13.Arnaud CH. Chem Eng News. 2007;85:34–35. [Google Scholar]
- 14.Mueller H, Kassack MU, Wiese M. J Biomol Screen. 2004;9:506–515. doi: 10.1177/1087057104265386. [DOI] [PubMed] [Google Scholar]
- 15.Kasibhatla S, Gourdeau H, Meerovitch K, Drewe J, Reddy S, Qiu L, Zhang H, Bergeron F, Bouffard D, Yang Q, et al. Mol Cancer Ther. 2004;3:1365–1374. [PubMed] [Google Scholar]
- 16.Dunn DA, Feygin I. Drug Discov Today. 2000;5:S84–S91. doi: 10.1016/s1359-6446(00)00064-7. [DOI] [PubMed] [Google Scholar]
- 17.Tan W, Desai TA. J Biomed Mat Res A. 2005;72:146–160. doi: 10.1002/jbm.a.30182. [DOI] [PubMed] [Google Scholar]
- 18.Mapili G, Lu Y, Chen S, Roy K. J Biomed Mater Res B. 2005;75:414–424. doi: 10.1002/jbm.b.30325. [DOI] [PubMed] [Google Scholar]
- 19.Chin VI, Taupin P, Sanga S, Scheel J, Gage FH, Bhatia SN. Biotechnol Bioeng. 2004;88:399–415. doi: 10.1002/bit.20254. [DOI] [PubMed] [Google Scholar]
- 20.Hubbell JA. Nat Biotechnol. 2004;22:828–829. doi: 10.1038/nbt0704-828. [DOI] [PubMed] [Google Scholar]
- 21.Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Proc Natl Acad Sci USA. 2006;103:2480–2487. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bailey SN, Sabatini DM, Stockwell BR. Proc Natl Acad Sci USA. 2004;101:16144–16149. doi: 10.1073/pnas.0404425101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torisawa Y-S, Shiku H, Yasukawa T, Nishizawa M, Matsue T. Biomaterials. 2005;26:2165–2172. doi: 10.1016/j.biomaterials.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 24.Lee M-Y, Park CB, Dordick JS, Clark DS. Proc Natl Acad Sci USA. 2005;102:983–987. doi: 10.1073/pnas.0406755102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown DA, MacLellan WR, Laks H, Dunn JCY, Wu BM, Beygui RE. Biotechnol Bioeng. 2007;97:962–975. doi: 10.1002/bit.21295. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi H, Higashiyama M, Minamigawa K, Tanisaka K, Takano T, Yokouchi H, Kodama K, Hata T. Jpn J Cancer Res. 2001;92:203–210. doi: 10.1111/j.1349-7006.2001.tb01083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knuefermann C, Lu Y, Liu B, Jin W, Liang K, Wu L, Schmidt M, Mills GB, Mendelsohn J, Fan Z. Oncogene. 2003;22:3205–3212. doi: 10.1038/sj.onc.1206394. [DOI] [PubMed] [Google Scholar]
- 28.Li D, Jang SH, Kim J, Wientjes MG, Au JLS. Pharm Res. 2003;20:45–50. doi: 10.1023/a:1022242607418. [DOI] [PubMed] [Google Scholar]
- 29.Kim IY, Han SY, Moon A. J Tox Environ Health A. 2004;67:2025–2035. doi: 10.1080/15287390490514750. [DOI] [PubMed] [Google Scholar]
- 30.Peirone M, Ross CJD, Hortelano G, Brash JL, Chang PL. J Biomed Mat Res. 1998;42:587–596. doi: 10.1002/(sici)1097-4636(19981215)42:4<587::aid-jbm15>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 31.Guengerich FP. Chem Res Toxicol. 2001;14:611–650. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- 32.Yu LJ, Matias J, Scudiero DA, Hite KM, Monks A, Sausville EA, Waxman DJ. Drug Metab Dispos. 2001;29:304–312. [PubMed] [Google Scholar]
- 33.Jover R, Ponsoda X, Castell JV, Gomezlechon MJ. Toxicol in Vitro. 1992;6:47–52. doi: 10.1016/0887-2333(92)90084-5. [DOI] [PubMed] [Google Scholar]
- 34.Raucy JL, Lasker JM, Lieber CS, Black M. Arch Biochem Biophys. 1989;271:270–283. doi: 10.1016/0003-9861(89)90278-6. [DOI] [PubMed] [Google Scholar]
- 35.Zhang WJ, Ramamoorthy Y, Kilicarslan T, Nolte H, Tyndale RF, Sellers EM. Drug Metab Dispos. 2002;30:314–318. doi: 10.1124/dmd.30.3.314. [DOI] [PubMed] [Google Scholar]
- 36.Fujita T, Washio K, Takabatake D, Takahashi H, Yoshitomi S, Tsukuda K, Ishibe Y, Ogasawara Y, Doihara H, Shimizu N. Int J Cancer. 2005;117:670–682. doi: 10.1002/ijc.21063. [DOI] [PubMed] [Google Scholar]
- 37.Dhiman HK, Ray AR, Panda AK. Biomaterials. 2004;26:979–986. doi: 10.1016/j.biomaterials.2004.04.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.