Abstract
Antibodies that bind protein antigens are indispensable in biochemical research and modern medicine. However, knowledge of RNA-binding antibodies and their application in the ever-growing RNA field is lacking. Here we have developed a robust approach using a synthetic phage-display library to select specific antigen-binding fragments (Fabs) targeting a large functional RNA. We have solved the crystal structure of the first Fab–RNA complex at 1.95 Å. Capability in phasing and crystal contact formation suggests that the Fab provides a potentially valuable crystal chaperone for RNA. The crystal structure reveals that the Fab achieves specific RNA binding on a shallow surface with complementarity-determining region (CDR) sequence diversity, length variability, and main-chain conformational plasticity. The Fab–RNA interface also differs significantly from Fab–protein interfaces in amino acid composition and light-chain participation. These findings yield valuable insights for engineering of Fabs as RNA-binding modules and facilitate further development of Fabs as possible therapeutic drugs and biochemical tools to explore RNA biology.
Keywords: antigen-binding fragments, x-ray crystallography
Antibodies are integral components of the immune system and represent a rapidly growing sector of the biotechnology industry (1, 2). Clinically, antibodies serve as diagnostic markers for disease antigens and play increasingly important roles as therapeutic agents for a wide range of diseases (3). Antibodies also provide invaluable biomedical research tools, serving to define the components and functions of macromolecular complexes, to establish cellular distributions of proteins, and to facilitate structural analysis as chaperones for crystallization of membrane proteins (4–6). Hybridoma and other technologies have yielded antibodies against a vast array of specific antigens (2). An enormous body of literature documents the molecular details of antibody interactions with a variety of antigens, including proteins (7), polysaccharides (8), and small haptens (9). However, much less information (and, in particular, no structural information) exists for antibody–RNA interactions.
The relative absence of antibodies that bind RNA from the immunologic repository is striking, especially considering that recent genome-wide analyses of the metazoan transcriptome have revealed the presence of vast numbers of noncoding RNAs, including silencing RNAs, riboswitches, catalytic RNAs, and a multitude of other functional RNA moleucles (10, 11). A large number of these RNAs adopt complex three-dimensional architectures that frequently act in complex with proteins to mediate their biological function (12, 13). Nevertheless, with the exception of a handful of examples, mostly isolated from the sera of autoimmune patients (14–17), we know little about anti-RNA antibodies and their recognition of nucleic acids. This dearth of information reflects our inability to elicit antibodies against RNA by using traditional approaches. RNA appears to lack immunogenic potency (18), and its susceptibility to nuclease degradation prohibits direct immunization of animals, which precludes the use of hybridoma technology for large structured RNAs. A robust platform for obtaining antibodies against RNA would enable the investigation of RNA biology by using approaches analogous to those that have proven to be extremely effective for the study and therapeutic manipulation of protein–protein interactions.
Using a phage platform for the display of libraries of synthetic antigen-binding fragments (Fabs), we have established a general approach to obtain Fabs that bind to RNA. As an RNA antigen for proof-of-concept experiments, we chose the ΔC209 P4-P6 domain derived from the Tetrahymena group I intron, which folds into a well defined three-dimensional structure (19, 20). We demonstrate that Fabs targeting the ΔC209 P4-P6 domain bind with high affinity and specifically recognize the RNA tertiary structure. Crystallization of the Fab2-ΔC209 P4-P6 complex yielded a structure at 1.95-Å resolution, revealing the molecular interactions within an RNA–antibody interface and demonstrating the feasibility of antigen-binding fragments as chaperones for RNA crystallization.
Results
Selection of ΔC209 P4-P6-Binding Fabs.
The design of our synthetic naïve library for RNA-binding Fab selection employs a “reduced genetic code” approach (21, 22), in which the solvent-accessible regions of light-chain CDR-L3 and heavy-chain CDR-H1 and H2 are randomized with a binary degenerate codon that encodes equal proportions of Tyr and Ser. For heavy-chain CDR-H3, the CDR that usually contributes most to specific antigen binding (23), we replaced the seven residues with diversified loops of variable lengths (6–17 residues) in which each position was a mixture of 20% Tyr, 15% Ser, 15% Gly, and 50% Z (referred to herein as the YSG library). Z represents an equimolar mixture of all natural amino acids except for Cys, Tyr, Ser, and Gly. We chose this library type as the starting design for RNA targets because it has yielded high-affinity Fabs for a wide variety of protein targets (21, 22, 24).
Initially, we carried out the selection according to the procedure described by Laird-Offringa and Belasco (25) for the U1A RNA binding protein. However, we observed severe enrichment of streptavidin-binding phages after three rounds of selection, presumably reflecting the large exposed streptavidin surface used in target immobilization (Fig. 1A). We devised two strategies to circumvent this background binding problem. First, Fab phage libraries were preincubated with streptavidin beads to remove streptavidin binders. Second, after incubation of these prescreened phages with the immobilized RNA target, we used biotinylated RNase A to elute the RNA-binding phages selectively from the beads. Biotinylated RNase A allows for convenient removal with streptavidin beads. Moreover, to increase the specificity of the selected Fabs after the first round of selection, we added 20 equivalents of an Escherichia coli tRNA mixture as competitors during target phage binding.
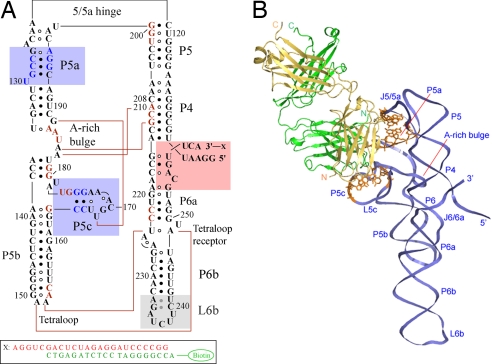
Fig. 1.
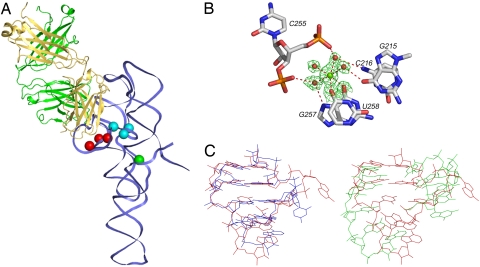
The RNA antigen: ΔC209 P4-P6 independently folding domain derived from Tetrahymena group I intron. (A) ΔC209 P4-P6 primary and secondary structure schematic. Residue numbering (102–261) corresponds to the Tetrahymena group I intron. Nucleotides in color within the secondary structure represent residues protected from hydroxyl radicals. Brown corresponds to protections arising from the ΔC209 P4-P6 tertiary structure; blue corresponds to additional protections induced by Fab2 binding. Blue boxes indicate the P5a and P5c arms involved in binding to Fab2. Loop L6b (residues 239–247; gray box) adopts a partially disordered conformation in the Fab2-ΔC209 P4-P6 crystals. Filled and open dots indicate canonical Watson–Crick and noncanonical pairs, respectively, as derived from the crystal structure of the Fab2-ΔC209 P4-P6 complex. Brown lines indicate long-range tertiary interactions. The pink box highlights the 5′ and 3′ terminal nucleotides, which adopt a conformation that is different from the previous ΔC209 P4-P6 structure [Protein Data Bank (PDB) ID code 1HR2] (20). The X moiety (see boxed area at the bottom) at the 3′ terminus represents the added RNA sequence (red), which, when hybridized to complimentary 19-nt 5′ biotinylated DNA oligonucleotide (green), allows immobilization on streptavidin-coated magnetic beads. (B) Ribbon representation of the crystal structure of Fab2-ΔC209 P4-P6 complex. Green and yellow ribbons indicate Fab2 heavy and light chains, respectively. The blue ribbon indicates the RNA backbone. Protein and RNA termini and RNA domains are labeled. Orange sticks show residues protected from hydroxyl radical footprinting upon Fab2 binding.
Using the strategies described above, we picked seven unique ΔC209 P4-P6-binding Fabs after three rounds of selection (Fig. 2). The overall complementarity-determining region (CDR) sequences of the seven clones show no obvious consensus. In the positions of CDR-L3, H1, and H2, which were diversified with a binomial combination of Tyr and Ser, we observed a preference for Ser (76% actual compared with 50% designed occupancy) over Tyr. Possibly, the importance of Tyr observed in the context of protein targets (22, 24, 26, 27) is diminished in selections against RNA targets. In CDR-H3, positively charged residues are slightly enriched. However, for each of the seven unique ΔC209 P4-P6-binding Fabs, there are no more than two positively charged residues present in CDR-H3, suggesting that these sequences do not simply act like the polyarginine/lysine peptides that often bind nucleic acids nonspecifically.
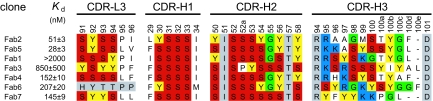
Fig. 2.
The dissociation constants (Kd) and CDR sequences of antigen-binding fragments selected against immobilized ΔC209 P4-P6. Numbering is according to the Kabat definition (37). Color scheme is as follows: gray, residues not randomized; yellow, Tyr; red, Ser; green, Gly; blue, positively charged residues Arg and Lys. Kd was determined by nitrocellulose filter binding. CDR-L3 of Fab6 has the same sequence as the parental Fab template, which is a result of the stop template not being used for CDR-L3 during construction of the library (47).
Affinity and Specificity of the ΔC209 P4-P6-Binding Fabs.
The selected Fabs were expressed as soluble proteins and purified (28). Two Fabs, Fab2 and Fab5, bind ΔC209 P4-P6 (without the 3′ tail sequence used for immobilization, Fig. 1A) with a Kd of 51 and 28 nM, respectively, as determined by nitrocellulose filter binding (10 mM MgCl2 and 150 mM NaCl; Fig. 2). Binding constants obtained by using independent methods [competitive phage ELISA (28), surface plasmon resonance, and hydroxyl radical footprinting] were within 2-fold of each other [supporting information (SI) Table 1]. Gel filtration experiments on complexes formed by Fab2 and ΔC209 P4-P6 in various ratios established that Fab2 and ΔC209 P4-P6 form a 1:1 complex (SI Fig. 7).
We then characterized the binding specificity of Fab2 and Fab5 by using three different RNAs (Fig. 3 A and B). The Fabs exhibited no detectable affinity for an unrelated RNA oligonucleotide derived from the sarcin/ricin loop of ribosomal RNA (29). Disruption of the P4-P6 fold by mutation of J5/5a (BP P4-P6), so as to form a duplex contiguous with P5 and P5a (30), reduced binding of Fab2 and Fab5 by at least an order of magnitude. The tertiary folding of ΔC209 P4-P6 requires magnesium, and Fab binding strongly depended on magnesium concentration. As the magnesium concentration decreased, binding of Fab2 and Fab5 to ΔC209 P4-P6 was attenuated and became undetectable in the absence of magnesium (Fig. 3 C and D). Taken together, these results show that Fabs recognize specifically the tertiary structure of ΔC209 P4-P6.
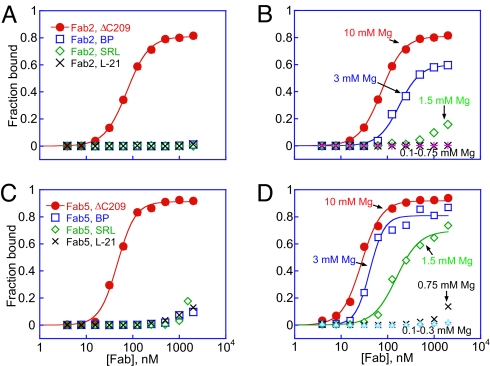
Fig. 3.
Fab2 and Fab5 bind specifically to the ΔC209 P4-P6 tertiary architecture. (A and B) Graphic representation of fraction RNA bound vs. Fab2 (A) or Fab5 (B) concentration. Fraction bound reflects the 5′ 32P-labeled RNA retained on a nitrocellulose filter (SI Methods). Binding reactions (23°C) contained 10 mM MgCl2, 150 mM NaCl, 10 mM phosphate buffer (pH 7.3), and ≈0.3 nM RNA. BP, a P4-P6 mutant designed to form Watson–Crick base pairs in the J4/5 hinge region, rendering P4 and P5 contiguous; SRL, an RNA oligonucleotide containing the sequence of the sarcin/ricin loop derived from the large ribosomal subunit RNA; L-21, a derivative of the Tetrahymena group I intron that lacks the first 21 nucleotides. Fab2- and Fab5-binding curves with BP, SRL, and L-21. (C and D) Binding of ΔC209 P4-P6 to Fab2 (C) and to Fab5 (D) depends on Mg2+ concentration. Binding curves were fit to the following equation: fraction bound = fmax × [Fab]n/([Fab]n + Kdn), where fmax is the maximum bound fraction at saturating Fab, Kd is the binding affinity, and n is the Hill coefficient, which is generally close to 1.
Strikingly, the Fabs exhibited no detectable affinity (even at concentrations as high as 2 μM) for the Tetrahymena group I intron (L-21), which contains the P4-P6 sequence. The marked discrimination against the intron does not arise from the presence of C209 because Fab2 binds wild-type P4-P6 and ΔC209 P4-P6 with similar affinities (SI Table 2). At lower salt concentration (50 mM NaCl and 10 mM MgCl2), hydroxyl radical footprinting shows that Fab2 retained some binding to BP P4-P6 but still did not bind the L-21 intron (SI Table 2). The inability of Fab2 to bind L-21 may reflect steric interference from peripheral element loop L2 according to Michel and Westhof's (31) model.
Crystal Structure of the Fab2-ΔC209 P4-P6 Complex.
We successfully crystallized the complex of Fab2 with ΔC209 P4-P6 (Fig. 1B), and the crystal structure was determined by molecular replacement (MR) at 1.95 Å (SI Table 3), an improvement over the previous ΔC209 P4-P6 structure at 2.25 Å (20). Although the final structure of the complex was solved by MR using both Fab and RNA models, we found that the Fab MR solution was sufficient to build the RNA model from the resulting electron density maps, demonstrating good phasing capacity of the Fab chaperone (SI Fig. 8). Comparing the Fab2 scaffold (excluding the six CDRs) and the corresponding part of the parent (32) Fab-4D5 gave a Cα rmsd value of 0.7 Å, indicating that the Fab scaffold remains essentially unchanged. Likewise, the RNA portion deviates from the previously determined free ΔC209 P4-P6 with a rmsd of 1.4 Å for C1′ atoms (SI Fig. 9), excluding the inherently flexible L6b, the internal loop J6/6a [which interacts with helix P3 in the full-length Tetrahymena group I intron (33)] and the end sequences of the RNA, which participate in very different crystal packing interactions compared with the original ΔC209 P4-P6 structure (20). This rmsd value is comparable with 1.2 Å for the two molecules in the asymmetric unit of the ΔC209 P4-P6 structure (20). Therefore, we conclude that the binding of Fab2 does not significantly alter the ΔC209 P4-P6 structure.
As an independent test, we probed the tertiary architecture of ΔC209 P4-P6 complexed with Fab2 in solution by hydroxyl radical footprinting and compared the results to published protection patterns of ΔC209 P4-P6 alone (30, 34). In the presence of Fab2, ΔC209 P4-P6 retains its native protections, suggesting that this RNA maintains its overall structure in the presence of Fab2 in solution and in the crystal. We observe four additional regions of protection in the presence of Fab2: nucleotides 194–196, 174–175, 165–166, and 127–130 (Fig. 1B and SI Fig. 10). These Fab-induced protection sites match the Fab epitope of ΔC209 P4-P6 as revealed by the crystal structure (Fig. 1B).
Binding Interface Between ΔC209 P4-P6 and Fab2.
Fab2 binds to ΔC209 P4-P6 with an exceptionally large buried surface area (Fig. 4A), 1,316 Å2 (on the Fab side alone), which is almost twice as large as that of most Fab–protein antigen interfaces (777 ± 135 Å2) (35). Consistent with the extensive surface burial, residues from all six CDRs and some scaffold residues form the interface (Fig. 4 and SI Fig. 11). Superposition of the variable domains reveals significant differences in the main chain conformations of CDR-H3, L2 and H2 compared with the parent (32) Fab-4D5, illustrating the conformational plasticity of the hypervariable regions (Fig. 4B). Within the Fab–RNA interface, CDR-H3 and L2 make direct contact with both P5a and P5c stems and contribute most to the buried surface area: 34% and 29%, respectively (Fig. 4C). This observation suggests that CDR-H3 and L2 likely play significant roles in the recognition of ΔC209 P4-P6, even though L2 was not diversified in the library.
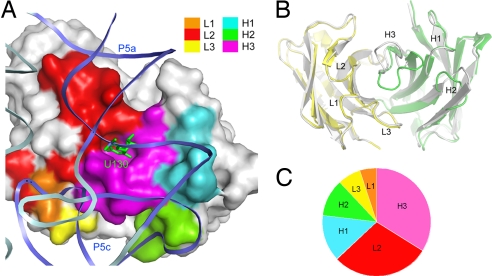
Fig. 4.
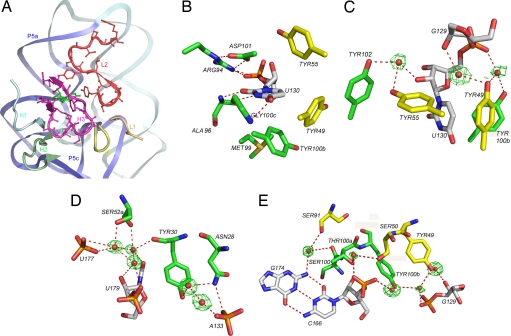
CDR contribution in Fab2-ΔC209 P4-P6 binding. (A) Molecular surface of the variable domain of Fab2 in the Fab2-ΔC209 P4-P6 structure. Color indicates CDR loop regions in the binding interface as defined by the color scheme legend. Ribbons represent the RNA backbone, with P5a and P5c colored blue and the rest colored light cyan. The green sticks protruding from the RNA ribbon represent the bulged U130. (B) Superposition of the variable domains of Fab2 (light chain, yellow; heavy chain, green) and humanized Fab-4D5 (1FVD, white). Note the large differences in CDR-H3, L2, and H2. (C) Pie chart showing relative contribution of each CDR to the total buried Fab2 surface area (1,316 Å2).
CDR-H3 resides in the center of the P5a/P5c helical interface at the three-helix junction (Fig. 5A). Residues Gly-98, Ser-100, and Thr100a interact with the wide and shallow minor groove of P5c via direct H bonds, and Tyr-102 contacts P5a by H bonding to U131 in the minor groove. Several CDR-H3 residues form a pocket (Fig. 5B) that encloses U130, the bulged uridine in P5a, and they use a variety of molecular strategies to interact with U130. Arg-94, Ala-96, Asp-101, and Gly100c form direct H bonds to the uracil base and sugar ring, whereas the side chains of Ala-96 and Met-99 embrace the nucleotide with nonpolar contacts. Tyr100b stacks perpendicularly with the base, and Arg-94 forms an N-O bridge-type of ion pair with the adjacent phosphate (36). Interestingly, although Arg-95 was selected from the library, it makes no interaction with the RNA in the structure; instead, its side chain points away from the RNA and makes hydrogen bonding interactions with the backbone carbonyl groups of Tyr100b and Met-99, possibly stabilizing the CDR-H3 loop conformation.
Fig. 5.
Fab2-ΔC209 P4-P6-binding interactions. (A) CDR-H3 (magenta) and L2 (red) dominate the Fab–RNA interface, whereas other CDRs occupy the peripheral region. Color scheme is the same as in Fig. 4A. (B) CDR-H3 and L2 form a binding pocket for the bulged uridine. On one side, heavy chain residues Arg-94, Ala-96, Asp-101, and Gly100c form direct hydrogen bonds (shown as red dashed lines) to the base, sugar, and adjacent phosphate of U130. On the other side, heavy-chain residues Met-99 and Tyr100b and light-chain residues Tyr-49 and Tyr-55 make nonpolar contacts. The side chain of Met-103 has dual occupancy on two conformations. Green, yellow, and white represent carbon atoms from residues in the heavy chain, light chain, and RNA, respectively. (C) Water-mediated hydrogen-bonding network involving four tyrosine residues on the nonpolar side of the U13-binding pocket. Red spheres represent water molecules, with corresponding electron density maps (green) contoured at 1.2 σ. (D and E) Water-mediated H bonding networks form a bridge between P5a and the three-helix junction (D) and between P5a and P5c (E).
CDR-L2 loop residues (defined as residues 50–56) (37) and the flanking framework residues interact extensively with RNA, mainly in the P5a minor groove. Residues Ser-56, Gly-57, and Ser-60 form direct H bonds to the 3′ strand of P5a, across from U130 near the hinge region (Fig. 5A). A possible long-range ion pair between Arg-61 and the phosphate moiety of C197 may stabilize this interaction. Direct H bonding between Ser-50 and the U167 phosphate links CDR-L2 to P5c, making L2 another CDR that binds both helical stems. Tyr-49 and Tyr-55 stack perpendicularly with U130 from the nonpolar side to seal the U130-binding pocket generated by CDR-H3 (Fig. 5B). Three water molecules organized by the four tyrosine residues surrounding U130 interact with the sugar–phosphate moiety, which may further strengthen the RNA-Fab interaction at the nonpolar interface of the U130 binding pocket (Fig. 5C).
The remaining four CDRs (H1, H2, L1, and L3) reside in the peripheral region of the Fab-RNA interface and contribute significantly less to surface burial (Figs. 4 and 5A). CDR-H1 binds to the minor groove of P5a on the same strand as U130, and H2 binds to the base of the three-helix junction. CDR-L3 and L1 bind to L5c and contribute the least to the buried surface area (Fig. 4C). Within the interface, more than a dozen water molecules mediate interactions between Fab2 and ΔC209 P4-P6, including two sets of hydrogen-bonding networks. Each utilizes five well defined water molecules in addition to several Fab residues, bridging either between P5a and the three-helix junction (Fig. 5D) or between P5a and P5c (Fig. 5E). Similar to the binding of CDR-H3 and CDR-L2, these interactions link RNA domains that are remote from one another in the secondary structure.
Changes in Magnesium Ion Coordination.
Within the previous ΔC209 P4-P6 crystal structure, six magnesium ions make innersphere coordination to the RNA (designated m1/2, m3/4, m5/6, m7/8, m9/10, and m11/12) (20). In the crystal structure of the Fab–RNA complex, we observe only three of these magnesium ions; two form the binuclear metal center in the A rich bulge (m1/2 and m3/4), and the third mediates interactions between the A rich bulge and P5c (m11/12). We do not observe the magnesium ions in P5c (m7/8 and m9/10) or at the base of the three-helix junction (m5/6) (20). The observed metal ions in the complex are buried in the ΔC209 P4-P6 core, whereas the ions apparently missing from the complex are partially solvent exposed and sit closer to the Fab epitope in the ΔC209 P4-P6 structure. It is possible that Fab2 supplants the role of the missing metal ions.
In addition to the three innersphere coordinated magnesium ions identified in the ΔC209 P4-P6 core, we observe a new innersphere coordinated magnesium ion near the 3′ terminus of the RNA (Fig. 6B). The presence of this magnesium ion accompanies restoration of the phylogenetically conserved G215-U258 wobble pair and C216-G257 Watson–Crick pair in the P6 helix. In the wild-type (19) and ΔC209 P4-P6 (20) structures, crystal contacts between L5c and J6/6a apparently preclude formation of these base pairs. In this region of the RNA, the Fab2-ΔC209 P4-P6 structure superposes with the Tetrahymena group I intron structure (33) better than does the original ΔC209 P4-P6 structure (20) (Fig. 6C).
Fig. 6.
Differences in innersphere magnesium ion coordination and a local conformation compared with the previous ΔC209 P4-P6 structure. (A) Innersphere coordinated magnesium ions in the ΔC209 P4-P6 core (cyan spheres) and 3′ end region (green sphere). Red spheres indicate missing metal ions observed in ΔC209 P4-P6, but not in the complex with Fab2. (B) A magnesium ion associates with P6 and J6/6a via innersphere coordination to U258 and G257 and outersphere coordination to C255, C216, G215, and G257. (C) Superposition of P6 and J6/6a from the full-length group I intron structure (red) with P6 and J6/6a from the complex (blue) or ΔC209 P4-P6 structure (green).
Statistical Comparison of Fab2-ΔC209 P4-P6 Binding Interface with Other Protein–RNA Interfaces.
The crystal structure of the Fab2-ΔC209 P4-P6 complex provides the first opportunity to study an antibody–RNA interface in detail. Fab residues that participate in direct or water-mediated hydrogen bonds with RNA occur in the following relative frequency: Ser > Tyr > Gly > Arg > Ala ≈ Thr ≈ Asn. The preference for Ser, Tyr, and Gly presumably reflects the enrichment prescribed by the library design. Beyond that, the relative frequency of amino acids does not differ significantly from the statistical amino acid composition found within other protein–RNA interfaces (38–40) (SI Methods). For the Fab2-ΔC209 P4-P6 complex, interactions with nucleobase edges and ribose atoms comprise 40% and 32% of the direct contacts, respectively, which is similar to the analysis by Lejeune et al. (40) of 49 RNA-binding proteins (35% and 43%). Main-chain atoms contribute 37% of the interactions on the protein side of the interface, which is similar to the analysis of Treger and Westhof (32%) (38). These comparisons suggest that, at the atomic level, the Fab–RNA interface resembles other protein–RNA binding interfaces.
Crystal Packing.
The Fab2-ΔC209 P4-P6 complex crystallizes in space group C2, and the adjacent molecules form homodimers across the 2-fold crystallographic axis, through both Fab–Fab and RNA–RNA interactions. The Fab–Fab interaction extends the antiparallel β-sheet of the heavy-chain constant domain (SI Fig. 12A). In the RNA–RNA interaction, the 5′ end of one molecule forms four intermolecular Watson–Crick base pairs with two nucleotides each from both the 5′ and 3′ ends of the adjacent RNA (SI Fig. 12B). Packing of this RNA homodimeric complex in the crystal lattice involves mainly Fab–RNA crystal contacts (SI Fig. 12C). Crystal contacts bury a total surface area of 3,641 Å2, 61% of which involves Fab2. Including the Fab paratope buried within the complex, Fab2 provides a total buried surface area of 3,538 Å2, which is significantly larger than the 1,942 Å2 for U1A-RBD in the HDV-U1A crystal structure (41, 42). Thus, as a crystallization chaperone, Fab2 plays an important role in crystal packing via interactions with both protein and RNA.
Discussion
Selection of Anti-RNA Fabs by Phage Display.
This work demonstrates the first successful selection of anti-RNA antibody fragments from a synthetic naïve library. Braun et al. (43) obtained antipolyinosine antibodies through immunization of mice. Beyond that, the only known examples of anti-RNA antibodies occur pathologically in autoimmune diseases such as systemic lupus erythematosus (14, 44) and rheumatoid arthritis (15). The successful isolation of synthetic Fabs that bind tightly to ΔC209 P4-P6 and to a diverse panel of other structured RNAs (J.-D.Y., Y. Koldobskaya, J. Min, and J.A.P., unpublished work) suggests that the paucity of available anti-RNA antibodies probably reflects nucleolytic instability of RNA during in vivo immunization, rather than some fundamental limitation in the ability of natural antibody repertoires to bind RNA. In vitro selection using phage display allows for fine adjustment of solution conditions so that the structural integrity of an RNA antigen can be maintained. Moreover, our selected Fabs recognize antigens in a specific manner, probably owing to the inclusion of negative selection with tRNA, which removes Fabs that interact nonspecifically with RNA.
The Fab as an RNA-Binding Protein Scaffold.
Despite statistical similarities at the atomic level (see above), Fab2 recognizes RNA in a manner that differs topologically from that of other RNA-binding proteins. RNA-binding proteins often consist of several modular domains that generally have topologically rugged surfaces and engulf regions of the RNA or penetrate into the grooves. In Fab2, the CDR loops form a relatively smooth, shallow surface that recognizes numerous structural features of ΔC209 P4-P6. The CDR-H3 and L2 loops span the binding interface to interact with the wide and flat minor grooves of two adjacent helices (P5a and P5c) at the three-helix junction. Together, these loops also form a binding pocket for the bulged uridine residue (U130) that protrudes from P5a. The remaining CDR loops give the shallow Fab surface a slightly convex curvature, contributing 37% to the buried surface area by recognizing both the ribose–phosphate backbone moiety and the unpaired nucleobases in the single-stranded regions. Possibly, these interactions enhance the binding specificity through both shape complementarity and specific nucleobase recognition.
With the exception of unstructured peptides or arginine-rich motifs, a given type of RNA-binding module usually binds to a distinct class of RNA structures. Considering the possible diversity in sequence composition, length, and main-chain conformation of the CDRs, Fabs may provide a highly versatile scaffold for binding to a wide range of RNAs. As a preliminary demonstration, we have obtained >100 Fabs in selections against 10 structurally diverse RNA targets (J.-D.Y., Y. Koldobskaya, J. Min, and J.A.P., unpublished work).
Fab2-ΔC209 P4-P6-Binding Interface Suggests Strategies to Improve the Library Design.
The Fab library used in this work successfully produced highly specific and tightly binding Fabs against ΔC209 P4-P6. However, the design of this library is based on bioinformatic analysis of antibodies binding to protein antigens (24). The CDR sequences of our ΔC209 P4-P6-binding Fabs and the Fab2–RNA interface suggest several strategies to improve the library design for RNA targets (1). With respect to CDR-H3, all of our selected clones showed an enrichment for positively charged amino acids, suggesting that increasing Arg content in CDR-H3 may yield a library better suited for selection targeting RNAs (2). The YSG library used herein contains no diversity in CDR-L1 and L2; yet Fab2 uses light-chain CDRs for ΔC209 P4-P6 recognition, especially CDR-L2, which seldom participates in protein antigen recognition. Thus, expanding synthetic diversity to include the L1 and L2 CDR loops might improve the RNA-binding repertoire further (3). With respect to the binary YS code used for CDR-L3, H1 and H2, we find that Ser and Tyr residues contribute 29% and 28%, respectively, to the buried surface area (SI Fig. 13). Twelve different amino acids contribute to the remaining 43%, with Gly and Arg residues contributing most among them. In contrast, Fabs selected against protein antigens from YS-enriched libraries (22, 26, 27) reveal a dominant role for tyrosine residues at the antigen interface, especially in the case of an anti-hVEGF Fab-D1 selected from the same library as used in our selections (24). In the Fab-D1 paratope, Tyr residues contribute 69% of the buried surface area, Ser residues contribute only an additional 18%, and four other residue types contribute the remaining 13%. These striking differences between the protein and RNA-binding paratopes suggest that the YS binary code may not be optimal for RNA antigens, and the inclusion of additional diversity may produce a more robust response against RNA antigens.
Fab Chaperones for the Crystallization of Structured RNA.
In this study we have established that an anti-RNA Fab can act as a chaperone to assist crystallization of a large folded RNA, enabling structure determination of the Fab2-ΔC209 P4-P6 complex at 1.95-Å resolution. Fab2 participates extensively in the crystal contacts, contributing 61% of the total surface area buried by crystal lattice interactions, within the range observed for the U1A protein chaperone-mediated lattice interactions (41, 45, 46) (HDV-U1A, 85%; hairpin ribozyme-U1A, 59%; Azoarcus-U1A, 28%). In contrast to the U1A chaperone, Fabs circumvent the need to engineer protein-binding sequences into the RNA. Additionally, the 4-fold-larger size of Fab compared with U1A may, in certain cases, have advantages in facilitating RNA crystallization and providing better phasing information (SI Fig. 8). Although the generality of the approach remains to be determined, this Fab-RNA cocrystal establishes proof-of-concept for using Fabs as RNA crystallization chaperones.
Concluding Remarks.
The synthetic Fab repertoire used herein has produced highly specific and tight-binding antibody fragments to an elaborately structured RNA. The ability to obtain antibodies that bind RNA targets with high affinity and specificity enables many new clinical and experimental applications in RNA structural, molecular, and cellular biology. Herein, we have demonstrated one such application for RNA crystallization, but others include, for example, the use of anti-RNA antibodies to identify unique RNA markers that certain disease states might carry. Direct immunoprecipitation and visualization of RNA for cellular biological investigations also becomes possible, either by using antibodies selected directly against the RNA of interest or by using an RNA antigen such as P4-P6 as an epitope tag.
Methods
Fab Selection and Production.
The Fab phage display library and general library manipulation methods have been described in refs. 24 and 47. Selection was done at room temperature by using the magnetic beads method (25). In the first round, 0.5 nmol of biotinylated ΔC209 P4-P6 (AGGUCGACUCUAGAGGAUCCCCGG was added to the 3′ end of ΔC209 P4-P6 and annealed to the 5′ biotinylated DNA oligonucleotide, 5′-ACCGGGGATCCTCTAGAGTC-3′) was immobilized on magnetic beads (Promega) and incubated with ≈1012-13 cfu of phages for 15 min in 1 ml of buffer A [PBS (8 mM Na2HPO4, 1.5 mM KH2PO4, 137 mM NaCl, and 3 mM KCl), 0.05% Tween 20, 2.5 mM EDTA, 12.5 mM MgCl2, and 5% glycerol, pH 7.2] supplemented with 0.1 mg/ml BSA, 0.1 mg/ml streptavidin, and 1 unit/μl RNase inhibitor (Promega). The solution was then removed, and the beads were washed twice with buffer A. In the subsequent rounds, purified phage pools were first incubated with streptavidin beads in buffer A for 30 min, and the supernatant was used in the subsequent selection on a KingFisher magnetic particle processor (Thermo Electron Corporation). Phages (≈1010-11 cfu) were incubated for 15 min with 50 nM biotinylated ΔC209 P4-P6 in 100 μl of buffer A, supplemented with 0.1 mg/ml BSA, 1 unit/μl RNase inhibitor, and 50 μg/ml E. coli tRNA mixture. Streptavidin magnetic beads were then added to the solution for 15 min to allow the capture of the biotinylated ΔC209 P4-P6 together with the bound phages. The beads were then blocked with 50 μM biotin, washed five times with buffer A, and eluted in 50 μl of elution buffer (PBS, 5% glycerol, and 1 μg/ml biotinylated RNase A). The biotinylated RNase A was removed from the resulting phage library by incubation with streptavidin beads. After each round of selection, recovered phages were amplified as described in ref. 47.
After selection, individual clones were grown in a 96-well format in 500 μl of 2YT broth supplemented with ampicillin and M13-KO7 helper phage. The culture supernatants were used in phage ELISA to detect positive clones that bound to antigen-coated plates, but not to control plates (47). Positive clones were sequenced. Competitive phage ELISA was used to estimate the binding affinities of the unique clones, and Fab proteins of interest were expressed and purified as described in ref. 28.
Filter binding analysis and Hydroxyl radical footprinting are described in SI Methods.
Crystallization.
ΔC209 P4-P6 (0.194 mM) was incubated at 60°C for 10 min in 10 mM Tris·HCl, pH 7.5, 25 mM MgCl2, and 50 mM NaCl and was slowly cooled to room temperature in ≈1.5 h. Fab2 protein (1 equivalent) and spermine-4HCl (0.5 mM final concentration) were added to ΔC209 P4-P6 while maintaining the concentration of MgCl2 and NaCl. The mixture was incubated at room temperature for 30 min and cooled to 4°C over a period of ≈0.5 h. Crystallization was set up by using the hanging-drop vapor diffusion method: two volumes of the complex were combined with 1 vol of reservoir buffer (34% 2-methyl-2,4-pentanediol, 0.1 M sodium citrate, pH 5.9, and 0.2 M NH4OAc) and equilibrated against 0.5 ml of reservoir buffer at 4°C. The crystal clusters appeared in 3 days and grew to their maximum size (≈0.1–0.15 mm) in ≈14 days. For data collection, the crystal clusters were broken into thin plates by using crystal manipulation tools (Hampton Research) and were flash-frozen in liquid nitrogen by using CryoLoops (Hampton Research) directly from the mother liquid. Data collection and structure determination are described in SI Methods.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Barbara Golden and the members of the J.A.P. Laboratory for helpful discussions and the staffs of the Structural Biology Center Collaborative Access Team, the Southeast Regional Collaborative Access Team, and the General Medicine and Cancer Institutes Collaborative Access Team for help with data collection. This work was supported by grants from the Howard Hughes Medical Institute (to J.A.P.) and from the U.S. National Institutes of Health (to A.A.K. and S. K.) and by the University of Chicago Cancer Research Center.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2R8S).
This article contains supporting information online at www.pnas.org/cgi/content/full/0709082105/DC1.
References
- 1.Schrama D, Reisfeld RA, Becker JC. Nat Rev Drug Discov. 2006;5:147–159. doi: 10.1038/nrd1957. [DOI] [PubMed] [Google Scholar]
- 2.Carter PJ. Nat Rev Immunol. 2006;6:343–357. doi: 10.1038/nri1837. [DOI] [PubMed] [Google Scholar]
- 3.Arrowhead Publishers. Monoclonal Antibody Therapies: Entering A New Competitive Era. Minneapolis: Arrowhead Publishers; 2004. [Google Scholar]
- 4.Nielsen UB, Geierstanger BH. J Immunol Methods. 2004;290:107–120. doi: 10.1016/j.jim.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 5.Hunte C, Michel H. Curr Opin Struct Biol. 2002;12:503–508. doi: 10.1016/s0959-440x(02)00354-8. [DOI] [PubMed] [Google Scholar]
- 6.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 7.Davies DR, Cohen GH. Proc Natl Acad Sci USA. 1996;93:7–12. doi: 10.1073/pnas.93.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucas AH, Reason DC. Immunol Rev. 1999;171:89–104. doi: 10.1111/j.1600-065x.1999.tb01343.x. [DOI] [PubMed] [Google Scholar]
- 9.Livesay DR, Subramaniam S. Protein Eng Des Sel. 2004;17:463–472. doi: 10.1093/protein/gzh058. [DOI] [PubMed] [Google Scholar]
- 10.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 11.Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 12.Winkler WC, Breaker RR. Annu Rev Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 13.Doudna JA, Cech TR. Nature. 2002;418:222–228. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]
- 14.van Venrooij WJ, Hoet R, Castrop J, Hageman B, Mattaj IW, van de Putte LB. J Clin Invest. 1990;86:2154–2160. doi: 10.1172/JCI114954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalmakoff J, Maskill WJ, Thongkrajai P, Palmer DG. Aust N Z J Med. 1981;11:173–178. doi: 10.1111/j.1445-5994.1981.tb04227.x. [DOI] [PubMed] [Google Scholar]
- 16.Ohosone Y, Matsumura M, Chiba J, Nagaoka S, Matsuoka Y, Irimajiri S, Mimori T. Clin Rheumatol. 1998;17:144–147. doi: 10.1007/BF01452262. [DOI] [PubMed] [Google Scholar]
- 17.Gilliam AC, Steitz JA. Proc Natl Acad Sci USA. 1993;90:6781–6785. doi: 10.1073/pnas.90.14.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pokkuluri PR, Bouthillier F, Li Y, Kuderova A, Lee J, Cygler M. J Mol Biol. 1994;243:283–297. doi: 10.1006/jmbi.1994.1654. [DOI] [PubMed] [Google Scholar]
- 19.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 20.Juneau K, Podell E, Harrington DJ, Cech TR. Structure (London) 2001;9:221–231. doi: 10.1016/s0969-2126(01)00579-2. [DOI] [PubMed] [Google Scholar]
- 21.Fellouse FA, Wiesmann C, Sidhu SS. Proc Natl Acad Sci USA. 2004;101:12467–12472. doi: 10.1073/pnas.0401786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fellouse FA, Li B, Compaan DM, Peden AA, Hymowitz SG, Sidhu SS. J Mol Biol. 2005;348:1153–1162. doi: 10.1016/j.jmb.2005.03.041. [DOI] [PubMed] [Google Scholar]
- 23.Wu TT, Johnson G, Kabat EA. Proteins. 1993;16:1–7. doi: 10.1002/prot.340160102. [DOI] [PubMed] [Google Scholar]
- 24.Fellouse FA, Esaki K, Birtalan S, Raptis D, Cancasci VJ, Koide A, Jhurani P, Vasser M, Wiesmann C, Kossiakoff AA, et al. J Mol Biol. 2007;373:924–940. doi: 10.1016/j.jmb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Laird-Offringa IA, Belasco JG. Methods Enzymol. 1996;267:149–168. doi: 10.1016/s0076-6879(96)67011-6. [DOI] [PubMed] [Google Scholar]
- 26.Fellouse FA, Barthelemy PA, Kelley RF, Sidhu SS. J Mol Biol. 2006;357:100–114. doi: 10.1016/j.jmb.2005.11.092. [DOI] [PubMed] [Google Scholar]
- 27.Koide A, Gilbreth RN, Esaki K, Tereshko V, Koide S. Proc Natl Acad Sci USA. 2007;104:6632–6637. doi: 10.1073/pnas.0700149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee CV, Liang WC, Dennis MS, Eigenbrot C, Sidhu SS, Fuh G. J Mol Biol. 2004;340:1073–1093. doi: 10.1016/j.jmb.2004.05.051. [DOI] [PubMed] [Google Scholar]
- 29.Korennykh AV, Piccirilli JA, Correll CC. Nat Struct Mol Biol. 2006;13:436–443. doi: 10.1038/nsmb1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy FL, Cech TR. Biochemistry. 1993;32:5291–5300. doi: 10.1021/bi00071a003. [DOI] [PubMed] [Google Scholar]
- 31.Michel F, Westhof E. J Mol Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 32.Eigenbrot C, Randal M, Presta L, Carter P, Kossiakoff AA. J Mol Biol. 1993;229:969–995. doi: 10.1006/jmbi.1993.1099. [DOI] [PubMed] [Google Scholar]
- 33.Guo F, Gooding AR, Cech TR. Mol Cell. 2004;16:351–362. doi: 10.1016/j.molcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 34.Uchida T, He Q, Ralston CY, Brenowitz M, Chance MR. Biochemistry. 2002;41:5799–5806. doi: 10.1021/bi020042v. [DOI] [PubMed] [Google Scholar]
- 35.Jones S, Thornton JM. Proc Natl Acad Sci USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar S, Nussinov R. Biophys J. 2002;83:1595–1612. doi: 10.1016/S0006-3495(02)73929-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kabat EA, Wu TT. Ann NY Acad Sci. 1971;190:382–393. doi: 10.1111/j.1749-6632.1971.tb13550.x. [DOI] [PubMed] [Google Scholar]
- 38.Treger M, Westhof E. J Mol Recognit. 2001;14:199–214. doi: 10.1002/jmr.534. [DOI] [PubMed] [Google Scholar]
- 39.Jeong E, Kim H, Lee SW, Han K. Mol Cells. 2003;16:161–167. [PubMed] [Google Scholar]
- 40.Lejeune D, Delsaux N, Charloteaux B, Thomas A, Brasseur R. Proteins. 2005;61:258–271. doi: 10.1002/prot.20607. [DOI] [PubMed] [Google Scholar]
- 41.Ferre-D'Amare AR, Zhou K, Doudna JA. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 42.Ferre-D'Amare AR, Doudna JA. J Mol Biol. 2000;295:541–556. doi: 10.1006/jmbi.1999.3398. [DOI] [PubMed] [Google Scholar]
- 43.Braun RP, Woodsworth ML, Lee JS. Mol Immunol. 1986;23:685–691. doi: 10.1016/0161-5890(86)90107-0. [DOI] [PubMed] [Google Scholar]
- 44.Teunissen SW, Stassen MH, Pruijn GJ, van Venrooij WJ, Hoet RM. RNA. 1998;4:1124–1133. doi: 10.1017/s1355838298980633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rupert PB, Ferre-D'Amare AR. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 46.Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. Nature. 2004;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- 47.Sidhu SS, Lowman HB, Cunningham BC, Wells JA. Methods Enzymol. 2000;328:333–363. doi: 10.1016/s0076-6879(00)28406-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.