Abstract
The aim of this study was to identify the endogenous retrovirus (ERV) sequences in a bovine genome. We subjected bovine genomic DNA to PCR with degenerate or ovine ERV (OERV) family-specific primers that aimed to amplify the retroviral pro/pol region. Sequence analysis of 113 clones obtained by PCR revealed that 69 were of retroviral origin. On the basis of the OERV classification system, these clones from degenerate PCR could be divided into the β3, γ4, and γ9 families. PCR with OERV family-specific primers revealed an additional ERV that was classified into the bovine endogenous retrovirus (BERV) γ7 family. In conclusion, here we report the results of a genome scale study of the BERV. Our study shows that the ERV family expansion in cattle may be somewhat limited, while more diverse family members of ERVs have been reported from other artiodactyls, such as pigs and sheep.
The endogenous retroviruses (ERVs) in mammals are classified into the retroviral β (B-/D-type) and γ (C-type) genera (3, 20). To date, analyses of the ERVs in the genomes of several mammals, including humans, pigs, and sheep, have revealed the presence of multiple different families (1, 2, 4, 5, 7, 10, 11, 13, 15, 16, 17, 19). For example, in the sheep genome, 12 different ovine ERV (OERV) families were detected. These were classified into three β families (β1 to β3) and nine γ families (γ1 to γ9) (11). However, except for one pro/pol sequence from murine leukemia virus-related retrovirus (MLVRT1-BoEV) (19), no other sequence information regarding ERVs residing in the cattle genome is available.
It is known that human ERVs (HERVs) comprise approximately 8% of the human genome (6). Thus, the identification of the ERVs in the bovine genome will be helpful for the annotation of the bovine genome; it will also improve our understanding of ERVs. We searched the bovine genome for the conserved pro/pol nucleotide sequences of ERVs by PCR using degenerate primers, which contain the active site motifs DTGA of protease (PR) protein and YMDD or YVDD of reverse transcriptase (RT) protein (7, 10, 11, 18). Since we found that all ERV clones obtained by degenerate PCR were closely related to three specific OERV families, we also subjected the bovine genome to PCR using OERV family-specific primers.
Degenerate PCR amplification of BERV sequences.
We isolated the genomic DNA from one Korean Yellow native female cow (Bos taurus) by a simple lysis method (14). The template DNA was subjected to PCR using Taq polymerase and six pairs of degenerate primers previously used for successful amplification of ERVs in pigs, sheep, and other vertebrates (7, 10, 11, 19). The two 5′ and three 3′ oligonucleotides, which consist of six pairs of primers, are 5′ primers against PR protein active site motifs, 5′-GT(T/G) TTI (G/T)TI GA(T/C) ACI GGI (G/T)C-3′ and 5′-(C/T)TI (T/G)TI GA(T/C) ACI GGI GCI (G/C)I-3′; and 3′ primers against RT protein active site motifs, 5′-AGI AGG TC(A/G) TCI AC(A/G) TA(C/G) TG-3′, 5′-ATI AGI A(G/T)(A/G) TC(A/G) TCI AC(A/G) TA-3′, and 5′-ATI AGI A(G/T)(A/G) TC(A/G) TCC AT(A/G) TA-3′ (I stands for inosine). PCRs consisted of 2 min at 80°C, followed by 35 cycles of a 45°C annealing step for 30 s, extension at 74°C for 60 s, denaturation at 94°C for 30 s, and finally, one cycle at 45°C for 3 min and 74°C for 10 min. Reaction conditions were as follows: 40 pmol of each primer, 200 μM deoxynucleoside triphosphates, PCR buffer (10 mM Tris [pH 8.3], 50 mM KCl, 1.5 mM MgCl2), 100 ng of genomic DNA, and 2 U of Taq polymerase in a 25-μl reaction volume (7, 18, 19). The amplified fragments were separated on 1.3% agarose gels, extracted, and cloned into the pCRII vector (Invitrogen, Carlsbad, CA). Cloned inserts were sequenced in both directions with an ABI Prism BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems) on ABI 3700 automated sequencers (Applied Biosystems). Sequencing results were analyzed by BLAST searches against the nr database at NCBI (http://www.ncbi.nlm.nih.gov/BLAST/) and the bovine genome database at Baylor College of Medicine (http://www.hgsc.bcm.tmc.edu/blast.hgsc). Full sequences of clone inserts from the PR-RT region were assembled by overlapping forward and reverse sequencing products. Ambiguous sequences or unique polymorphisms were confirmed by resequencing the region in question using the same clone. Of the 105 unique sequences, 64 (61%) of a length of 0.7 to 1.0 kb contained the retroviral PR-RT characteristic motifs and showed significant matches (expected [E] value, <10−10) with other ERVs from GenBank BLAST analysis, indicating that these sequences were of retroviral origin. Among the 68 clones from retroviral origin, 8 clones of four different pairs showed redundancy, thereby indicating a low rate of PCR errors in the amplification process, as reported previously (17).
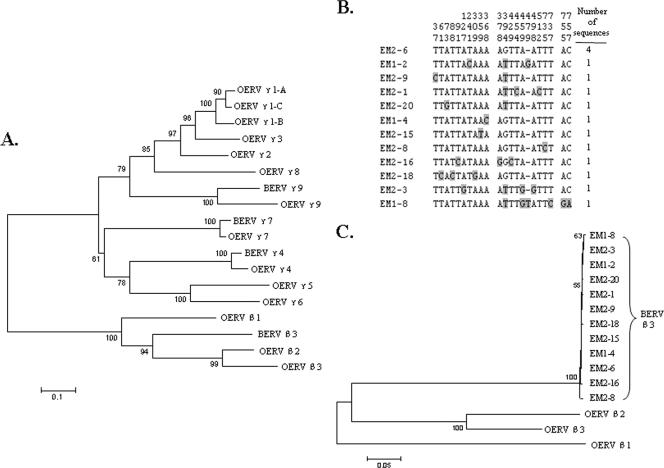
We then compared the BERV sequences to the pro/pol regions of the OERV families. The neighbor-joining tree constructed with the BERV and OERV nucleotide sequences using the MEGA (version 3.1) package (12) revealed that 52 of the 64 BERV clones from degenerate PCR could be classified into the BERV γ family, while the remaining 12 clones could be classified into the BERV β family (Fig. 1A). More specifically, since the 12 BERV clones clustered most closely with the OERV β3 family (Fig. 1C), we classified the 12 clones as belonging to the BERV β3 family. The 52 BERV γ family clones were classified into the BERV γ4 and γ9 families. We identified 370 bovine bacterial artificial chromosome clones containing the cloned pro/pol sequences by BLAST search against the bovine genome database. When the flanking regions of the matched pro/pol sequences within the available bacterial artificial chromosome sequences were analyzed, long terminal repeat, gag, and env genes, which are the characteristic of ERV genomic structure (3, 5), were identified (21). The pro/pol clones, having the fewest nonsense mutations and being most closely related to potentially infectious ERV (17), were selected as representatives of each family and deposited into GenBank. Nomenclature of the families was carried out according to the results of studies of sheep (11).
FIG. 1.
Analysis of the pro/pol nucleotide sequences of the 12 BERV β3 clones. (A) Classification of the BERV β3 and γ families relative to the pro/pol nucleotide sequences of the OERV β and γ families. A neighbor-joining tree was constructed using 65 BERV and 14 OERV sequences. The BERV γ4, γ7, and γ9 families cluster closely with the corresponding OERV γ families, while the BERV β3 family is closely related to the OERV β2 and β3 families. The nucleotide sequences of OERV families used here have the following GenBank accession numbers: AY193896 (OERV γ1A), AY193898 (OERV γ1B), AY193899 (OERV γ1C), AY193900 (OERV γ2), AY193903 (OERV γ3), AY193905 (OERV γ5), AY193906 (OERV γ6), and AY193908 (OERV γ8). (B) Comparison of the 22 nucleotides containing variable bases among 12 BERV β3 790-bp pro/pol sequences. Individual nucleotides that are polymorphic are shown in shades, and the nucleotide positions containing variable bases in the sequence are indicated at the top. The names of the amplified BERV β3 clones are marked on the left. (C) Relationship of the 12 BERV β3 sequences to the three OERV β families. A neighbor-joining tree was constructed using the pro/pol nucleotide sequences of the 12 BERV β clones and the three OERV β families (β1, accession no. AY266332; β2, accession no. AY193894; and β3, accession no. AY193895). Bootstrap values of above 50 from 500 replicates are indicated at the branch nodes.
Characterization of the BERV β3 clones.
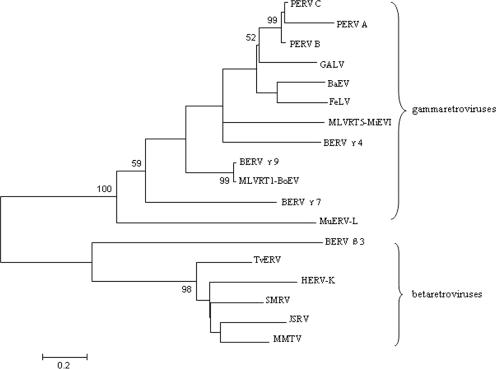
We then compared the 12 BERV β3 pro/pol clones. Of the 790 bp examined, only 22 nucleotide positions (2.8%) were variable (Fig. 1B). In contrast, of 51 BERV γ4 pro/pol sequences with 933 bp examined, 508 nucleotides positions (54.4%) were variable (data not shown), which indicates that the β3 family is less variable than the γ4 family, although the sequence lengths and identified numbers of β3 clones are less than those of the γ4 family. The BERV β3 clones clustered separately from the OERV β families and converged into a single family in the neighbor-joining tree (Fig. 1C). Phylogenetic analysis using the Pro/Pol amino acid sequences from multiple retroviruses showed that the BERV β3 family clustered with the betaretroviruses (Fig. 2). In contrast to BERV γ4, the lower intersequence variation of BERV β3 is consistent with the identification of a single β family member in the bovine genome from our analysis.
FIG. 2.
Phylogenetic analysis of previously identified retroviruses and BERVs based on the amino acid sequences of the pro/pol region using the neighbor joining method. Numbers at the branch nodes denote the bootstrap values (>50) from 500 replicates. The BERV β and γ families clustered with betaretroviruses (jaagsiekte sheep retrovirus [JSRV], mouse mammary tumor virus [MMTV], simian sarcoma virus [SMRV], Trichosurus vulpecula retrovirus [TvERV], and HERV-K) and gammaretroviruses (PERV-A, PERV-B, PERV-C, gibbon ape leukemia virus [GALV], feline leukemia virus [FeLV], baboon endogenous virus [BaEV], MLVRT1-BoEV, and MLVRT5-MiEVI), respectively. The viruses and GenBank accession numbers of the sequences used in the phylogenetic tree are as follows: JSRV, AAA89182; MMTV, AAA46542; SMRV, AAA66453; TvERV, AAF36395; HERV-K, CAB56603; and murine ERV-L (MuERV-L), CAA73251.
Characterization of the BERV γ4 clones.
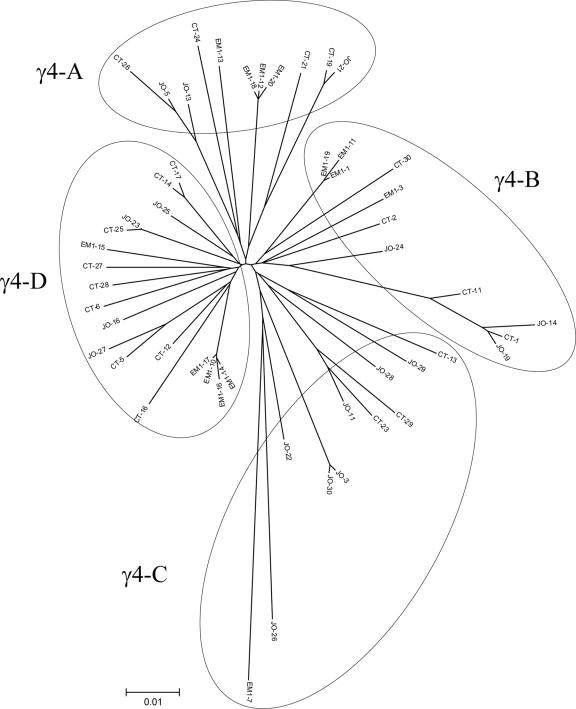
Of the 57 BERV γ family clones, 51 were classified as members of the BERV γ4 family as they showed more than 90% homology to the OERV γ4 pro/pol sequence (GenBank accession no. AY193904). Phylogenetic analysis indicated that these clones could be assigned to four distinct subfamilies, namely, γ4-A, γ4-B, γ4-C, and γ4-D (11, 11, 11, and 18 clones, respectively) (Fig. 3).
FIG. 3.
Phylogenetic analysis of the BERV γ4 pro/pol nucleotide sequences. The tree was created by using the neighbor-joining method with 600 bootstrap replicates. The genetic distance (0.01) is defined. BERV γ4 sequences were further classified into the γ4-A, γ4-B, γ4-C, and γ4-D subfamilies.
Phylogenetic analysis of the Pro/Pol amino acid sequences from multiple retroviruses showed that the BERV γ4 family was grouped with porcine ERV (PERV)-A (GenBank accession no. AAL87853), PERV-B (CAB65341), PERV-C (AAC16764), gibbon ape leukemia virus (accession no. AAA46810), feline leukemia virus (accession no. AAA93092), baboon endogenous virus strain M7 (accession no. BAA89659), and a murine leukemia-related virus strain (MLVRT5-MiEVI; accession no. X99928) (Fig. 2). The result of the phylogenetic analysis on the respective pro/pol nucleotide sequences was also consistent with that of amino acids (data not shown).
Characterization of the BERV γ9 clone.
The remaining clone belonged to the BERV γ9 family because it showed 74% and 62% homology with the OERV γ9 pro/pol nucleotide and amino acid sequences, respectively (GenBank accession no. AY193909). Upon comparison with the amino acid sequences of the pro/pol region from multiple retroviruses, the BERV γ9 clone matched that of murine leukemia virus-related retrovirus MLVRT1-BoEV (GenBank accession no. X99924), with 99% nucleotide identity. Endogenous MLVRT1-BoEV has also been shown to have a close relationship with OERV γ OvEVII (GenBank accession no. X99932), indicating the presence of strong sequence similarity between ERVs in cattle and sheep (19).
Identification of an additional BERV family by PCR using OERV family-specific primers.
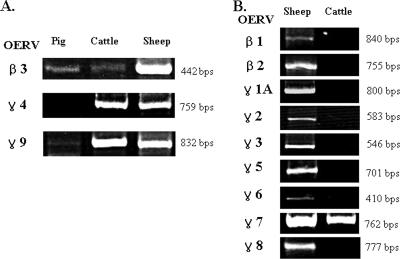
To confirm the results of degenerate PCR analysis, family-specific primers of ERVs were designed and genomic DNAs from pigs, cattle, and sheep, which belong to the mammal order Artiodactyla, were subjected to PCR using the primers. As shown in Fig. 4A, PCR with degenerate primers revealed that the BERV families were highly homologous to OERVs. However, either very weak or no amplification resulted from PCR using pig DNA. Consequently, we speculated that we might be able to identify more BERV families by amplifying the bovine genome with primers that are specific for the pro/pol sequences of the OERV β1 and β2, γ1A to γ3, and γ5 to γ8 families. Thus, nine pairs of primers with slight degeneracy in their 3′ ends were designed. Each PCR mixture contained 50 ng genomic DNA, 4.5 pmol each primer, and 25 μM deoxynucleoside triphosphate. The quality of genomic DNA was confirmed by efficient PCR amplification with a control primer. Only the OERV γ7 (GenBank accession no. AY193907) primers generated a specific PCR product (Fig. 4B). Five clones from this product were sequenced. Sequence analysis revealed that they had 94% nucleotide identity to OERV γ7 and also clustered closely with OERV γ7 in phylogenetic analysis (Fig. 1A). Thus, the bovine genome also contains a BERV γ7 family ERV.
FIG. 4.
Detection of a new ERV family in the bovine genome by OERV family-specific PCR amplification. (A) ERVs detected in porcine, bovine, and ovine genomes by degenerate PCR amplifying the pro/pol region. The β3 primers were designed according to sequence homology between pig and sheep ERVs, while the γ4 and γ9 primers were designed according to sequence homology between cattle and sheep. Pig genomic DNA was not amplified and weakly amplified by the γ4-specific (γ4-sb) and γ9-specific (γ9-sb) primers, respectively, due to nucleotide mismatches within the primer regions. The nucleotide sequences for primers were 5′-GTAGCCACTGCTCAAATTCC-3′ and 5′-GYAASACTGTCCATTGATAA-3′ for β-p5s3 (β3-specific), 5′-CTCCTCCCAAACCTGTACCA-3′ and 5′-AATACTGTCCAAGTCATCTG-3′ for γ4-sb, and 5′-AACCTGTGGCATCACTCTCN-3′ and 5′-GGAGTCCAGATGAGCTGTTN-3′ for γ9-sb. (B) PCR with OERV family-specific primers revealed a new BERV family, namely, BERV γ7. The nucleotide sequences of the family-specific primers were 5′-TTCCGTGGAGTGCTTGATAN-3′ and 5′-CTCTCCATTGATAGCGTTGN-3′ for OERV-β1, 5′-TGTTTCGGTTATTTCTCTCC-3′ and 5′-TGTCTCACAGTAATAAGAGC-3′ for OERV-β2, 5′-GATATTACTGGACTACTA-3′ and 5′-GGTACTCACAGAGATCTT-3′ for OERV-γ1A, 5′-GGCATCCTCTTGAGGTCTTN-3′ and 5′-CGAGTCCATGTCAGCTGCAN-3′ for OERV-γ2, 5′-TATAGACTGTACCAGCCGAN-3′ and 5′-GAGGTGAGTCCAGGTTAGTN-3′ for OERV-γ3, 5′-GGTATAGATGGCACGTCTTN-3′ and 5′-AGGACTGTCCACGTTAGCTN-3′ for OERV-γ5, 5′-GGCCTTCCTAATGCTATTGN-3′ and 5′-CCTTGAGGTCAATAACGGTN-3′ for OERV-γ6, 5′-TGACTTCTCTGTTCTTCCTT-3′ and 5′-TGTTCCCAGGTCCCACCACT-3′ for OERV-γ7, and 5′-CCGAAGATCAGTGGACTTGN-3′ and 5′-TCCTCATCGAAGATGGTTGN-3′ for OERV-γ8.
Identification of BERVs from the bovine genome database.
Since multiple ERV families have been reported for the pig and sheep genomes (10, 11, 17), we examined whether the current Btau_3.1 bovine genome assembly with approximately 7.1× coverage (an approximately 95% genome representation rate) (ftp://ftp.hgsc.bcm.tmc.edu/pub/data/Btaurus/fasta/Btau20060815-freeze/ReadMeBovine.3.1.txt) contains sequences that are homologous to the pro/pol sequences of all known OERVs and PERVs. The bovine genome assembly used for bioinformatics analysis comes from DNA of a Hereford breed animal. Apart from the β3, γ4, γ7, and γ9 families, which were all identified by our PCR analyses, other ERV sequences were not identified. This shows that the degenerate PCR approach can successfully identify diverse ERV families in vertebrates, which confirms what has been observed previously (7, 10, 11). Thus, it appears that the bovine genome may contain less diverse ERVs than those in the porcine or ovine genome.
Estimation of the copy number of BERVs.
To estimate the copy numbers of BERVs, we directly analyzed the insertion sites of BERVs in the bovine genome using the Btau_3.1 bovine genome assembly by BLAST search using the BERV γ4, γ7, γ9, and β3 pro/pol sequences and counted insertion sites for each BERV family in each chromosome. An E value of 0.0 was used as the criterion for BLAST analysis. Since the nucleotide sequence identity of the pro/pol region between different BERV families is less than 90%, insertion sites of each BERV family were clearly distinguished. With 163 insertions, the γ4 family was the most abundant (see Table S1 in the supplemental material). The BERV γ7, γ9, and β3 families had 7, 3, and 57 insertions, respectively. None of the BLAST matches were shared between different BERV families, indicating that the results are specific for each family. BERVs are almost evenly distributed through the genome, depending on the length of the chromosome (see Table S1 in the supplemental material). Considering that the current bovine genome assembly covers 95% of the cow genome, a total of 242 BERV insertion sites (230/0.95) could be present in the genome of the reference animal.
In pigs, the residing ERV families and copy numbers in the genome were consistent across breeds (10). Similarly, our experimental analysis of a Korean native cow and our bioinformatics analysis of a Hereford cow genome showed consistent patterns in residing ERV families in the genome. In fact, the genetic distances based on mitochondrial DNA D-loop sequences among European, Japanese, and Korean native cattle were not much different from their intrapopulation distances (9). However, the pigs indigenous to Asia, including China, Korea, and Japan, are similar in their mitochondrial sequences, but different from European-type pigs (8). Interestingly, unlike ERVs in other artiodactyls, such as pigs and sheep, the ERV family expansion in cattle may be somewhat limited since only four different BERV families were identifiable in our study.
Nucleotide sequence accession numbers.
The GenBank accession numbers of representatives of the BERV families are DQ889607 for BERV β3, DQ889608 for γ4-A, DQ889609 for γ4-B, DQ889610 for γ4-C, DQ889611 for γ4-D, DQ889612 for γ7, and DQ889613 for γ9.
Supplementary Material
Acknowledgments
This work was supported by grants (20050301-034-467-006-03-00 and 20070401034029) from the Biogreen 21 Program, Rural Development Administration, Republic of Korea.
Footnotes
Published ahead of print on 24 October 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Akiyoshi, D. E., M. Denaro, H. Zhu, J. L. Greenstein, P. Banerjee, and J. A. Fishman. 1998. Identification of a full-length cDNA for an endogenous retrovirus of miniature swine. J. Virol. 724503-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baillie, G. J., L. N. van de Lagemaat, C. Baust, and D. L. Mager. 2004. Multiple groups of endogenous betaretroviruses in mice, rats, and other mammals. J. Virol. 785784-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coffin, J. M., S. H. Hughes, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 4.Ericsson, T., B. Oldmixon, J. Blomberg, M. Rosa, C. Patience, and G. Andersson. 2001. Identification of novel porcine endogenous beta retrovirus sequences in miniature swine. J. Virol. 752765-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gifford, R., and T. Michael. 2003. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes 26291-315. [DOI] [PubMed] [Google Scholar]
- 6.Griffiths, D. J. 2001. Endogenous retroviruses in the human genome sequence. Gen. Biol. 21017.1-1017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herniou, E., J. Martin, K. Miller, J. Cook, M. Wilkinson, and M. Tristem. 1998. Retroviral diversity and distribution in vertebrates. J. Virol. 725955-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim, K. I., J. H. Lee, K. Li, Y. P. Zhang, S. S. Lee, J. Gongora, and C. Moran. 2002. Phylogenetic relationships of Asian and European pig breeds determined by mitochondrial DNA D-loop sequence polymorphism. Anim. Genet. 3319-25. [DOI] [PubMed] [Google Scholar]
- 9.Kim, K. I., J. H. Lee, S. S. Lee, and Y. H. Yang. 2003. Phylogenetic relationships of northeast Asian cattle to other cattle populations determined using mitochondrial DNA D-loop sequence polymorphism. Biochem. Genet. 4191-98. [DOI] [PubMed] [Google Scholar]
- 10.Klymiuk, N., M. Müller, G. Brem, and B. Aigner. 2002. Characterization of porcine endogenous retrovirus γ pro-pol nucleotide sequences. J. Virol. 7611738-11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klymiuk, N., M. Müller, G. Brem, and B. Aigner. 2003. Characterization of endogenous retroviruses in sheep. J. Virol. 7711268-11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 13.Löwer, R., J. Lower, and R. Kurth. 1996. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc. Natl. Acad. Sci. USA 935177-5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller, S. A., D. D. Dykes, and H. F. Polesky. 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 161215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palmarini, M., C. Hallwirth, D. York, C. Murgia, T. de Oliveira, T. Spencer, and H. Fan. 2000. Molecular cloning and functional analysis of three type D endogenous retroviruses of sheep reveal a different cell tropism from that of the highly related exogenous jaagsiekte sheep retrovirus. J. Virol. 748065-8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmarini, M., M. Mura, and T. E. Spencer. 2004. Endogenous betaretroviruses of sheep: teaching new lessons in retroviral interference and adaptation. J. Gen. Virol. 851-13. [DOI] [PubMed] [Google Scholar]
- 17.Patience, C., W. M. Switzer, Y. Takeuchi, D. J. Griffiths, M. E. Goward, W. Heneine, J. P. Stoye, and R. A. Weiss. 2001. Multiple groups of novel retroviral genomes in pigs and related species. J. Virol. 752771-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tristem, M. 1996. Amplification of divergent retroelements by PCR. Biotechniques 20608-612. [DOI] [PubMed] [Google Scholar]
- 19.Tristem, M., P. Kabat, L. Lieberman, S. Linde, A. Karpas, and F. Hill. 1996. Characterization of a novel murine leukemia virus-related subgroup within mammals. J. Virol. 708241-8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Regenmortel, M. H., C. M. Fauquet, D. H. L. Bishop, E. B. Carsten, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner. 2000. Virus taxonomy: the classification and nomenclature of viruses. Academic Press, San Diego, CA.
- 21.Xiao, R., J. Kim, H. Choi, K. Park, H. Lee, and C. Park. Characterization of the bovine endogenous retrovirus β3 genome. Mol. Cell, in press. [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.