Abstract
We report the microbiological, biochemical, and molecular characterization of an unusual Brucella strain (BO1) isolated from a breast implant wound in a 71-year-old woman with clinical symptoms consistent with brucellosis. Initial phenotypic analysis, including biochemical and antimicrobial susceptibility testing, cellular fatty acid analysis, and molecular analysis based on DNA-DNA reassociation and the presence of multiple copies of IS711 element suggested that the isolate was a Brucella-like organism, but species determination using microbiological algorithms was unsuccessful. Furthermore, molecular data based on 16S rRNA gene sequencing and multilocus sequence analysis demonstrated that BO1 was an unusual Brucella strain and not closely related to any currently described Brucella species. However, comparison with equivalent sequences in Ochrobactrum spp. confirms that the isolate is much more closely related to Brucella than to Ochrobactrum spp., and thus the isolate likely represents an atypical and novel strain within the genus Brucella.
Brucellosis is primarily a zoonotic disease caused by members of the genus Brucella, which consists of six recognized species based on pathogenicity and host preferences: Brucella abortus (cattle), Brucella canis (dogs), Brucella melitensis (goats or sheep), Brucella suis (swine), Brucella ovis (rams), and Brucella neotomae (desert rats), as well as recently identified strains from marine mammals (5, 10, 13, 26, 34). Early studies on genomic DNA hybridization demonstrate a high degree of homology among the brucellae (40) and thus suggest the genus Brucella as one species, Brucella melitensis with several biovars (14), which has been confirmed by a number of molecular approaches, including 16S rRNA gene sequencing (21), multilocus enzyme electrophoresis (19), and whole-genome sequence analyses (15, 24, 36). However, because of the lack of widespread support the Brucella Taxonomy Subcommittee has recently returned to the traditional classification of the six Brucella nomenspecies with recognized biovars, along with two presumptive Brucella spp. from marine mammals: B. cetaceae and B. pinnipediae (34). Brucellae are intracellular facultative pathogens that infect many organs and soft tissues, including mammary glands, and frequently result in abortion, low milk production, and fetal death in animals (12, 45).
Most human disease is caused by B. abortus, B. suis, B. melitensis, and B. canis (13, 22, 35) and is most frequently associated with the consumption of unpasteurized dairy products or direct contact with infected animals or animal products (38, 46). Brucellosis is also an occupational hazard of laboratory scientists infected by the inhalation of aerosols in a microbiology laboratory setting (32, 37). In humans, brucellosis is a systemic, febrile illness and can be associated with chronic debilitating infection of major organ systems that may include bone and the kidney, brain, epididymis, liver, ovary, and gallbladder (7, 8, 18, 27). However, association of Brucella with infection of mammary glands, including prosthetic devices, is a rare phenomenon in humans (11, 16, 17, 23, 30, 33). Three cases of human brucellosis have been reported associated with B. melitensis infection of breast implant patients. These involved patients who had either consumed unpasteurized cheese or sniffed B. melitensis cultures in a clinical laboratory (1, 3, 20, 31).
We report the isolation and identification of a novel Brucella strain from a breast implant wound in a 71-year-old woman from Oregon who developed symptoms consistent with brucellosis.
MATERIALS AND METHODS
Case report.
In early June 2005, a 71-year-old female patient was hospitalized with fever, hypotension, leukocytosis, and inflammation around her right breast implant. She was treated initially with cephalexin unsuccessfully, followed by cefazolin until late June 2005. The patient underwent successful removal of both breast implants in early July 2005. Blood and breast implant wound fluid specimens were analyzed at the local hospital.
The patient first received silicone implants in the early 1970s. In 1999 or 2000, she had the silicone implants removed and had them replaced by saline implants in a local hospital. The patient recalled being lethargic with myalgias in late January and early February 2005. Two months after that, she had fever greater than 103°F for 2 days, which she self treated with acetaminophen. Other family members also reported similar symptoms. In late May and early June, she reported again fever at approximately 104°F and noted inflammation of her right breast. The patient denied common risk factors associated with human brucellosis; however, she traveled to Ireland for a few weeks in July 2004. Up to late January or February 2005, the patient had two dogs in good health and no contact with kennels or cattle. She had a rabbit that died (cause unknown) 6 months prior to her illness. She often visited horse shows over the previous year. The patient has lived in the Portland area in Oregon for the past 39 years.
A gram-negative coccobacillus was isolated on routine chocolate agar (Remel, Lenexa, KS) from cultures of the right breast implant wound fluid of the patient. The isolate was initially tested according to standard microbiological procedures and was identified as a Brucella species. The culture was forwarded to the Centers for Disease Control and Prevention (CDC), Atlanta, GA, for confirmatory identification. The isolate was designated BO1 and stored at −70°C in defibrinated rabbit blood until testing.
Microbiological and biochemical tests.
Phenotypic identification of the BO1 isolate was performed by standard microbiologic procedures, which include species and biovar identification tests: susceptibility to Tbilisi phage lysis and biochemical and serological criteria (42). Monospecific rabbit anti-A and anti-M antisera and Tbilisi phage were obtained from the National Veterinary Services Laboratories (Ames, IA). All type and reference strains included in the present study are part of the Brucella culture collection at the CDC.
The antimicrobial susceptibility testing of BO1 was conducted using the broth microdilution procedure for susceptibility testing of Brucella spp. in accordance with Clinical and Laboratory Standards Institute (CLSI) protocol (9). Briefly, the isolate was tested using in-house prepared MIC panels containing brucella broth. Panels were incubated at 35°C in ambient air supplemented with 5% CO2 for 2 days.
The total cellular fatty acid (CFA) composition of BO1 was determined by using gas-liquid chromatography as described previously (42). Briefly, 48-h-old BO1 cells were harvested and saponified, and the liberated fatty acids were analyzed by gas-liquid chromatography using the Sherlock microbial identification system software package (CDC library) (42).
Molecular tests.
The BO1 cells were grown on Trypticase soy agar with 5% defibrinated sheep blood agar (SBA) or rabbit blood agar (RBA) (BBL Microbiology Systems, Cockeysville, MD), and the cell-lysate DNA template was prepared as described previously (21). To analyze the 16S rRNA gene sequence, the cell lysate DNA was amplified by using the eubacterial primers, F8 and R1492 (21). The 16S rRNA amplicon was sequenced with a panel of eubacterial primers by using the BigDye terminator cycle sequencing kit (ABI, Foster City, CA) as described previously (21). Computer analysis of the 16S rRNA gene sequence was performed by using the GCG Wisconsin software package (version 10.2; Accelrys, San Diego, CA) and by using MEGA 3.1 (28). The accession number of the B01 16S rRNA sequence is EU053207.
To analyze the DNA-DNA association, BO1 cells were grown on Trypticase soy agar with defibrinated SBA (BBL Microbiology) for 2 days at 37°C, and genomic DNA was prepared by the sodium dodecyl sulfate-phenol extraction method as described previously (4, 40). The purified BO1 genomic DNA was sonicated, labeled with [32P]dCTP by nick translation (4), and hybridized with B. melitensis 16M and B. suis 1330 DNA templates, respectively, as described previously (40). The relative binding ratios were determined at both 60 and 75°C.
To detect the Brucella-specific insertion sequence IS711 element (842 bp) (25), cell lysate DNA from B. melitensis 16M and B. ovis (ATCC 25840) and BO1 were amplified by using forward and reverse primers as described previously (6, 25). The amplicons were analyzed by electrophoresis using a 2% E-Gel agarose gel (Invitrogen Corp., Carlsbad, CA) as described by the manufacturer.
Multilocus sequence analysis (MLSA) involving the characterization of nine independent genetic loci to determine the relationship of BO1 to classical Brucella species was carried out as described previously (44). Sequences of the nine loci were concatenated and compared to sequences of the 27 sequence types (STs) identified in a study of 160 Brucella isolates representing the breadth of the known genetic diversity of Brucella (44). Genetic distances were calculated by using the Jukes-Cantor method, and an unrooted phylogenetic tree was constructed by using the neighbor-joining approach implemented in the MEGA3.1 package (28).
To assess the relationship of BO1 with the nearest known neighbors of the brucellae, the MLSA primer sets and primer sets corresponding to an additional five housekeeping genes were used to attempt to amplify the equivalent gene products from the type strains of five Ochrobactrum spp. (O. tritici [LMG18957T], O. intermedium [LMG3301T], O. gallinifaecis [DSM15295T], O. anthropi [LMG3331T], and O. grignonense [LMG18954T]). Where amplification and sequencing proved possible, phylogenetic trees were constructed as described above, and the robustness of branching was assessed by performing 1,000 bootstrap analyses of the data. In this manner the relationship of BO1 with the type strain of each Brucella spp. and Ochrobactrum spp. (where amplification was possible) was determined for each of eight distinct genetic fragments. Sequences examined originated from dnaK, gap, and gyrB as described previously (44), as well from as fumC (fumarate hydratase C), fbaA (fructose-bisphosphate aldolase), prpE (propionate-coenzyme A ligase), soxA (sarcosine oxidase, alpha subunit), and csdB (cysteine desulfhydrase). All sequences have been submitted to GenBank. The nine BO1 fragments included in the MLSA were assigned the accession numbers AM884761 (dnaK), AM884787 (gap), AM884790 (gyrB), AM884813 (aroA), AM884814 (glk), AM884815 (trpE), AM884816 (cobQ), AM884817 (omp25), and AM884818 (int-hyp). Ochrobactrum sequences of dnaK, gap, and gyrB were assigned the accession numbers AM884762 to AM884763, AM884788 to AM884789, and AM884790 to AM884791, respectively. Additional Brucella sequences included in these analyses have been reported previously (43). The sequences of both Ochrobactrum and Brucella representing genes not examined previously by us were assigned the accession numbers AM884752 to AM884760 (csdB), AM884764 to AM884774 (fbaA), AM884775 to AM884786 (fumC), AM884793 to AM884802 (prpE), and AM884803 to AM884812 (soxA).
RESULTS
Microbiological and biochemical characterization.
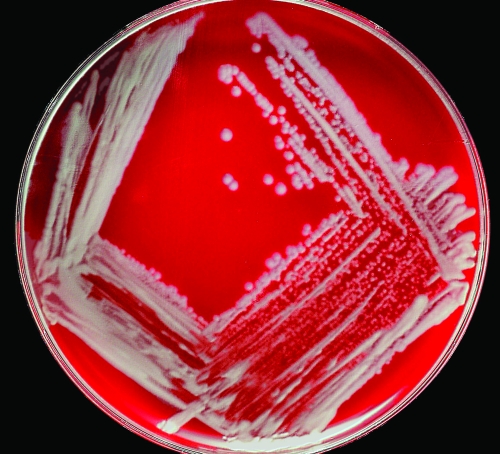
The BO1 cells grown on SBA or RBA plates with or without 5% CO2 at 35 to 37°C for 24 to 48 h were small gram-negative coccoid rods. Isolated colonies were circular, convex, entire, smooth, opaque, and 1 to 2 mm in diameter in 48 h (Fig. 1). BO1 was positive for growth on MacConkey agar, oxidase, catalase, nitrate reduction with production of gas, growth in nutrient broth with or without 6% NaCl, fast urease reaction (<5 min), and growth at 25, 37, and 42°C. All biochemical tests were performed in aerobic conditions. Hemolysis was not observed after 24 h of incubation at 37°C. BO1 was nonmotile and negative for hydrolysis of esculin, gelatin liquefaction, production of indole, citrate utilization, growth on centrimide, and SS agar. Acid production was observed in King's oxidation-fermentation base from d-glucose and d-xylose, whereas no acid production was observed in King's oxidation-fermentation base from mannitol, lactose, sucrose, and maltose. Growth of BO1 was not inhibited in the presence of thionine (1:25,000, 1:50,000, and 1:100,000 dilutions) or basic fuchsin (1:50,000 and 1:100,000 dilutions) dyes and did not form a gel in phenolized saline. The BO1 strain showed weak or no agglutination with monospecific anti-A and anti-M antisera, respectively, and was not susceptible to lysis by Tbilisi phage at the two routine test dilutions (1× and 4×) tested (2).
FIG. 1.
Colony morphology of BO1 strain after 48 h of growth on SBA incubated at 37°C in 5% CO2.
The antimicrobial susceptibility pattern of BO1 was determined and compared to reference Brucella strains. Using the CLSI interpretive criteria for Brucella spp., BO1 was determined to be susceptible to the following antimicrobial agents: doxycycline (0.12 μg/ml), tetracycline, (0.25 μg/ml), streptomycin (2 μg/ml), gentamicin, (1 μg/ml), and trimethoprim-sulfamethoxazole (<0.5 and 9.5 μg/ml). The antimicrobial susceptibility pattern of BO1 was similar to profiles of other Brucella spp. isolates described previously (26a).
CFA analysis was performed, and the profile of BO1 was compared to the profiles of reference Brucella strains. The BO1 isolate whole-cell CFA profile was similar to those of B. abortus, B. melitensis, B. neotomae, B. ovis, and B. suis characterized by the presence of major amounts (5 to 46%) of C19:0cyc11-12, C18:1ω7c, C18:0, and C16:0 and smaller amounts (1 to 4%) of Br-C19:1.
Molecular characterization.
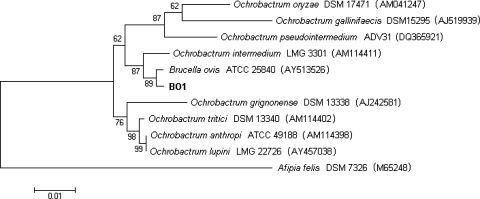
Standard microbiological characterization, including antimicrobial susceptibility and CFA profiles, suggested that BO1 was a Brucella strain, but reliable species identification could not be made. Therefore, we performed 16S rRNA gene sequencing to confirm identification as a Brucella species. The 16S rRNA gene was amplified by PCR from BO1 genomic DNA, and both forward and reverse strands were sequenced. A near-full-length amplicon (1,412 bp) of the 16S rRNA gene was generated and compared to the Brucella consensus 16S rRNA gene sequence as well as to the sequence for the Ochrobactrum intermedium type strain (GenBank accession no. AM114411T) (21). BESTFIT analysis indicated five base differences at positions 167 to 170 and 234 between BO1 and the Brucella consensus sequence, resulting in 99.6% identity. Comparing the 16S rRNA gene sequence of BO1 to that of O. intermedium indicated 14 base differences, which resulted in 99.0% identity by BESTFIT analysis. A dendrogram prepared using the neighbor-joining algorithm indicates that BO1 and the Brucella consensus sequence (represented by the 16S rRNA gene sequence for B. ovis) cluster together (Fig. 2).
FIG. 2.
Phylogenetic tree based on analysis of 16S rRNA gene sequence data indicating relationships between BO1 and other Brucella (represented by the B. ovis type strain) and Ochrobactrum species. MEGA 3.1 was used to generate the dendrogram using the neighbor-joining algorithm with Kimura two-parameter correction and a 1,000-step bootstrap. The bar indicates 0.01 substitutions per nucleotide position. The sequence from the Afipia felis type strain was used as an outgroup. GenBank accession numbers are in parentheses.
DNA-DNA reassociation studies were performed to examine the level of genomic relatedness of BO1 to two reference strains: B. melitensis 16M and B. suis 1330 (21). BO1 genomic DNA was labeled with 32P by nick translation, and the relative binding ratios were determined at both 60 and 75°C. The observed DNA relatedness was 78 to 80% at 60°C and 81 to 83% at 75°C with a divergence (ΔTm) of <1.0 (Table 1). These hybridization values indicate that BO1 is a member of the genus Brucella (39, 40).
TABLE 1.
Genomic DNA relatedness analysis of BO1 with B. melitensis 16M and B. suis 1330 strains using DNA-DNA reassociation
| Strain | Relative binding ratio (%)a at:
|
% Divergenceb | |
|---|---|---|---|
| 60°C | 75°C | ||
| BO1 | 100 | 100 | 0.0 |
| B. melitensis 16M | 80 | 83 | 1.0 |
| B. suis 1330 | 78 | 81 | 1.0 |
The relative binding ratio is the amount of double-stranded DNA formed between labeled and unlabeled DNAs from different strains divided by the amount of double-stranded DNA formed between labeled and unlabeled DNA from the same strain and is expressed as a percentage.
The divergence within related sequences is calculated on the assumption that each 1°C decrease in the thermal stability of a DNA duplex is caused by 1% of unpaired bases within that duplex. The ΔTm was calculated to the nearest 0.5%.
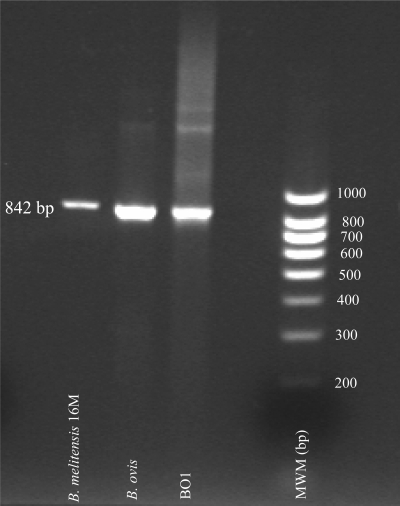
The repetitive DNA insertion sequence, IS711 (842 bp), is found in all Brucella spp. tested to date and is recognized as a unique marker for the genus (23). Therefore, we performed PCR to determine whether the IS711 element was present in the BO1 genome. An IS711 amplicon was generated using BO1 genomic DNA that was approximately the same size and intensity as that of the B. melitensis 16M and B. ovis (ATCC 25840) strains. In addition, several large amplicons (>1,000 bp) were observed for both BO1 and B. ovis (Fig. 3).
FIG. 3.
Detection of the IS711 element in BO1, B. ovis, and B. melitensis 16M by PCR. The amplicons were separated by electrophoresis using a 2% E-Gel agarose gel.
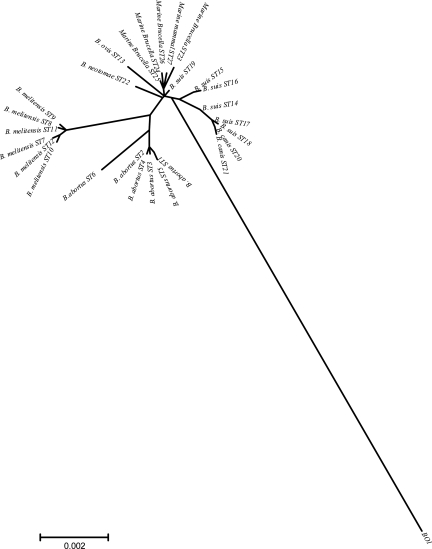
A nine-locus MLSA scheme that divides a panel of 160 Brucella isolates representing all known species and biovars into 27 distinct STs was recently described (44). The relationship of BO1 with the classical Brucella spp. was determined by inclusion in this analysis. BO1 was found to possess novel alleles at all nine loci and to be 1.67% divergent from ST1 when considering all nine fragments, with diversity at each locus relative ranging from 0.23 to 2.84%. In contrast, divergence from ST1 among Brucella over all 4,396 bp examined ranged from a minimum of 0.02% (1 change) up to a maximum of 0.41% (18 changes). This is reflected in the phylogenetic tree based on this analysis showing that BO1 is an obvious outgroup relative to currently described Brucella spp. (Fig. 4).
FIG. 4.
Unrooted phylogenetic reconstruction of the relationships between Brucella STs and BO1. The tree was constructed with concatenated sequence data representing nine distinct genetic loci (4,396 bp) using the neighbor-joining approach.
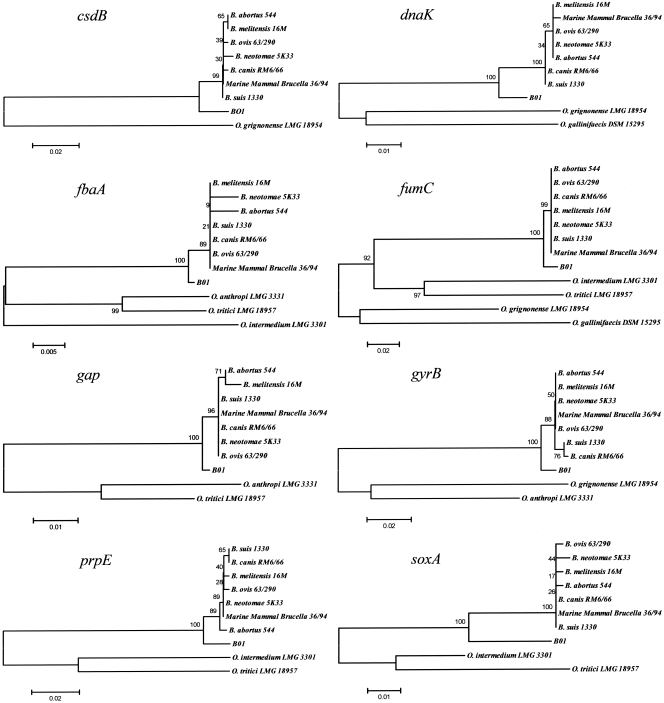
In light of this we next addressed the relationship of BO1 with the organisms currently accepted as being those most closely related to Brucella spp. by sequencing fragments equivalent to three of the genes included in the MLSA scheme (44), as well as fragments from five additional housekeeping genes. Attempts were made to amplify the equivalent regions from five type strains representing different Ochrobactrum spp. Amplification of products from Ochrobactrum spp. was sporadic, likely reflecting the extensive genetic diversity previously noted within this group (29, 43). However, suitable quality products representing between one and four of the Ochrobactrum spp. were obtained and sequenced for eight distinct genetic loci, and relationships between the Brucella reference strains (B. abortus 544, B. melitensis 16M, B. ovis 63/290, B. neotomae 5K33, B. canis RM6/66, B. suis 1330, and marine mammal Brucella 36/94), Ochrobactrum type strains (O. grignonense LMG18954T, O. gallinifaecis DSM 15295T, O. anthropi LMG 3331T, O. tritici LMG 18957T, and O. intermedium LMG 3301T), and isolate BO1 are shown in Fig. 5. The figure demonstrates a consistent relationship for all eight loci where BO1 is distinct and well separated from any of the classical Brucella spp. but is much more closely related to Brucella than to members of the Ochrobactrum group as currently described.
FIG. 5.
Relationships between Brucella, BO1, and Ochrobactrum sequences at eight distinct housekeeping genes. The trees were constructed by using the neighbor-joining method, and percent bootstrap confidence levels of internal branches were calculated from 1,000 resamplings of the original data.
DISCUSSION
We describe here the identification of an unusual Brucella strain (BO1) isolated from a breast implant wound of a 71-year-old patient. Colony morphology, Gram stain appearance, and standard microbiological tests indicated that the BO1 isolate was a nonmotile, gram-negative coccobacillus. The results from standard biochemical tests used for biotyping were inconclusive; these included the lack of reactivity with antisera specific for the lipopolysaccharide O antigens of most common members of the genus Brucella (e.g., B. abortus, B. melitensis, and B. suis), gel formation (B. canis), and lysis by Tbilisi phage (B. abortus and B. suis). However, BO1 rapidly hydrolyzed urea (like B. canis and B. suis biovars 1, 2, 3, 4, and 5), produced H2S (common to B. abortus biovars 1, 2, 3, 4, and 7 and B. suis biovar 1), and grew well in all thionine and fuchsin dye media (similar to B. melitensis biovars 1, 2, and 3 and B. suis biovars 3 and 4), which are characteristics of some of the Brucella spp. Thus, the results of the biotyping procedures suggested that the BO1 isolate was more closely related to B. suis than to other members of the genus Brucella. The antimicrobial susceptibility and CFA profiles of this strain were similar to those of other Brucella spp. The presence of multiple copies of IS711 element, hallmarks of Brucella spp. (25), and the DNA-DNA hybridization studies were also all consistent with members of the genus Brucella (40).
Results of DNA sequencing of the full-length 16S rRNA gene of BO1 demonstrated that the 16S rRNA gene sequence of BO1 was 99.6% identical to the consensus sequence of Brucella spp. Considering that the 16S rRNA gene sequences of all other Brucella spp. determined to date by our group are identical (21), we suggest that BO1 is an atypical Brucella strain. Comparative 16S rRNA gene sequence analysis indicates that BO1 is different from 17 close relatives of Brucella, including O. intermedium and O. anthropi. The DNA-DNA hybridization results also clearly indicate that BO1 is a member of the genus Brucella using the current threshold of 70% (39-41).
Characterization of isolate BO1 by a newly described MLSA method allowed us to determine unequivocally that the isolate is distinct from any currently described Brucella group. Indeed, based on Fig. 4, the isolate appears to be very distant from currently described Brucella spp. However, this is somewhat misleading and reflects the relative homogeneity within the currently described genus: overall, BO1 is only 1.67% divergent from B. abortus 544 at the nucleotide level. When the respective sequences of BO1 are compared to both Brucella and Ochrobactrum isolates (Fig. 5), it is clear that the isolate, while distinct, is much more closely related to the extant Brucella group than to any of the Ochrobactrum type strains. Based on this genetic analysis the most appropriate identification of BO1 currently appears as an atypical member of the Brucella spp.
The collective set of microbiological, biochemical, and molecular analyses of the breast implant isolate have demonstrated that BO1 exhibits the general characteristics of the genus Brucella but is distinct from any previously described member of this species. This is a first case of human brucellosis caused by an unusual Brucella isolate of unknown origin. The natural ecological niche of this organism remains unclear and requires further investigation.
Acknowledgments
We thank Laura A. Jevitt and Jean B. Patel, Division of Healthcare Quality Promotion, CDC, for performing antimicrobial susceptibility testing and Stephen Shankster for technical support at the Veterinary Laboratories Agency (VLA). Brucella research at the VLA is funded by the Department of Environment, Food, and Rural Affairs of the United Kingdom.
Footnotes
Published ahead of print on 31 October 2007.
REFERENCES
- 1.Al Abdely, H. M., M. A. Halim, and T. M. Amin. 1996. Breast abscess caused by Brucella melitensis. J. Infect. 33219-220. [DOI] [PubMed] [Google Scholar]
- 2.Alton, G. G., L. M. Jones, and D. E. Pietz (ed.). 1975. Laboratory techniques in brucellosis, 2nd ed., p. 33-153. Monograph 55. World Health Organization, Geneva, Switzerland. [PubMed]
- 3.Ariza, J., O. Servitje, R. Pallares, P. Fernandez-Viladrich, G. Rufi, J. Peyri, and F. Gudiol. 1989. Characteristic cutaneous lesions in patients with brucellosis. Arch. Dermatol. 125380-383. [PubMed] [Google Scholar]
- 4.Brenner, D. J., A. C. McWhorter, J. K. Knutson, and A. G. Steigerwalt. 1982. Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds. J. Clin. Microbiol. 151133-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bricker, B. J., D. R. Ewalt, A. P. MacMillan, G. Foster, and S. Brew. 2000. Molecular characterization of Brucella strains isolated from marine mammals. J. Clin. Microbiol. 381258-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bricker, B. J., and S. M. Halling. 1994. Differentiation of Brucella abortus bv. 1, 2, and 4, Brucella melitensis, Brucella ovis, and Brucella suis bv. 1 by PCR. J. Clin. Microbiol. 322660-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerri, D., V. V. Ebani, A. Pedrini, R. Nuvoloni, G. Renzoni, E. Andreani, and R. Farina. 1999. Epididymitis by Brucella ovis: experimental infection in virgin ram lambs. New Microbiol. 22227-231. [PubMed] [Google Scholar]
- 8.Chu, M., and R. S.Weyant. 2003. Francisella and Brucella, p. 789-808. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. ASM Press, Washington, DC.
- 9.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing; 16th informational supplement. M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 10.Cloeckaert, A., M. Grayon, O. Grepinet, and K. S. Boumedine. 2003. Classification of Brucella strains isolated from marine mammals by infrequent restriction site-PCR and development of specific PCR identification tests. Microbes Infect. 5593-602. [DOI] [PubMed] [Google Scholar]
- 11.Cokca, F., A. Azap, and O. Meco. 1999. Bilateral mammary abscess due to Brucella melitensis. Scand. J. Infect. Dis. 31318-319. [DOI] [PubMed] [Google Scholar]
- 12.Collier, L., A. Balows, and M. Sussman. 1998. Topley & Wilson's microbiology and microbial infections, 9th ed., vol. 3. Oxford University Press, London, England.
- 13.Corbel, M. J. 1997. Brucellosis: an overview. Emerg. Infect. Dis. 3213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corbel, M. J. 1988. International Committee on Systemic Bacteriology Subcommittee on the Taxonomy of Brucella: report of the meeting, 5 September 1986, Manchester, England. Int. J. Syst. Bacteriol. 38450-452. [Google Scholar]
- 15.DelVecchio, V. G., V. Kapatral, R. J. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, I. Anderson, A. Bhattacharyya, A. Lykidis, G. Reznik, L. Jablonski, N. Larsen, M. D'Souza, A. Bernal, M. Mazur, E. Goltsman, E. Selkov, P. H. Elzer, S. Hagius, D. O'Callaghan, J. J. Letesson, R. Haselkorn, N. Kyrpides, and R. Overbeek. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dowden, R. V. 1994. Periprosthetic bacteria and the breast implant patient with systemic symptoms. Plast. Reconstr. Surg. 94300-305. [DOI] [PubMed] [Google Scholar]
- 17.Erdem, G., H. M. Karakas, F. Yetkin, A. Alkan, A. K. Firat, and B. Kahraman. 2006. Brucellar breast abscess. Breast 15554-557. [DOI] [PubMed] [Google Scholar]
- 18.Fenkci, V., S. Cevrioglu, and M. Yilmazer. 2003. Ovarian abscess due to Brucella melitensis. Scand. J. Infect. Dis. 35762-763. [DOI] [PubMed] [Google Scholar]
- 19.Gandara, B., A. L. Merino, M. A. Rogel, and E. Martinez-Romero. 2001. Limited genetic diversity of Brucella spp. J. Clin. Microbiol. 39235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gasser, I., B. Almirante, F. Fernandez-Perez, and C. Mendoza. 1991. Bilateral mammary abscess and uveitis caused by Brucella melitensis: report of a case. Infection 1944-45. [DOI] [PubMed] [Google Scholar]
- 21.Gee, J. E., B. K. De, P. N. Levett, A. M. Whitney, R. T. Novak, and T. Popovic. 2004. Use of 16S rRNA gene sequencing for rapid confirmatory identification of Brucella isolates. J. Clin. Microbiol. 423649-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorvel, J. P., and E. Moreno. 2002. Brucella intracellular life: from invasion to intracellular replication. Vet. Microbiol. 90281-297. [DOI] [PubMed] [Google Scholar]
- 23.Gurleyik, E. 2006. Breast abscess as a complication of human brucellosis. Breast J. 12375-376. [DOI] [PubMed] [Google Scholar]
- 24.Halling, S. M., B. D. Peterson-Burch, B. J. Bricker, R. L. Zuerner, Z. Qing, L. L. Li, V. Kapur, D. P. Alt, and S. C. Olsen. 2005. Completion of the genome sequence of Brucella abortus and comparison to the highly similar genomes of Brucella melitensis and Brucella suis. J. Bacteriol. 1872715-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halling, S. M., F. M. Tatum, and B. J. Bricker. 1993. Sequence and characterization of an insertion sequence, IS711, from Brucella ovis. Gene 133123-127. [DOI] [PubMed] [Google Scholar]
- 26.Jahans, K. L., G. Foster, and E. S. Broughton. 1997. The characterisation of Brucella strains isolated from marine mammals. Vet. Microbiol. 57373-382. [DOI] [PubMed] [Google Scholar]
- 26a.Jevitt, L. A., L. M. Weigel, B. De, T. Popovic, and J. B. Patel. 2005. Abstr. C-357. Abstr. 105th Gen. Meet. Am. Soc. Microbiol. 2005. American Society for Microbiology, Washington, DC.
- 27.Kalelioglu, M., S. Ceylan, I. Koksal, K. Kuzeyli, and F. Akturk. 1990. Brain abscess caused by Brucella abortus and Staphylococcus aureus in a child. Infection 18386-387. [DOI] [PubMed] [Google Scholar]
- 28.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 29.Lebuhn, M., S. Bathe, W. Achouak, A. Hartmann, T. Heulin, and M. Schloter. 2006. Comparative sequence analysis of the internal transcribed spacer 1 of Ochrobactrum species. Syst. Appl. Microbiol. 29265-275. [DOI] [PubMed] [Google Scholar]
- 30.Marbach, F., L. Saiah, J. F. Fischer, J. Huismans, and A. Cometta. 2007. Prosthetic joint infection of the knee due to Brucella spp. Rev. Med. Suisse 31007-1009. (In French.) [PubMed] [Google Scholar]
- 31.Memish, Z. A., M. Alazzawi, and R. Bannatyne. 2001. Unusual complication of breast implants: Brucella infection. Infection 29291-292. [DOI] [PubMed] [Google Scholar]
- 32.Miller, C. D., J. R. Songer, and J. F. Sullivan. 1987. A twenty-five year review of laboratory-acquired human infections at the National Animal Disease Center. Am. Ind. Hyg. Assoc. J. 48271-275. [DOI] [PubMed] [Google Scholar]
- 33.Navarro, V., J. Solera, E. Martinez-Alfaro, L. Saez, E. Escribano, and J. C. Perez-Flores. 1997. Brucellar osteomyelitis involving prosthetic extra-articular hardware. J. Infect. 35192-194. [DOI] [PubMed] [Google Scholar]
- 34.Osterman, B., and I. Moriyon. 2006. Report of the International Committee on Systematics Prokaryotes, Subcommittee on the Taxonomy of Brucella. Int. J. Syst. Evol. Microbiol. 561173-1175. [Google Scholar]
- 35.Pappas, G., P. Papadimitriou, N. Akritidis, L. Christou, and E. V. Tsianos. 2006. The new global map of human brucellosis. Lancet Infect. Dis. 691-99. [DOI] [PubMed] [Google Scholar]
- 36.Paulsen, I. T., R. Seshadri, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, T. D. Read, R. J. Dodson, L. Umayam, L. M. Brinkac, M. J. Beanan, S. C. Daugherty, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, W. C. Nelson, B. Ayodeji, M. Kraul, J. Shetty, J. Malek, S. E. Van Aken, S. Riedmuller, H. Tettelin, S. R. Gill, O. White, S. L. Salzberg, D. L. Hoover, L. E. Lindler, S. M. Halling, S. M. Boyle, and C. M. Fraser. 2002. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. USA 9913148-13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Staszkiewicz, J., C. M. Lewis, J. Colville, M. Zervos, and J. Band. 1991. Outbreak of Brucella melitensis among microbiology laboratory workers in a community hospital. J. Clin. Microbiol. 29287-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trout, D., T. M. Gomez, B. P. Bernard, C. A. Mueller, C. G. Smith, L. Hunter, and M. Kiefer. 1995. Outbreak of brucellosis at a United States pork packing plant. J. Occup. Environ. Med. 37697-703. [DOI] [PubMed] [Google Scholar]
- 39.Verger, J. M., M. Grayon, A. Cloeckaert, M. Lefevre, E. Ageron, and F. Grimont. 2000. Classification of Brucella strains isolated from marine mammals using DNA-DNA hybridization and ribotyping. Res. Microbiol. 151797-799. [DOI] [PubMed] [Google Scholar]
- 40.Verger, J. M., F. Grimont, P. A. D. Grimont, and M. Grayon. 1985. Brucella, a monospecific genus as shown by deoxyribonucleic acid hybridization. Int. J. Syst. Bacteriol. 35292-295. [Google Scholar]
- 41.Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. I. Krichevsky, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, and M. P. Starr. 1987. Report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Int. J. Syst. Bacteriol. 37463-464. [Google Scholar]
- 42.Weyant, R. S., C. W. Moss, R. E. Weaver, D. G. Hollis, J. J. Jordon, E. C. Cook, and M. I. Daneshvar. 1996. Identification of unusual pathogenic gram-negative aerobic and facultative anaerobic bacteria, 2nd ed. The Williams & Wilkins Co., Baltimore, MD.
- 43.Whatmore, A. M., T. J. Murphy, S. Shankster, E. Young, S. J. Cutler, and A. P. Macmillan. 2005. Use of amplified fragment length polymorphism to identify and type Brucella isolates of medical and veterinary interest. J. Clin. Microbiol. 43761-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whatmore, A. M., L. L. Perrett, and A. P. MacMillan. 2007. Characterisation of the genetic diversity of Brucella by multilocus sequencing. BMC Microbiol. 734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wright, P. W. 1987. Brucellosis. Am. Fam. Physician 35155-159. [PubMed] [Google Scholar]
- 46.Young, E. J., and U. Suvannoparrat. 1975. Brucellosis outbreak attributed to ingestion of unpasteurized goat cheese. Arch. Intern. Med. 135240-243. [PubMed] [Google Scholar]