Abstract
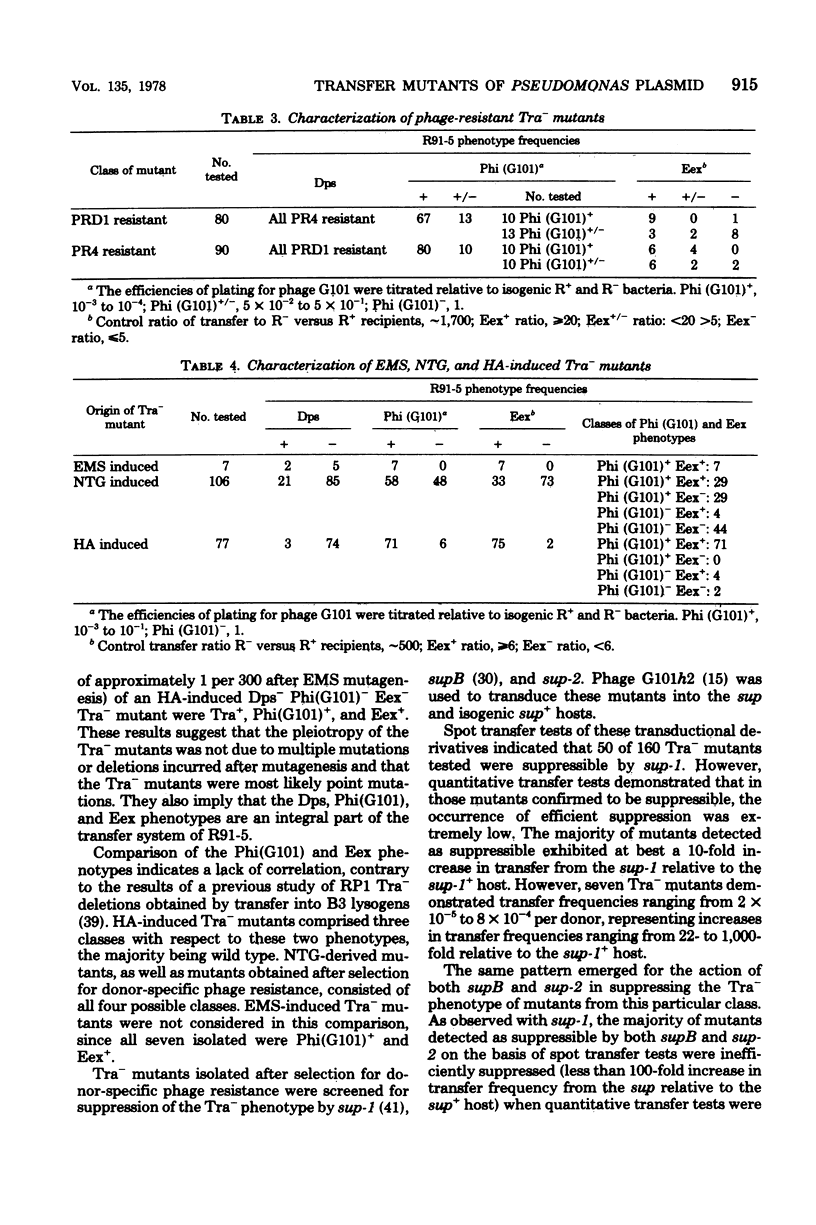
Three methods have been successful in the isolation of transfer-deficient mutants of the narrow-host-range R plasmid R91-5 of Pseudomonas aeruginosa: (i) selection for donor-specific phage resistance; (ii) direct screening after mutagenic treatment with either ethyl methane sulfonate or N-methyl-N'-nitro-N-nitrosoguanidine; (iii) in vitro mutagenesis of plasmid DNA by hydroxylamine followed by transformation and direct screening. The majority of transfer-deficient mutants were donor-specific phage resistant, supporting the view that sex pili and other surface components are essential for conjugal transfer (since the phages PRD1 and PR4 adsorb to these sites). Some of the transfer-deficient mutants were also unable to inhibit the replication of phage G101 or lost entry exclusion or both phenotypes. The ability to revert these pleiotropic mutants to wild type implicates the latter two functions in R91-5 transfer. Suppressor mutations in P. aeruginosa enabled the detection of suppressor-sensitive, transfer-deficient mutants. Such mutants should prove useful in conjugational complementation tests for the identification of the transfer cistrons of R91-5.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achtman M., Kennedy N., Skurray R. Cell--cell interactions in conjugating Escherichia coli: role of traT protein in surface exclusion. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5104–5108. doi: 10.1073/pnas.74.11.5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achtman M., Willetts N., Clark A. J. Beginning a genetic analysis of conjugational transfer determined by the F factor in Escherichia coli by isolation and characterization of transfer-deficient mutants. J Bacteriol. 1971 May;106(2):529–538. doi: 10.1128/jb.106.2.529-538.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony W. M., Deonier R. C., Lee H. J., Hu S., Otsubo E., Davidson N., Broda P. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. IX. Note on the deletion mutant of F, F delta(33-43). J Mol Biol. 1974 Nov 15;89(4):647–650. doi: 10.1016/0022-2836(74)90041-2. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J., Low K. B., Taylor A. L. Recalibrated linkage map of Escherichia coli K-12. Bacteriol Rev. 1976 Mar;40(1):116–167. doi: 10.1128/br.40.1.116-167.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth P. T., Grinter N. J., Bradley D. E. Conjugal transfer system of plasmid RP4: analysis by transposon 7 insertion. J Bacteriol. 1978 Jan;133(1):43–52. doi: 10.1128/jb.133.1.43-52.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth P. T., Grinter N. J. Map of plasmid RP4 derived by insertion of transposon C. J Mol Biol. 1977 Jul 5;113(3):455–474. doi: 10.1016/0022-2836(77)90233-9. [DOI] [PubMed] [Google Scholar]
- Bradley D. E. Adsorption of bacteriophages specific for Pseudomonas aeruginosa R factors RP1 and R1822. Biochem Biophys Res Commun. 1974 Apr 8;57(3):893–900. doi: 10.1016/0006-291x(74)90630-5. [DOI] [PubMed] [Google Scholar]
- Bradley D. E. Adsorption of the R-specific bacteriophage PR4 to pili determined by a drug resistance plasmid of the W compatibility group. J Gen Microbiol. 1976 Jul;95(1):181–185. doi: 10.1099/00221287-95-1-181. [DOI] [PubMed] [Google Scholar]
- Chandler P. M., Krishnapillai V. Characterization of Pseudomonas aeruginosa derepressed R-plasmids. J Bacteriol. 1977 May;130(2):596–603. doi: 10.1128/jb.130.2.596-603.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler P. M., Krishnapillai V. Isolation and properties of recombination-deficient mutants of Pseudomonas aeruginosa. Mutat Res. 1974 Apr;23(1):15–23. doi: 10.1016/0027-5107(74)90155-9. [DOI] [PubMed] [Google Scholar]
- Chandler P. M., Krishnapillai V. Phenotypic properties of R factors of Pseudomonas aeruginosa: R factors readily transferable between Pseudomonas and the Enterobacteriaceae. Genet Res. 1974 Jun;23(3):239–250. doi: 10.1017/s0016672300014890. [DOI] [PubMed] [Google Scholar]
- Chandler P. M., Krishnapillai V. Phenotypic properties of R factors of Pseudomonas aeruginosa: R factors transferable only in Pseudomonas aeruginosa. Genet Res. 1974 Jun;23(3):251–257. doi: 10.1017/s0016672300014907. [DOI] [PubMed] [Google Scholar]
- Freifelder D. Preparation of covalently closed and open circular DNA molecules of phage lambda. Biochim Biophys Acta. 1976 Apr 15;432(1):113–117. doi: 10.1016/0005-2787(76)90047-2. [DOI] [PubMed] [Google Scholar]
- Guerola N., Ingraham J. L., Cerdá-Olmedo E. Induction of closely linked multiple mutations by nitrosoguanidine. Nat New Biol. 1971 Mar 24;230(12):122–125. doi: 10.1038/newbio230122a0. [DOI] [PubMed] [Google Scholar]
- Haas D., Holloway B. W. R factor variants with enhanced sex factor activity in Pseudomonas aeruginosa. Mol Gen Genet. 1976 Mar 30;144(3):243–251. doi: 10.1007/BF00341722. [DOI] [PubMed] [Google Scholar]
- Hashimoto T., Sekiguchi M. Isolation of temperature-sensitive mutants of R plasmid by in vitro mutagenesis with hydroxylamine. J Bacteriol. 1976 Sep;127(3):1561–1563. doi: 10.1128/jb.127.3.1561-1563.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway B. W. Genetics of Pseudomonas. Bacteriol Rev. 1969 Sep;33(3):419–443. doi: 10.1128/br.33.3.419-443.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys G. O., Willshaw G. A., Smith H. R., Anderson E. S. Mutagenesis of plasmid DNA with hydroxylamine: isolation of mutants of multi-copy plasmids. Mol Gen Genet. 1976 Apr 23;145(1):101–108. doi: 10.1007/BF00331564. [DOI] [PubMed] [Google Scholar]
- Isaac J. H., Holloway B. W. Control of pyrimidine biosynthesis in Pseudomonas aeruginosa. J Bacteriol. 1968 Nov;96(5):1732–1741. doi: 10.1128/jb.96.5.1732-1741.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy N., Beutin L., Achtman M., Skurray R., Rahmsdorf U., Herrlich P. Conjugation proteins encoded by the F sex factor. Nature. 1977 Dec 15;270(5638):580–585. doi: 10.1038/270580a0. [DOI] [PubMed] [Google Scholar]
- Krishnapillai V. Superinfection inhibition by prophage B3 of some R plasmids in Pseudomonas aeruginosa. Genet Res. 1977 Feb;29(1):47–54. doi: 10.1017/s0016672300017109. [DOI] [PubMed] [Google Scholar]
- Krishnapillai V. The use of bacteriophages for differentiating plasmids of Pseudomonas aeruginosa. Genet Res. 1974 Jun;23(3):327–334. doi: 10.1017/s0016672300014968. [DOI] [PubMed] [Google Scholar]
- Mindich L., Cohen J., Weisburd M. Isolation of nonsense suppressor mutants in Pseudomonas. J Bacteriol. 1976 Apr;126(1):177–182. doi: 10.1128/jb.126.1.177-182.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Clowes R. C., Cohen S. N., Curtiss R., 3rd, Datta N., Falkow S. Uniform nomenclature for bacterial plasmids: a proposal. Bacteriol Rev. 1976 Mar;40(1):168–189. doi: 10.1128/br.40.1.168-189.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R. H., Siak J. S., Gray R. H. Characteristics of PRD1, a plasmid-dependent broad host range DNA bacteriophage. J Virol. 1974 Sep;14(3):689–699. doi: 10.1128/jvi.14.3.689-699.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsubo E., Nishimura Y., Hirota Y. Transfer-defective mutants of sex factors in Escherichia coli. I. Defective mutants and complementation analysis. Genetics. 1970 Feb;64(2):173–188. doi: 10.1093/genetics/64.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palchaudhuri S., Maas W. K. Physical mapping of genes on the F plasmid of Escherichia coli responsible for inhibition of growth of female-specific bacteriophages. J Bacteriol. 1977 Nov;132(2):740–743. doi: 10.1128/jb.132.2.740-743.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards G. M. Modifications of the diphenylamine reaction giving increased sensitivity and simplicity in the estimation of DNA. Anal Biochem. 1974 Feb;57(2):369–376. doi: 10.1016/0003-2697(74)90091-8. [DOI] [PubMed] [Google Scholar]
- Shahrabadi M. S., Bryan L. E., Van Den Elizen H. M. Further properties of P-2 R-factors of Pseudomonas aeruginosa and their relationship to other plasmid groups. Can J Microbiol. 1975 May;21(5):592–605. doi: 10.1139/m75-086. [DOI] [PubMed] [Google Scholar]
- Stanisich V. A., Bennett P. M. Isolation and characterisation of deletion mutants involving the transfer genes of P-group plasmids in Pseudomonas aeruginosa. Mol Gen Genet. 1976 Dec 8;149(2):211–216. doi: 10.1007/BF00332891. [DOI] [PubMed] [Google Scholar]
- Stanisich V. A., Bennett P. M., Richmond M. H. Characterization of a translocation unit encoding resistance to mercuric ions that occurs on a nonconjugative plasmid in Pseudomonas aeruginosa. J Bacteriol. 1977 Mar;129(3):1227–1233. doi: 10.1128/jb.129.3.1227-1233.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanisich V. A. The properties and host range of male-specific bacteriophages of Pseudomonas aeruginosa. J Gen Microbiol. 1974 Oct;84(2):332–342. doi: 10.1099/00221287-84-2-332. [DOI] [PubMed] [Google Scholar]
- Watson J. M., Holloway B. W. Suppressor mutations in Pseudomonas aeruginosa. J Bacteriol. 1976 Mar;125(3):780–786. doi: 10.1128/jb.125.3.780-786.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willetts N., Achtman M. Genetic analysis of transfer by the Escherichia coli sex factor F, using P1 transductional complementation. J Bacteriol. 1972 Jun;110(3):843–851. doi: 10.1128/jb.110.3.843-851.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]


