Abstract
Members of the genus Brucella are known worldwide as pathogens of wildlife and livestock and are the most common organisms of zoonotic infection in humans. In general, brucellae exhibit a range of host specificity in animals that has led to the identification of at least seven Brucella species. The genomes of the various Brucella species are highly conserved, which makes the differentiation of species highly challenging. However, we found single-nucleotide polymorphisms (SNPs) in housekeeping and other genes that differentiated the seven main Brucella species or clades and thus enabled us to develop real-time PCR assays based around these SNPs. Screening of a diverse panel of 338 diverse isolates with these assays correctly identified each isolate with its previously determined Brucella clade. Six of the seven clade-specific assays detected DNA concentrations of less than 10 fg, indicating a high level of sensitivity. This SNP-based approach places samples into a phylogenetic framework, allowing reliable comparisons to be made among the lineages of clonal bacteria and providing a solid basis for genotyping. These PCR assays provide a rapid and highly sensitive method of differentiating the major Brucella groups that will be valuable for clinical and forensic applications.
Brucella spp. are pathogenic bacteria that infect a wide variety of mammalian hosts worldwide, often causing reproductive failure. The genus Brucella has classically been divided into six species based on host specificity, including B. abortus (cattle and bison), B. melitensis (goats and sheep), B. suis (pigs), B. canis (dogs), B. neotomae (desert woodrat), and B. ovis (sheep) (12). Two new species have been discovered recently in marine mammals (B. cetaceae in dolphins and whales and B. pinnipediae in seals) (10). Taxonomic limits of the marine clade, however, are not fully defined, and this group may represent one to three species (8, 18). B. abortus, B. melitensis, B. suis, and B. canis are well-characterized zoonotic pathogens, annually infecting >500,000 people worldwide (26). In the United States, the first three of these species are defined as select agents due to their pathogenicity and potential use as biological weapons (11).
Despite host-based segregation, Brucella spp. have proven challenging to differentiate using molecular techniques. Brucella genomes are highly conserved, with >90% homology among species based on DNA-DNA hybridization (35), identical 16S rRNA sequences among all species (15), and >90% of genes sharing >98% sequence identity (16, 27). Serological methods and biochemical testing of isolates allow differentiation of species and biovars. However, PCR-based methods have been used increasingly due to their accuracy, sensitivity, and speed of identification and the ability to work with DNA as opposed to highly infectious live cultures. A wide array of genetic polymorphisms can be assayed for the differentiation of Brucella spp., including the insertion element IS711 (2, 3, 29, 31) and genes of outer membrane proteins (7, 10, 20), and other assay techniques may be used, such as whole-gene differentiation (30), infrequent restriction site PCR (6), and amplified fragment length polymorphisms (37). PCR-based assays for identifying Brucella were recently reviewed (1).
Improved resolution among Brucella isolates for the purposes of genotyping and epidemiology has been obtained by using more rapidly evolving markers. For example, variable-number tandem repeats (VNTR) incorporated into multilocus VNTR analysis (MLVA) successfully differentiate even closely related isolates and provide fairly accurate species-level resolution (1a, 17, 21, 39). Rapidly evolving VNTR markers often suffer from homoplasy, i.e., the appearance of the same genetic alteration in two or more branches of a phylogenetic tree. These phenomena can disrupt and confound the accurate phylogenetic placement of all isolates within a single MLVA tree and prevent the accurate species-level designation of some isolates.
For distinguishing bacteria with clonally derived population structures (such as Brucella [38]), single-nucleotide polymorphisms (SNPs) can be used to accurately describe the phylogenetic framework of a species (28, 33). SNPs can be discovered through either whole-genome comparisons or multilocus sequence typing (MLST) of housekeeping genes. In Brucella, these stable and slowly evolving markers provide the initial resolution within taxonomic trees. A nested hierarchical approach involving SNP analysis to define the major branches, followed by analysis using more rapidly evolving VNTR markers, will generate unambiguous differentiation of isolates into major clades, with maximum resolution at the individual isolate level (19). SNP-based differentiation of clades can then be incorporated into real-time PCR assays, providing a quick method for determining specific groupings (23, 32, 34).
We discovered SNPs that use nucleotide sequences from housekeeping genes and published gene sequences, and we developed real-time PCR assays to identify the seven main Brucella species. These TaqMan assays contained probes specific to each allele and were screened against a large and diverse collection. Our assays provide a reliable and rapid method for the identification of Brucella species that can readily be incorporated into clinical, forensic, or evolutionary applications.
MATERIALS AND METHODS
SNP discovery.
To identify species-specific SNPs for the development of real-time PCR assays, we initially selected five housekeeping gene products: the ABC (abc) transporter ATP-binding protein, shikimate 5-dehydrogenase and shikimate dehydrogenase (aroE and gdh) family proteins, and the chaperonin (groEL), proline iminopeptidase (pip), and sulfate ABC transporter (cysW) proteins. Our primary goal was not to run a full MLST analysis but to find suitable SNPs for species differentiation. We also utilized SNPs in genes coding for an outer membrane protein (omp25), the RNA polymerase beta-subunit (rpoB), and the anthranilate synthase (trpE) (polymorphisms previously described in references 22, 36, and 38, respectively). We designed PCR primers to amplify a portion of each gene (Table 1).
TABLE 1.
Primers used to amplify portions of housekeeping genes and other loci in Brucella
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) | Fragment size (bp) |
|---|---|---|---|
| abc | TAGGCCGAATAATTGCCTTC | GACCGCTACAACGAGCTGAT | 419 |
| aroE | ATGGAAGGCAAGATCGTCAA | CTGGCACAGTTCGTCAACAG | 498 |
| cysW | CTTCCCTGCACTTCCATCAG | ACCAATCTCATCCAGGCAAG | 546 |
| gdh | GATGGTGTCGGTTTTGTGC | GATATGCTGGTGCATTGTGG | 557 |
| groEL | CTGGACGACAGCTTCGAGA | GGCTTCCAAGACCAACGATA | 485 |
| pip | CGGTCAGGCGCTTGTAAT | CGCATTCATGTCGAGCAAT | 484 |
| omp25 | TGGTGGCTATACCGGTCTTT | AGGATGTTGTCCGTCAGCTT | 384 |
| omp25 | AAGTCAAGCAGGGCTTTGAA | ACCGGATGCCTGAAATCCTTa | 395 |
Same primer designated as 25B and used by Cloeckaert et al. (9). A portion of the omp25 gene was sequenced with two overlapping primer sets. The fragment size is approximate due to a tandem repeat at the 3′ end of the sequence.
Cellular DNAs were extracted using heat soaks or genomic preparations and were diluted to roughly 0.1 to 1 ng/μl for assay screening. We amplified fragments in 10-μl PCR mixtures consisting of 1× PCR buffer, 2.8 mM MgCl2, 0.6 μM of each primer, 0.8 mM of each deoxynucleotide triphosphate, and 1 U of Platinum Taq (Invitrogen, Carlsbad, CA). We used the following cycling profile at 94°C for 5 min: 30 cycles of denaturation at 94°C for 30 s, annealing at 62°C for 30 s, and extension at 72°C for 1 min, with a final extension of 72°C for 7 min. We cycle sequenced 1 μl of product in a 10-μl reaction mixture with Applied Biosystems Big Dye 3.1 and then purified with 2.5 μl of 125 mM EDTA and followed with washes of 100% and 70% ethanol. Sequences were run on an AB 3730 model automated sequencer. We edited and aligned sequences using SEQUENCHER 4.6 software (Gene Codes Corporation, Ann Arbor, MI). Sequences were then compared in silico to those of whole-genome sequences from GenBank or unpublished sources from B. abortus 2308 (4), B. abortus 9-941 (16), B. melitensis 16 M (13), B. suis 1330 (27), B. ovis 63/290 (The Institute for Genomic Research accession numbers CP000708 and CP000709), and B. canis RM6/66 (Los Alamos National Laboratory, NM).
Real-time PCR development.
Real-time PCR assays were developed with six genes, containing species-specific SNPs: abc, cysW, omp25, pip, rpoB, and trpE. We designed primers and probes with Primer Express TaqMan minor groove binding (MGB) for Allelic Discrimination version 3.0 (Applied Biosystems, Foster City, CA) software. Primers amplified the region containing the SNP, which binds to either of the two probes for the specific allele of the SNP state (Table 2). Assays were run on an ABI Prism 7900HT sequence detection system (Applied Biosystems). Each 10-μl PCR mixture contained 1× TaqMan Universal master mixture (Applied Biosystems), 0.9 μM of each primer, and 0.2 μM of each probe. In addition, 0.2 U of Platinum Taq (Invitrogen) per reaction mixture was added to increase the efficiency of the amplification, except for the rpoB assay, which used 0.25 U of Platinum Taq. For the trpE assay, the PCR was slightly modified to reduce amplification of the nonmarine mammal probe. The quantity of the probes was set to 0.06 μM of probe 1 (using 6-carboxyfluorescein [FAM] dye) and 0.14 μM of probe 2 (using VIC dye) to optimize the reaction. We ran each assay under standard conditions consisting of a 2-min inactivation at 50°C and a 10-min hot start at 95°C, followed by 40 cycles of a 15-s denaturation and 1 min of annealing at 60°C.
TABLE 2.
Oligonucleotide sequences for primers and TaqMan MGB probes for Brucella clade identification
| SNP clade | Assay name | Primers (5′-3′) | Probes (5′-3′)a | Genome positionb | SNP identityc |
|---|---|---|---|---|---|
| B. abortus | abc_205 | F-GTTTCCGCATCCAAATGGTT | FAM-AAGCAGAATCTTGCACA | 573136 | T, B. abortus |
| R-TTCGGGCGGTGAAAAGC | VIC-AAAGCAGAAGCTTG | G, other spp. | |||
| B. suis/B. canis | abc_246 | F-AGCCTGACCTGCTGCTTCTC | FAM-ATGAGCCCACCAAC | 573177 | C, B. suis/B. canis |
| R-GAGCCAGGCTGTGGTTTCC | VIC-ATGAGCCGACCAAC | G, other spp. | |||
| B. ovis | cysW_234 | F-CCGGGAAAGCGGAATTTC | VIC-CGAAAGCGATGTTGAT | 696894 | G, B. ovis |
| R-GCTGACCGCAATCGTTGTC | FAM-CGAAAGCCATGTTGAT | C, other spp. | |||
| B. melitensis | cysW_288 | F-GGAAAAAGGTATCTCCACGAAGGT | VIC-AGCCTGCGTCCGGG | 696948 | G, B. melitensis |
| R-CGTGGCTGGTGACGAAATT | FAM-TGAGGAGCCTTCG | T, other spp. | |||
| B. canis | omp25_256 | F-GGCTGGCGCCTTTGCT | FAM-AACTTCCAGAAGGAC | 1297911 | A, B. canis |
| R-GGCCCAGGAATAACCTGCAT | VIC-ACTTCCAGCAGGACC | C, other spp. | |||
| B. neotomae | rpoB_2673 | F-CATCCTGGCGACATTCTTGTC | VIC-AAGATCACGCCGAAGG | 776263 | G, B. neotomae |
| R-GGCGTCATCGGGCTTTC | FAM-TCACGCCTAAGGG | T, other spp. | |||
| Marine mammals | trpE_290 | F-ACGAGGATTCCTTCGTCCATAC | FAM-CCAATTATTTCCACCAGAC | 468289 | A, marine mammal |
| R-AACGCACGGTGGAAACCTT | VIC-TTGCCAATTATTTCCGCCA | G, other spp. |
TaqMan probes with a minor groove binder (MGB). Each probe was fluorescently labeled with either VIC or FAM dye.
Based on the genome of B. melitensis 16M chromosome I (GenBank accession no. AE008917).
The SNP identities are listed for the primer letters in bold.
We tested the limits and sensitivity of each assay, using serial 10-fold dilutions. Starting at a concentration of 1 ng/μl, we progressively diluted down to 1 fg/μl. Each sample was run in duplicate, and five samples were tested for each assay. We also quantified the efficiency of the reaction at 1 ng/μl with these 10 samples, as expressed by the cycle threshold values. DNA concentrations were quantified by UV spectroscopy by NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE) and PicoGreen assays (Molecular Probes, Eugene, OR).
Brucella DNA samples.
We screened a diverse collection of 338 Brucella DNA samples for each assay. These samples included 165 isolates of B. melitensis, 85 isolates of B. abortus, 53 isolates of B. suis, 11 marine mammal isolates (from eight seals and three dolphins), 10 isolates of B. canis, 8 isolates of B. ovis, and 6 isolates of B. neotomae (Table 3). Samples were known to contain all recognized biovars except for B. abortus biovar 3 and B. suis biovars 3 and 5. We also included four samples from Ochrobactrum anthropi, a closely related soil bacterium, to test the specificity of the assays. All Brucella samples had initially been identified using phenotypic, biochemical, and serological tests, including tests such as Gram stain morphology, lack of motility, oxidase positivity, and agglutination in Brucella antiserum (5). Over 75% of the samples had also undergone PCR testing for species designation by following procedures described in reference 3.
TABLE 3.
Brucella species isolates used in screening assaysa
| Species | No. of isolates of indicated biovar or strek
|
Total | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 9 | Type strain | None | ||
| B. abortus | 41 | 2 | 2 | 4 | 2 | 1 | 1 | 1 | 2 | 29 | 85 |
| B. canis | 1 | 9 | 10 | ||||||||
| B. melitensis | 49 | 15 | 20 | 1 | 80 | 165 | |||||
| B. neotomae | 2 | 4 | 6 | ||||||||
| B. ovis | 1 | 7 | 8 | ||||||||
| B. suis | 28 | 1 | 4 | 1 | 19 | 53 | |||||
| Marine mammal | 11 | 11 | |||||||||
Brucella species isolates (n = 338) used to screen assays are shown. The biovar is given when available.
Nucleotide sequence accession numbers.
We deposited all sequences in GenBank under accession numbers EU001373 to EU001657.
RESULTS
SNP identification.
We compared sequences for the presence of six housekeeping genes for at least 36 isolates across all seven Brucella species. Nineteen SNPs were identified, including four specific to B. abortus, three for B. canis/B. suis, two for B. ovis and one for B. melitensis. Four of these SNPs were developed into species-specific assays for each species/clade, including two SNPs in the abc gene and two SNPs in the cysW gene. Additional genes were necessary for species-specific assays for B. canis, B. neotomae, and the marine mammal clade. No SNPs were found that defined B. suis exclusively.
Allelic discrimination and assay screening.
All seven SNP assays correctly identified all samples (n = 338) belonging to the appropriate species. However, one sample we believe contained mixed DNA, although we did not have the cell culture to test this hypothesis. In this instance, both species-specific (B. melitensis and B. suis/B. canis) assays indicated a mixed sample, with both alleles amplifying almost equally, suggesting equal amounts of DNA from the corresponding species. We identified the correct species/clade of 11 blind samples and 6 incorrectly labeled samples whose identities were discovered only after the analyses. These samples were identified with their species by conventional biochemical testing and/or were assigned to species by MLVA (17).
Six of the seven assays exhibited rapid amplification of the allele containing the corresponding SNP, with the alternate allele either failing to amplify or weakly amplifying (Table 4). Amplification of the alternate allele above the minimum cycle threshold (CT) value occurred only in the assays for B. abortus and marine mammals. The difference in the CT values between probes (ΔCT) in the B. abortus assay was 12.3 (n = 8), providing easy differentiation. In our initial screening of the marine mammal assay, using standard probe concentrations (0. 9 μM, each probe), the allele for nonmarine mammal samples rapidly amplified at a CT value of 20.2 ± 0.9 (n = 5), and the alternate allele was not detected when nonmarine mammal samples were run. However, cross-hybridization of probes occurred when marine mammal samples were run (ΔCT = 1.6). Thus, we optimized probe concentrations to improve the differentiation of alleles. When the altered concentrations were used with the marine mammal assay, the ΔCT was 4.7 between the primary and the alternate probes for marine mammals and 9.9 for samples from nonmarine mammals (n = 14).
TABLE 4.
Cycle threshold values and detection concentrations for seven Brucella species assaysa
| SNP clade | Assay name |
CT value ± SD
|
Detection concn (fg/μl)b | |
|---|---|---|---|---|
| Allele 1 | Allele 2 | |||
| B. abortus | abc_205 | 21.1 ± 1.4 | 19.5 ± 0.8 | >10 |
| B. suis/B. canis | abc_246 | 19.3 ± 1.1 | 20.1 ± 0.9 | >10 |
| B. ovis | cysW_234 | 17.5 ± 1.3 | 18.4 ± 1.3 | >10 |
| B. melitensis | cysW_288 | 20.9 ± 0.1 | 21.2 ± 0.5 | >10 |
| B. canis | omp25_256 | 19.3 ± 0.7 | 19.2 ± 1.1 | >10 |
| B. neotomae | rpoB_2673 | 21.3 ± 0.1 | 26.3 ± 1.2 | >100 |
| Marine mammals | trpE_290 | 20.2 ± 0.9 | 21.2 ± 0.2 | >10 |
CT values ± standard deviations and detection concentrations are shown for seven Brucella species assays. CT values are from assays run at a concentration of 1 ng/μl (n = 8 to 10 isolates).
We considered the positive detection of a sample when the majority of samples successfully amplified within 40 cycles. For the marine mammal assay, detection concentrations were assessed at standard (equal) probe concentrations.
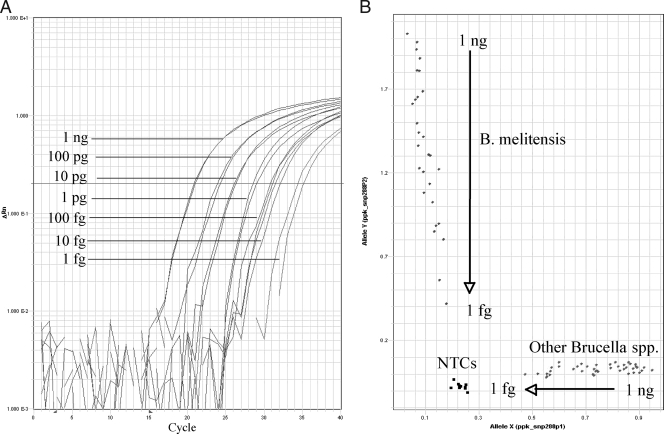
All assays detected at least one sample with a DNA concentration of less than 10 fg/μl, with reliable amplification for all assays at 100 fg/μl and greater (Fig. 1). In fact, the majority of samples at a concentration of 1 fg/μl were successfully amplified in six of the seven assays (the assay for B. neotomae was less sensitive). In every instance, the amplification was clearly distinguishable from the no-template controls. Samples from the closely related soil bacterium Ochrobactrum anthropi failed to amplify in five of the seven assays. In the B. abortus assay (abc_205), the alternate allele (i.e., all non-B. abortus samples) amplified strongly, and in the B. neotomae assay (rpoB_2673), the alternate allele amplified weakly.
FIG. 1.
Real-time PCR and allelic discrimination plots from TaqMan MGB assays for an SNP defining Brucella melitensis. Ten samples (4 B. melitensis and 6 other Brucella species samples) were run at concentrations decreasing from 1 ng/μl to 1 fg/μl. (A) Amplification curves for B. melitensis samples run in duplicate. (B) Allelic discrimination plots for all samples. Samples at the top left are B. melitensis and at the bottom right are other Brucella spp., and at the bottom left, squares near the plot origin are no-template controls (NTCs; n = 8). Numbers on the axes represent degrees of differentiation of points and are based on the threshold values of the reactions.
DISCUSSION
The taxonomic description of the Brucella species has been accomplished using a broad range of microbiological and molecular approaches. In a clinical setting, working with Brucella requires the handling of highly infectious agents, expertise with culturing bacteria on a variety of media, and at least 5 to 7 days under standard microbiological practices. Current molecular genetic techniques for species-level identification are technically challenging and/or are limited to only a few species. Thus, development of quick and reliable methods for identifying Brucella species from limited amounts of DNA is crucial for today's clinical and epidemiological applications. Furthermore, determination of the Brucella species will allow implementation of appropriate public health interventions in a timely manner.
Our real-time PCR assays provide rapid identification of the seven major Brucella clades: B. abortus, B. melitensis, B. canis, B. suis/B. canis, B. neotomae, B. ovis, and Brucella in marine mammals. SNPs defining B. suis exclusively have yet to be found, but this species can be identified using the B. suis/B. canis assay to assign a sample to this clade and a B. canis assay to rule out that it is B. canis. Each assay that we have presented is binary within the Brucella genus; that is, either the sample is in a particular Brucella clade or it is one of the other Brucella species. One of the closest known relatives, Ochrobactrum anthropi, served as a good negative control and never amplified as the primary allele in any assay. Although the strains used were biochemically similar to those of other fastidious gram-negative coccobacilli, such as Bordetella bronchiseptica, Oligella ureolytica, or other Brucella “mimics” (5), GenBank BLAST searches of the real-time PCR sequences failed to produce significant homology to any genus besides Brucella. Thus, it is very unlikely that broader specificity testing with other bacteria would amplify the primary allele in any of the assays.
The consistent sensitivity of the assays at 10 fg/μl or less represents detection of less than three genome equivalents of DNA. In fact, most of our assays detected DNA at concentrations of 1 fg/μl (less than a genome's equivalent), which is close to the theoretical limit of detection. Similar results have been achieved with B. abortus assays with detections of roughly two genome copies of DNA (25). At extremely low concentrations of DNA, a failure of at least 37% of reactions is expected based on Poisson statistics and the likelihood of sampling error (14). Low-level detectability is essential for forensic applications, as well as for environmental sampling and clinical detection. Detecting minute quantities in blood is important for clinical work relating to Brucella infections, where small amounts of bacteria may be circulating in the blood. Diagnostic PCR assays from blood must deal with PCR inhibitors such as heme and/or leukocyte DNA, but this can be alleviated through specific lysis and washing procedures (24). We believe our assays can be modified and incorporated into clinical tests to detect and subtype Brucella DNAs from human or animal blood samples. Because a false negative could have serious consequences regarding laboratory exposure, when running our assays to test for the presence of Brucella, a 16S rRNA control should be included to confirm the presence of bacterial DNA.
Sequencing of whole genomes, housekeeping genes for MLST, and specific other loci have provided the SNPs necessary for the differentiation of various Brucella species. To increase our confidence that the SNPs chosen could accurately place each isolate into its appropriate species or clade, assays were tested against a large and diverse collection of 338 isolates. Each additional sample that is screened increases the confidence in the assay. Providing that the assays successfully amplify each allele with minimal cross-hybridization of probes, SNP-based real-time PCR assays are a very reliable method for differentiating species. The conserved distribution of the SNPs used in this study with a relatively large collection of diverse Brucella subtypes suggests that these sites are nonhomoplastic. This idea is supported by previous studies that indicate sparse mutation density and rare recombination events and suggests a primary clonal population structure for brucellae (16, 27, 38). The conserved Brucella phylogeny is reminiscent of the extensively characterized Bacillus anthracis genome, the status of 1,000 SNPs in 27 diverse isolates revealed only a single homoplastic event (33). A more definitive estimate of the extent of homoplasy in Brucella organisms can eventually be obtained by similar comparative genomic analysis and SNP discovery in the Brucella genomes. The hypothesis that a limited number of SNPs (canonical SNPs) along a conserved phylogenetic branch can represent a large subset of SNPs in B. anthracis (19, 28, 33) appears to be similar to the scenario for Brucella, for which limited but powerful SNP-based assays provide a strong phylogenetic framework and, combined with MLVA, provide a highly resolved genotyping scheme across a broad spectrum of isolates. Again, SNPs resolve the major Brucella species, and MLVA provides finer-scale resolution and genotyping that can be used for epidemiological purposes (1a).
Our samples may or may not have contained isolates from the biovars B. abortus biovar 3 (Tulya) and B. suis biovars 3 and 5. Of these, B. suis biovar 5 represents the most likely challenge to our B. suis/B. canis assay because strong differentiation of this biovar from other B. suis species has been shown by both MLVA (21, 39) and MLST (38). These papers also suggest that B. suis is the most diverse clade within the Brucella genus. Furthermore, finding SNPs that separate B. canis from B. suis is challenging due to a high degree of sequence homology that indicates a recent split between these species.
In conclusion, we present an efficient method of determining all currently recognized Brucella species that should have broad clinical and forensic applications. Furthermore, samples can be distinguished near the limits of detection. Multiplexing of reactions appears possible in the future so that discrimination of Brucella can be achieved with one test. Nonetheless, in many areas of the world, the Brucella species that cause infection are likely known, and this test will rapidly confirm species identification.
Acknowledgments
We thank numerous contributors of DNA to our Brucella collection, including Bryan Bellaire, Robert Burgess, Ted Hadfield, Wally Buchholz, Brian Bell, and William Slanta. Tom Brettin of the Los Alamos National Laboratory allowed access to the unpublished B. canis genome. James Schupp, Judy Lee, Dawn Birdsell, Jodi Beaudry, Ray Auerbach, Shalamar Georgia, and Tal Pearson provided invaluable technical support.
Funding was provided by an HSARPA grant to Northern Arizona University by the U.S. Department of Homeland Security Science and Technology Directorate.
Footnotes
Published ahead of print on 21 November 2007.
REFERENCES
- 1.Bricker, B. J. Molecular diagnostics of animal brucellosis: a review of PCR-based assays and approaches, p. 24-51. In I. López-Goñi and I. Moriyón (ed.), Brucella: molecular and cellular biology. Horizon Bioscience, Norfolk, United Kingdom.
- 1a.Bricker, B. J., D. R. Ewalt, and S. M. Halling. 2003. Brucella “HOOF-Prints”: strain typing by multi-locus analysis of variable number tandem repeats (VNTRs). BMC Microbiol. 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bricker, B. J., D. R. Ewalt, A. P. MacMillan, G. Foster, and S. Brew. 2000. Molecular characterization of Brucella strains isolated from marine mammals. J. Clin. Microbiol. 381258-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bricker, B. J., and S. M. Halling. 1994. Differentiation of Brucella abortus bv. 1, 2, and 4, Brucella melitensis, Brucella ovis, and Brucella suis bv. 1 by PCR. J. Clin. Microbiol. 322660-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chain, P. S. G., D. J. Comerci, M. E. Tolmasky, F. W. Larimer, S. A. Malfatti, L. M. Vergez, F. Aguero, M. L. Land, R. A. Ugalde, and E. Garcia. 2005. Whole-genome analyses of speciation events in pathogenic brucellae. Infect. Immun. 738353-8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu, M., and R. S. Weyant. 2003. Brucella. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed., vol. I. American Society for Microbiology, Washington, DC.
- 6.Cloeckaert, A., M. Grayon, O. Grepinet, and K. S. Boumedine. 2003. Classification of Brucella strains isolated from marine mammals by infrequent restriction site-PCR and development of specific PCR identification tests. Microbes Infect. 5593-602. [DOI] [PubMed] [Google Scholar]
- 7.Cloeckaert, A., J. M. Verger, M. Grayon, and O. Grepinet. 1995. Restriction site polymorphism of the genes encoding the major 25 Kda and 36 Kda outer-membrane proteins of Brucella. Microbiology 1412111-2121. [DOI] [PubMed] [Google Scholar]
- 8.Cloeckaert, A., J. M. Verger, M. Grayon, J. Y. Paquet, B. Garin-Bastuji, G. Foster, and J. Godfroid. 2001. Classification of Brucella spp. isolated from marine mammals by DNA polymorphism at the omp2 locus. Microbes Infect. 3729-738. [DOI] [PubMed] [Google Scholar]
- 9.Cloeckaert, A., J. M. Verger, M. Grayon, M. S. Zygmunt, and O. Grepinet. 1996. Nucleotide sequence and expression of the gene encoding the major 25-kilodalton outer membrane protein of Brucella ovis: evidence for antigenic shift, compared with other Brucella species, due to a deletion in the gene. Infect. Immun. 642047-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cloeckaert, A., and N. Vizcaino. 2004. DNA polymorphism and taxonomy of Brucella species, p. 1-24. In I. Lopez-Goni and I. Moriyon (ed.), Brucella: molecular and cellular biology. Horizon Bioscience, Norfolk, United Kingdom.
- 11.Code of Federal Regulations, U.S. 2005. Multiple parts: 7 CFR Part 331, 9 CFR Part 121, and 42 CFR Part 73.
- 12.Corbel, M. J., and W. J. Brinley-Morgan. 1984. Genus Brucella Meyer and Shaw 1920, 173AL. Williams and Wilkins, Baltimore, MD.
- 13.DelVecchio, V. G., V. Kapatral, R. J. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, I. Anderson, A. Bhattacharyya, A. Lykidis, G. Reznik, L. Jablonski, N. Larsen, M. D'Souza, A. Bernal, M. Mazur, E. Goltsman, E. Selkov, P. H. Elzer, S. Hagius, D. O'Callaghan, J. J. Letesson, R. Haselkorn, N. Kyrpides, and R. Overbeek. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diaco, R. 1995. Practical considerations for the design of quantitative PCR assays, p. 84-108. In M. A. Innis, D. H. Gelfand, and J. J. Sninsky (ed.), PCR strategies. Academic Press, New York, NY.
- 15.Gee, J. E., B. K. De, P. N. Levett, A. M. Whitney, R. T. Novak, and T. Popovic. 2004. Use of 16S rRNA gene sequencing for rapid confirmatory identification of Brucella isolates. J. Clin. Microbiol. 423649-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halling, S. M., B. D. Peterson-Burch, B. J. Bricker, R. L. Zuerner, Z. Qing, L. L. Li, V. Kapur, D. P. Alt, and S. C. Olsen. 2005. Completion of the genome sequence of Brucella abortus and comparison to the highly similar genomes of Brucella melitensis and Brucella suis. J. Bacteriol. 1872715-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huynh, L. Y., M. N. Van Ert, T. Hadfield, W. S. Probert, B. H. Bellaire, M. Dobson, R. J. Burgess, R. S. Weyant, T. Popovic, S. Zanecki, D. M. Wagner, and P. Keim. Multiple locus variable number tandem repeat (VNTR) analysis (MLVA) of Brucella spp. identifies species-specific markers and provides insights into phylogenetic relationships. In V. St. Georgiev (ed.), Frontiers in research, in press. Humana Press, Totowa, NJ.
- 18.Jahans, K. L., G. Foster, and E. S. Broughton. 1997. The characterization of Brucella strains isolated from marine mammals. Vet. Microbiol. 57373-382. [DOI] [PubMed] [Google Scholar]
- 19.Keim, P., M. N. Van Ert, T. Pearson, A. J. Vogler, L. Y. Huynh, and D. M. Wagner. 2004. Anthrax molecular epidemiology and forensics: using the appropriate marker for different evolutionary scales. Infect. Genet. Evol. 4205-213. [DOI] [PubMed] [Google Scholar]
- 20.Leal-Klevezas, D. S., A. Lopez-Merino, and J. P. Martinez-Soriano. 1995. Molecular detection of Brucella spp.: rapid identification of B. abortus biovar 1 using PCR. Arch. Med. Res. 26263-267. [PubMed] [Google Scholar]
- 21.Le Fleche, P., I. Jacques, M. Grayon, S. Al Dahouk, P. Bouchon, F. Denoeud, K. Nockler, H. Neubauer, L. A. Guilloteau, and G. Vergnaud. 2006. Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC Microbiol. 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marianelli, C., F. Ciuchini, M. Tarantino, P. Pasquali, and R. Adone. 2006. Molecular characterization of the rpoB gene in Brucella species: new potential molecular markers for genotyping. Microbes Infect. 8860-865. [DOI] [PubMed] [Google Scholar]
- 23.Moorhead, S. M., G. A. Dykes, and R. T. Cursons. 2003. An SNP-based PCR assay to differentiate between Listeria monocytogenes lineages derived from phylogenetic analysis of the sigB gene. J. Microbiol. Methods 55425-432. [DOI] [PubMed] [Google Scholar]
- 24.Morata, P., M. I. Quiepo-Ortuno, and J. Dios Colmenero. 1998. Strategy for optimizing DNA amplification in a peripheral blood PCR assay used for diagnosis of human brucellosis. J. Clin. Microbiol. 362443-2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newby, D. T., T. L. Hadfield, and F. F. Roberto. 2003. Real-time PCR detection of Brucella abortus: a comparative study of SYBR green I, 5′-exonuclease, and hybridization probe assays. Appl. Environ. Biol. 694753-4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pappas, G., P. Papadimitriou, N. Akritidis, L. Christou, and E. V. Tsianos. 2006. The new global map of human brucellosis. Lancet Infect. Dis. 691-99. [DOI] [PubMed] [Google Scholar]
- 27.Paulsen, I. T., R. Seshadri, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, T. D. Read, R. J. Dodson, L. Umayam, L. M. Brinkac, M. J. Beanan, S. C. Daugherty, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, W. C. Nelson, B. Ayodeji, M. Kraul, J. Shetty, J. Malek, S. E. Van Aken, S. Riedmuller, H. Tettelin, S. R. Gill, O. White, S. L. Salzberg, D. L. Hoover, L. E. Lindler, S. M. Halling, S. M. Boyle, and C. M. Fraser. 2002. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. USA 9913148-13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearson, T., J. D. Busch, J. Ravel, T. D. Read, S. D. Rhoton, J. M. U'Ren, T. S. Simonson, S. M. Kachur, R. R. Leadem, M. L. Cardon, M. N. Van Ert, L. Y. Huynh, C. M. Fraser, and P. Keim. 2004. Phylogenetic discovery bias in Bacillus anthracis using single-nucleotide polymorphisms from whole-genome sequencing. Proc. Natl. Acad. Sci. USA 10113536-13541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Probert, W. S., M. N. Schrader, N. Y. Khuong, S. L. Bystrom, and M. H. Graves. 2004. Real-time multiplex PCR assay for detection of Brucella spp., B. abortus, and B. melitensis. J. Clin. Microbiol. 421290-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ratushna, V. G., D. M. Sturgill, S. Ramamoorthy, S. A. Reichow, Y. Q. He, R. Lathigra, N. Sriranganathan, S. M. Halling, S. M. Boyle, and C. J. Gibas. 2006. Molecular targets for rapid identification of Brucella spp. BMC Microbiol. 613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Redkar, R., S. Rose, B. Bricker, and V. DelVecchio. 2001. Real-time detection of Brucella abortus, Brucella melitensis and Brucella suis. Mol. Cell. Probes 1543-52. [DOI] [PubMed] [Google Scholar]
- 32.U'Ren, J. M., M. N. Van Ert, J. M. Schupp, W. R. Easterday, T. S. Simonson, R. T. Okinaka, T. Pearson, and P. Keim. 2005. Use of a real-time PCR TaqMan assay for rapid identification and differentiation of Burkholderia pseudomallei and Burkholderia mallei. J. Clin. Microbiol. 435771-5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Ert, M. N., W. R. Easterday, L. Y. Huynh, R. T. Okinaka, M. E. Hugh-Jones, J. Ravel, S. R. Zanecki, T. Pearson, T. S. Simonson, J. M. U'Ren, S. M. Kachur, R. R. Leadem-Dougherty, S. D. Rhoton, G. Zinser, J. Farlow, P. R. Coker, K. L. Smith, B. Wang, L. J. Kenefic, C. M. Fraser-Liggett, D. M. Wagner, and P. Keim. 2007. Global genetic population structure of Bacillus anthracis. PLoS ONE 2e461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Ert, M. N., W. R. Easterday, T. S. Simonson, J. M. U'Ren, T. Pearson, L. J. Kenefic, J. D. Busch, L. Y. Huynh, M. Dukerich, C. B. Trim, J. Beaudry, A. Welty-Bernard, T. Read, C. M. Fraser, J. Ravel, and P. Keim. 2007. Strain-specific single-nucleotide polymorphism assays for the Bacillus anthracis Ames strain. J. Clin. Microbiol. 4547-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verger, J. M., F. Grimont, P. A. D. Grimont, and M. Grayon. 1985. Brucella, a monospecific genus as shown by deoxyribonucleic acid hybridization. Int. J. Syst. Bacteriol. 35292-295. [Google Scholar]
- 36.Vizcaino, N., P. Caro-Hernandez, A. Cloeckaert, and L. Fernandez-Lago. 2004. DNA polymorphism in the omp25/omp31 family of Brucella spp.: identification of a 1.7-kb inversion in Brucella cetaceae and of a 15.1-kb genomic island, absent from Brucella ovis, related to the synthesis of smooth lipopolysaccharide. Microbes Infect. 6821-834. [DOI] [PubMed] [Google Scholar]
- 37.Whatmore, A. M., T. J. Murphy, S. Shankster, E. Young, S. J. Cutler, and A. P. MacMillan. 2005. Use of amplified fragment length polymorphism to identify and type brucella isolates of medical and veterinary interest. J. Clin. Microbiol. 43761-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whatmore, A. M., L. L. Perrett, and A. P. MacMillan. 2007. Characterisation of the genetic diversity of Brucella by multilocus sequencing. BMC Microbiol. 734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whatmore, A. M., S. J. Shankster, L. L. Perrett, T. J. Murphy, S. D. Brew, R. E. Thirlwall, S. J. Cutler, and A. P. MacMillan. 2006. Identification and characterization of variable-number tandem-repeat markers for typing of Brucella spp. J. Clin. Microbiol. 441982-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]