Abstract
Pseudomonas aeruginosa is a life-threatening opportunist human pathogen frequently associated with lung inflammatory diseases, namely, cystic fibrosis. Like other species, this gram-negative bacteria is increasingly drug resistant. During the past decade, intensive research efforts have been focused on the identification of natural innate defense molecules with broad antimicrobial activities, collectively known as antimicrobial peptides. Human pre-elafin, best characterized as a potent inhibitor of neutrophil elastase with anti-inflammatory properties, was also shown to possess antimicrobial activity against both gram-positive and gram-negative bacteria, including P. aeruginosa. Its mode of action was, however, not known. Using full-length pre-elafin, each domain separately, and mutated variants of pre-elafin with attenuated antipeptidase activity toward neutrophil elastase, we report here that both pre-elafin domains contribute, through distinct mechanisms, to its antibacterial activity against Pseudomonas aeruginosa. Most importantly, we demonstrate that the whey acidic protein (WAP) domain specifically inhibits a secreted peptidase with the characteristics of arginyl peptidase (protease IV). This is the first demonstration that a human WAP-motif protein inhibits a secreted peptidase to prevent bacterial growth in vitro. Since several WAP-motif proteins from various species demonstrate antimicrobial function with variable activities toward bacterial species, we suggest that this mechanism may be more common than initially anticipated.
Pseudomonas aeruginosa is an opportunistic pathogen that is life-threatening for immunocompromised individuals and for patients suffering from chronic respiratory diseases such as cystic fibrosis (CF) (10, 27). Chronic lung P. aeruginosa infection is the primary cause of morbidity and mortality in CF patients, and its prevalence increases with age. P. aeruginosa is also the predominant bacteria associated with nosocomial infections, and acute P. aeruginosa infection may result in sepsis and death (26). P. aeruginosa is characterized by the expression of numerous virulence factors, reflected by the unusually large genome of this bacteria and has developed sophisticated mechanisms to escape the host immune defenses (10, 15, 27, 33). In addition, due to low membrane permeability, active efflux of drugs, and the presence of β-lactamase activity, P. aeruginosa infections are becoming increasingly difficult to treat (17). Hence, the development of novel antimicrobial agents, preferably acting on targets exposed at the bacterial cell surface, is urgently needed.
Among the P. aeruginosa virulence factors, the secreted peptidases are thought to play a critical role in tissue invasion, nutrient accessibility, and degradation of the innate host defense molecules and therefore constitute potential therapeutic targets (18-20). Because opportunistic pathogens are taking advantage of a weakened host protective shield, augmentation therapy with lung natural defense molecules also appears to be a promising avenue for combating P. aeruginosa infections (5, 11, 44). In addition to cationic peptides known as defensins and cathelicidins, which possess broad bactericidal activity, other resident lung molecules associated with the innate and adaptive host immune system such as secretory-leukocyte-proteinase inhibitor (SLPI) and pre-elafin were also shown to possess antimicrobial action against P. aeruginosa (22, 31).
Pre-elafin is a 95-amino-acid peptide characterized by an N-terminal domain (amino acids 1 to 38) termed cementoin and a C-terminal domain (amino acids 39 to 95) with homology to the whey acidic protein (WAP) (24). It was first purified and characterized from human psoriatic skin (29, 38). In the lung, pre-elafin is expressed by alveolar macrophages and epithelial type II cells (1, 23). The WAP or elafin domain possesses antipeptidase activity toward the neutrophilic peptidases myeloblastin (formerly named proteinase 3) and neutrophil elastase. The role of the cementoin domain is not fully understood, but it may act as a substrate for tissue transglutaminase (30). The antipeptidase and anti-inflammatory properties of pre-elafin and elafin have been much studied (see references 36 and 40 for reviews). In contrast, the antimicrobial activity of this peptide has received less attention. It was initially reported that both N-terminal (pre-elafin residues 1 to 50) and C-terminal (pre-elafin residues 51 to 95) moieties of pre-elafin possess bactericidal activity against gram-negative and gram-positive bacteria (31). Using the elafin domain alone (i.e., pre-elafin residues 39 to 95), Meyer-Hoffert et al. (22) later confirmed an antimicrobial activity for this peptide against P. aeruginosa, but not on another gram-negative bacteria such as Escherichia coli. The mode of action of pre-elafin and derived-peptides against P. aeruginosa has, however, never been addressed.
In the present study, we used full-length pre-elafin, the elafin and cementoin domains alone, and mutated variants of pre-elafin with attenuated antipeptidase activity to investigate the mechanism by which pre-elafin exerts its antimicrobial activity against P. aeruginosa. We report here that while both domains of pre-elafin contribute to its antimicrobial activity, their mechanisms of action are distinct and additive. At low concentrations (i.e., <8 μM), the antimicrobial activity of pre-elafin against P. aeruginosa relies predominantly on its antipeptidase function. This activity prevents bacterial growth on complex media. The growth-inhibiting property of the elafin domain is not observed in a P. aeruginosa strain devoid of arginyl peptidase (AP; formerly named protease IV) activity and is markedly reduced when a P. aeruginosa strain with AP activity is incubated in the presence of pre-elafin mutants with attenuated antipeptidase function. We therefore conclude that while pre-elafin may exert antimicrobial activity against both gram-negative and gram-positive bacteria, as previously reported, its increased activity against P. aeruginosa appears to be directly related to the inhibition of a secreted serine-peptidase, one of the numerous virulence factors of this opportunistic pathogen.
MATERIALS AND METHODS
Bacterial and yeast strains and growth conditions.
P. aeruginosa strains ATCC 27853 and ATCC 33348, kindly provided by C. Duchaine (Université Laval) and Joseph Lam (University of Guelph), were used in all functional assays with the pre-elafin and derived peptides. These strains were routinely grown at 37°C with (250 rpm) or without agitation in peptone soy broth or, where indicated, in Mueller-Hinton medium. E. coli strains BL21(DE3) (Novagen, Mississauga, Ontario, Canada), MC1061, and CJ236 (Δdut Δung F′) were used for the recombinant production of the cementoin peptide, routine subcloning experiments, and site-directed mutagenesis, respectively. The strains were grown at 37°C in 2YT medium containing glucose (1%) and supplemented with the appropriate antibiotics, as needed. The Saccharomyces cerevisiae yeast strain YBAD1 (yps1Δ::HIS3) was used for the recombinant production of pre-elafin and mutated variants. Yeast transformants were grown at 30°C in selective media as previously described (2).
Plasmids, oligonucleotides, and DNA manipulations.
DNA manipulations and bacterial and yeast transformations were all carried out according to standard procedures (14, 28). Unless otherwise indicated, all restriction and DNA-modifying enzymes were purchased from New England Biolabs, Ltd. (Pickering, Ontario, Canada), and the oligonucleotides were obtained from Operon Biotechnologies, Inc. (Huntsville, AL). The plasmid pET32a(+) (Novagen) was chosen for the bacterial expression and purification of the cementoin domain. First, the full-length pre-elafin coding sequence was PCR amplified from pVT-Ela2 (see below) with the oligonucleotides: 5′-AACATGCCATGGGCTGTCACGGAGTTCCTGTTAAAGGG-3′ (forward) and 5′-CGGGATCCTCTAGTTGTGCAACGG-3′ (reverse). The resulting amplicon was cloned into pET32a(+) at the NcoI and BamHI sites. Next, the elafin domain was removed by loop-out mutagenesis, using the procedure described by Kunkel (16), with the oligonucleotide 5′-PO4−-TTGTCGACGGAGCTCGAATTGACGTCTCATTTGACTTTATCTTGACC (Qiagen Canada, Inc., Mississauga, Ontario, Canada). The resulting plasmid, named pET32-cem, expresses an N-terminally tagged fusion protein (6 × His and S-tag) composed of the bacterial thioredoxin and the cementoin domain, a cleavage site for enterokinase separating these two peptides. Yeast expression plasmids for the recombinant production of pre-elafin and the mutated variants pre-elafinM25K and pre-elafinM25G were described previously (2, 8).
Production and purification of recombinant pre-elafin and cementoin.
Recombinant His-tagged pre-elafin, pre-elafinM25K and pre-elafinM25G were purified to apparent homogeneity from the yeast culture supernatants, first by affinity chromatography on Ni2+/NTA resin (Qiagen Canada, Inc.), followed by cation-exchange chromatography, as described previously (2). Bacterial expression of the cementoin peptide in fusion with thioredoxin (BL21 transformed with pET32-cem) was induced, from 1 liter of culture, by 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). After centrifugation, the bacterial pellet was sonicated and the crude protein extract was applied to Ni2+/NTA resin, essentially as described in the manufacturer's brochure (Novagen). After extensive washes, the Ni2+/NTA resin, containing the cementoin-thioredoxin protein fusion, was incubated for 24 h at 22°C in the presence of ∼400 U of enterokinase (New England Biolabs, Ltd.) to release the cementoin peptide. Diafiltration of the solution (stirred-cell using a 1-kDa cutoff Amicon membrane; Millipore Canada Ltd., Nepean, Ontario, Canada) was used for buffer exchange, and the protein mixture in 50 mM sodium acetate (pH 4.5) was next subjected to cation-exchange chromatography under the conditions described previously for pre-elafin (2). The purified recombinant peptide solution was finally concentrated in deionized water using a stirred cell as described above, and aliquots were lyophilized and stored at −80°C until use. Recombinant human elafin was purchased from Sigma-Aldrich Canada, Ltd. (Oakville, Ontario, Canada).
Antimicrobial and enzymatic assays.
The antimicrobial activity of the various recombinant peptides was assessed essentially as described previously (22, 31). Briefly, bacteria (∼106 cells/ml), obtained from a growing culture at the mid-logarithmic phase, were incubated for 3 h at 37°C in 10 mM phosphate buffer (pH 7.4) in the presence or absence of the indicated recombinant peptide. Serial dilutions of the bacterial cultures were then plated for the determination of the CFU.
All enzymatic assays were performed with the crude supernatant obtained from peptone soy broth (100 ml) inoculated with a single colony of P. aeruginosa and incubated 16 h at 37°C, as described previously (3). Briefly, the overnight bacterial cultures were centrifuged, and supernatants were filtered twice on 0.45- and 0.22-μm-pore-size filters (Millipore Canada, Ltd., Nepean, Ontario, Canada). The cleared supernatants were then concentrated (∼200-fold) on Amicon membranes (10-kDa cutoff; Millipore Canada, Ltd.). Enzymatic assays were conducted essentially as described by Caballero et al. (3), except the volumes were adjusted to fit 96-well titer plates for colorimetric assays. The substrates used, all purchased from Sigma-Aldrich Canada, Ltd., included N-p-tosyl-Gly-Pro-Lys-pNA (Chromozym PL), elastin Congo red, poly-l-lysine, and azocasein. Poly-l-lysine digestion products were analyzed by thin-layer chromatography using Silica gels purchased from EMD Chemicals, Inc. (Gibbstown, NJ).
Preparation of artificial membranes and binding experiments with pre-elafin and derived peptides.
To prepare large unilamellar vesicles, lipids were dissolved in chloroform-methanol (4:1) at a concentration of 5 to 20 mg/ml and stored at −20°C. Phosphatidylcholine, phosphatidylglycerol, and lipid A from E. coli were obtained from Sigma-Aldrich Canada, Ltd. Aliquots of the required lipids were combined in the desired molar ratio, i.e., pure phosphatidylcholine, phosphatidylcholine-phosphatidylglycerol (3:1), pure phosphatidylglycerol, and pure lipid A. Large unilamellar vesicles were prepared by hydration and extrusion as previously described (37). The vesicles were extruded through polycarbonate filters (Nuclepore, Clifton, NJ, purchased from VWR Canlab, Mississauga, Ontario, Canada) by using a mini-extruder (Avanti-Polar Lipids, Inc., Alabaster, AL). Samples were successively subjected to 19 passages through 0.4- and 0.1-μm-pore-size polycarbonate membranes.
Binding experiments of the various pre-elafin-derived peptides to large unilamellar vesicles were carried out as follows. Liposome suspensions and the peptide solution (10 μM) in 10 mM Tris-HCl (pH 7.4)-0.1 mM EDTA were mixed and incubated at room temperature 1 h. Large unilamellar vesicles were then subjected to ultracentrifugation at 160,000 × g for 40 min in a Beckman Airfuge to separate free and bound peptides. After centrifugation, supernatants were carefully removed and transferred to separate tubes, and pellets were resuspended in 10 mM Tris-HCl (pH 7.4)-0.1 mM EDTA. The amount of protein in supernatants and pellets was determined by using a bicinchoninic acid protein assay kit (Pierce, Rockford, IL) with bovine serum albumin as a standard.
Fluorescence microscopy of P. aeruginosa with fluorescein-conjugated pre-elafin.
Pre-elafin was fluorescently labeled by using a dye reagent (Fluorescein-EX; Invitrogen Canada, Inc., Burlington, Ontario, Canada), which has a reactive succinimidyl ester moiety reacting with the primary amines of proteins, according to the manufacturer's procedure. Free dye was removed from the pre-elafin-fluorescein conjugate through filtration on an Amicon membrane (Millipore) with a molecular mass cutoff of 5 kDa.
Binding experiments of the fluorescein-labeled pre-elafin to P. aeruginosa (ATCC 27853 and ATCC 33348) were performed with bacterial cultures (5 ml) grown to the mid-logarithmic phase. Bacteria were first harvested by centrifugation, washed three times with 10 mM phosphate buffer (pH 7.4), and resuspended in the same buffer. Bacterial cell suspensions were then incubated with fluorescein-labeled pre-elafin (30 μg/ml) at 37°C for 1 h in the dark. After several washes in 10 mM phosphate buffer, bacterial cells were mounted on a glass slide, and microscopic observations were made with a Leica fluorescence microscope (DMRB) equipped with a CoolSNAP-Pro monochrome camera. Fluorescent images were obtained with a fluorescein filter set.
RESULTS
Pre-elafin mutants with attenuated antipeptidase function have reduced antimicrobial activity against P. aeruginosa.
It was previously postulated that the antimicrobial activity of pre-elafin and derived synthetic peptides was likely associated to their polycationic nature (31). Intriguingly, the least positively charged peptide (pre-elafin 51-95), which has a net positive charge of +2 compared to +5 for pre-elafin 1-50, was the most active moiety in antimicrobial assays against P. aeruginosa. In addition, the antibacterial activity of elafin (pre-elafin 39-95), the naturally cleaved product of pre-elafin, was later described as nonbactericidal against P. aeruginosa (22). This contrasts with the usual mode of action of polycationic peptides, which frequently leads to cell lysis, and it prompted us to investigate whether the antipeptidase function of elafin affects P. aeruginosa growth. To this end, we compared the CFU of P. aeruginosa cultures incubated 3 h at 37°C in the presence of pre-elafin, the cementoin and elafin domains alone, and two pre-elafin mutants with attenuated antipeptidase function relative to P. aeruginosa cultures incubated in buffer alone. These mutants were recently described and are characterized by a slight (∼7-fold; pre-elafinM25K) or marked (∼400-fold; pre-elafinM25G) increase in Ki, for the inhibition of human neutrophil elastase relative to wild-type pre-elafin (8).
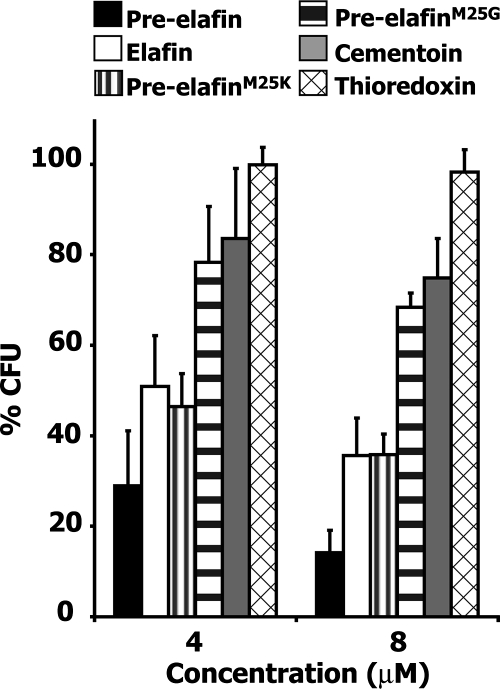
As shown in Fig. 1, when CFU counts were scored on peptone soy broth solid media, pre-elafin was the most effective peptide in reducing P. aeruginosa proliferation (ATCC 33348), with 71% ± 12% less CFU and 86% ± 5% less CFU at 4 and 8 μM, respectively, relative to that observed in the absence of peptides. At the same concentrations, elafin led to 49% ± 11% and 64% ± 8% decreases in CFU compared to 16% ± 15% and 25% ± 9% for the cementoin peptide. Therefore, both the elafin and the cementoin peptides appeared to contribute, apparently additively, to the overall antimicrobial activity of pre-elafin. In contrast, at the same concentrations, no effect on the percent CFU was observed when the unrelated thioredoxin protein was incubated with P. aeruginosa. Remarkably, and as expected for the involvement of the elafin antipeptidase function in controlling bacterial growth, both pre-elafin mutants were less efficient than wild-type pre-elafin at reducing the CFU counts. At 4 and 8 μM, the percent CFU values were 46 ± 7 and 36 ± 5 for pre-elafinM25K and 78 ± 12 and 68 ± 3 for pre-elafinM25G, respectively, compared to 29 ± 12 and 14 ± 5 for pre-elafin. For the most severely attenuated mutant (i.e., pre-elafinM25G), the relative CFU values were in fact very similar to that observed for the cementoin peptide alone (84 ± 15 and 75 ± 9), indicating a role for the antipeptidase function in controlling P. aeruginosa growth.
FIG. 1.
Antimicrobial assays of P. aeruginosa preincubated with wild type, mutated pre-elafin variants, and pre-elafin separate domains. The effect of the indicated peptides on P. aeruginosa growth (strain ATCC 33348) is expressed as the percent CFU relative to bacterial cultures incubated in phosphate buffer alone (see Materials and Methods). The error bars represent the standard deviation of the mean determined from three independent experiments performed in triplicate.
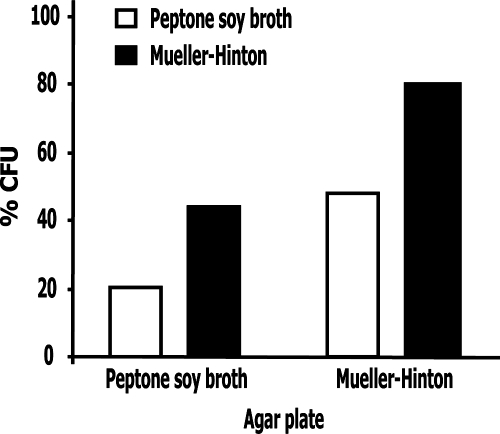
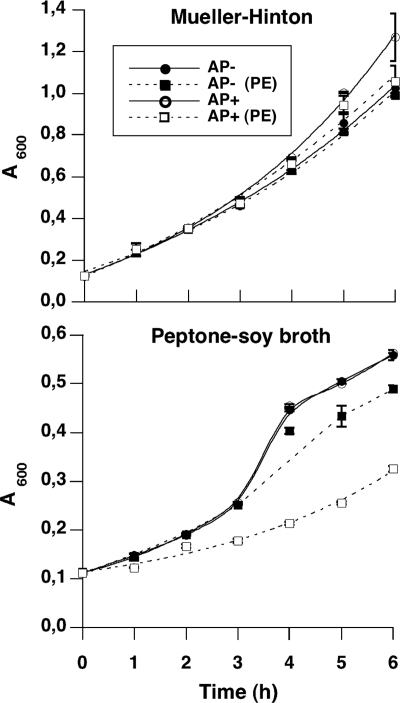
The simplest explanation for this finding is that by inhibiting a key P. aeruginosa peptidase, bacteria can no longer hydrolyze peptides and proteins from the culture medium to get their essential nitrogen and carbon sources. A corollary would be that this activity is medium dependent. To test this hypothesis, P. aeruginosa was first cultured in media either containing readily available carbon and nitrogen sources (i.e., Mueller-Hinton broth) or not (peptone soy broth). Next, 106 bacterial cells were washed and incubated in phosphate buffer for 3 h in the presence or absence of 8 μM pre-elafin prior to plating onto both Mueller-Hinton and peptone soy broth solid media for CFU determinations. As shown in Fig. 2, when bacteria were grown and plated on Mueller-Hinton medium, pre-elafin reduced the CFU counts by 20% relative to the control sample. As observed earlier, this reduction in CFU counts by pre-elafin was ∼80% when bacteria were cultured and plated on peptone soy broth. In contrast, intermediate CFU counts were noted when bacteria were first cultured in Mueller-Hinton and plated onto peptone soy broth (52% decrease) or cultured in peptone soy broth and plated onto Mueller-Hinton medium (58% decrease). Importantly, all CFU counts were identical after 16 and 48 h of growth at 37°C, indicating the absence of slow-growing colonies.
FIG. 2.
Sensitivity of P. aeruginosa to pre-elafin treatment is influenced by the growing and plating culture media. Antimicrobial assays with pre-elafin (8 μM) were conducted essentially as described in Fig. 1, except the P. aeruginosa strains were first precultured in the indicated media prior to pre-elafin treatment in phosphate buffer for 3 h and then plated on the indicated agar plates. The percent CFU was determined as described in Fig. 1 and represents the mean from an experiment performed in duplicate. In each case, duplicate sample variation was <10%.
Pilot experiments established that two to three cell divisions occurred, depending on the growth medium used, during the 3-h incubation period in phosphate buffer without pre-elafin prior to plating. Because more pronounced decreases in CFU counts were observed when bacteria were first cultured in peptone soy broth prior to pre-elafin treatment, it suggests that pre-elafin reduced proliferation in phosphate buffer presumably by preventing utilization of cell-associated complex nutrients through peptidase inhibition. This action of pre-elafin also appears not to be limited to the 3-h incubation period in phosphate buffer because fewer colonies grew in peptone soy broth compared to Mueller-Hinton solid media, irrespective of the liquid medium used (Fig. 2). However, pre-elafin also clearly affected P. aeruginosa growth independently from peptidase inhibition, as evidenced by the 20% decrease in CFU counts when bacteria were cultured and plated onto rich medium (Mueller-Hinton broth; Fig. 2). This is supported by the previous finding that the cementoin peptide alone led to a similar reduction in CFU counts (Fig. 1) and also by the presence of morphologically aberrant bacteria that do not develop into colonies upon microscopic examination of plated bacterial cultures subjected to pre-elafin treatment (results not shown). Overall, these data therefore support the hypothesis that the action of pre-elafin on P. aeruginosa proliferation is the sum of two activities; one is medium dependent (i.e., the antipeptidase function of the elafin domain) and the other not.
Pre-elafin and elafin inhibit a bacterial peptidase with the characteristics of AP.
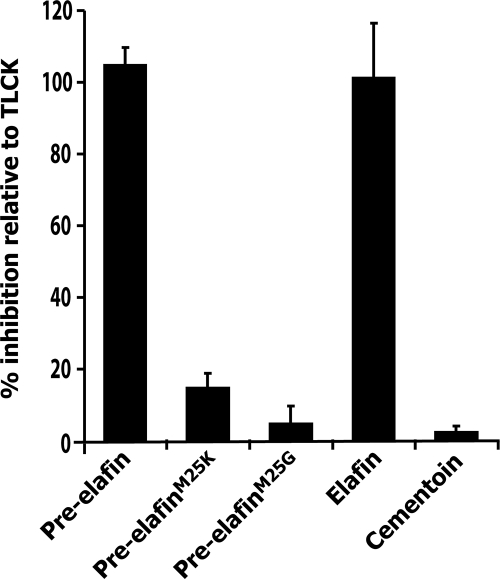
Elafin is a tight-binding inhibitor with narrow specificity for myeloblastin, neutrophil elastase, and pancreatic elastase, three serine-peptidases. We therefore hypothesized that the P. aeruginosa peptidase(s) inhibited by elafin and pre-elafin would belong to this family. Among the known P. aeruginosa-secreted peptidases, only AP is classified into this family (3). We therefore first tested whether the different elafin-derived peptides could inhibit hydrolysis of Chromozym PL, an AP-specific substrate (3), from the concentrated supernatant of P. aeruginosa cultures. As shown in Fig. 3 and for a better comparison, we expressed the inhibition of Chromozym PL hydrolysis by the various elafin-derived peptides relative to that of TLCK (Nα-p-tosyl-l-lysine chloromethyl ketone), which specifically and irreversibly inhibits AP (3). We found that both pre-elafin and elafin inhibit AP as efficiently as TLCK. In contrast, compared to that observed for TLCK, the relative inhibition by pre-elafinM25K was only 15% ± 3%, and only negligible inhibition (<5%) was measured with both pre-elafinM25G and cementoin. We therefore concluded that pre-elafin and elafin inhibit a P. aeruginosa peptidase with the characteristics of AP. Furthermore, the inefficient (pre-elafinM25K) and very poor (pre-elafinM25G) inhibitions of this peptidase by the pre-elafin mutants correlate with their reduced antimicrobial properties against P. aeruginosa, as estimated by the CFU counts in peptone soy broth agar plates (see Fig. 1).
FIG. 3.
Inhibition of Chromozym PL hydrolysis by pre-elafin-derived peptides. Concentrated culture supernatants prepared from an AP+ P. aeruginosa strain (ATCC 33348) was preincubated with the indicated peptides (50 μM), TLCK (50 μM) or with buffer alone, and the residual Chromozym PL hydrolytic activity was monitored. Inhibition by the pre-elafin-derived peptides is expressed relative to that observed for TLCK, which was set at 100%. The error bars represent the standard deviation of the mean determined from five independent experiments performed in triplicate.
To evaluate whether other P. aeruginosa-secreted peptidases could be inhibited by pre-elafin, we next assayed the major enzyme activities present in the culture supernatant of two P. aeruginosa strains, one possessing (AP+; ATCC 33348) and the other lacking (AP−; ATCC 27853) AP activity, in the presence or absence of pre-elafin, EDTA, and TLCK. The results, expressed semiquantitatively, are summarized in Table 1. Pseudolysin (formerly known as LasB), and to some extent staphylolysin (previously named LasA), contribute to the total elastolytic activity of P. aeruginosa, measured here by elastin Congo red hydrolysis, whereas staphylolysin is responsible for the staphylolytic activity. These two enzyme activities were detected in the culture supernatant of both strains, albeit reduced levels were found with the AP− strain. In each case, however, and in agreement with their classification as metallopeptidases, both activities were strongly inhibited by EDTA but not by pre-elafin or TLCK. Poly-l-lysine is a substrate for both aeruginolysin (formerly called alkaline protease) and AP. As expected for the AP− strain, the total poly-l-lysine hydrolytic activity was reduced compared to the AP+ strain. Furthermore, EDTA (100 mM), which targets aeruginolysin, completely inhibited this activity in the AP− strain, and neither pre-elafin nor TLCK had measurable effects. In contrast, about half the total hydrolytic activity found in the AP+ strain was inhibited by EDTA (100 mM), while the other half was inhibited with equal efficiency by both TLCK and pre-elafin (50 μM each). These two inhibitors also similarly reduced the total hydrolytic activity of the AP+ strain measured with azocasein as a substrate. Since AP, aeruginolysin, staphylolysin, and pseudolysin can all hydrolyze this substrate, the azocasein total hydrolytic activity from the AP− strain was completely abolished by EDTA, whereas a residual activity (∼25%) was detected with the AP+ strain. Overall, these inhibitory profiles indicated that, among the P. aeruginosa exopeptidases, AP is the main if not the sole target for pre-elafin-directed inhibition.
TABLE 1.
Inhibitory profiles of the major P. aeruginosa-secreted hydrolytic activities
| Strain | Inhibitor | Activitya
|
|||
|---|---|---|---|---|---|
| Elastin Congo red | Staphylolysis | Poly-l-lysine | Azocasein | ||
| AP− | None | +++ | ++ | ++ | ++ |
| Pre-elafin | +++ | ++ | ++ | ++ | |
| TLCK | +++ | ++ | ++ | ++ | |
| EDTA | + | − | − | − | |
| AP+ | None | ++++ | ++++ | ++++ | ++++ |
| Pre-elafin | ++++ | ++++ | ++ | +++ | |
| TLCK | ++++ | ++++ | ++ | +++ | |
| EDTA | + | − | ++ | + | |
The activity was graded on a scale from <20% activity (-) to >80% activity (++++). AP−, P. aeruginosa strain ATCC 27853; AP+, P. aeruginosa strain ATCC 33348.
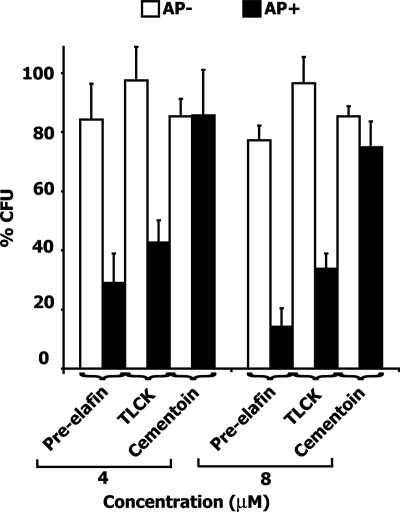
We would therefore predict that a P. aeruginosa strain lacking AP activity should not be more affected by a preincubation with pre-elafin than by a preincubation with the cementoin peptide alone. In addition, the action of pre-elafin on an AP+ strain should parallel that of TLCK. As shown in Fig. 4, when an AP− strain cultured in peptone soy broth was first preincubated with pre-elafin at 4 and 8 μM, the percent CFU determined on the same culture media was only modestly affected (77 ± 12 at 8 μM). As predicted, this small antimicrobial activity was also observed with the cementoin peptide alone (85 ± 6 at 8 μM), and TLCK had no effect on proliferation with this strain. In contrast, TLCK significantly diminished the percent CFU (34 ± 7 at 8 μM) determined with the AP+ strain. This effect was similar to that observed with pre-elafin (14 ± 6 at 8 μM), if we exclude the contribution of the cementoin domain (cementoin alone reduced the percent CFU by 25 ± 9 at 8 μM) and closely paralleled that observed with elafin (36 ± 8, see Fig. 1). To assess more directly the action of pre-elafin on the growth of these two P. aeruginosa strains, we next compared their growth curves in rich (Mueller-Hinton) and complex (peptone soy broth) liquid media in the presence or absence of 8 μM pre-elafin. As shown in Fig. 5, growth curves of the AP− and AP+ strains in the absence of pre-elafin were very similar in both liquid media. Although this was expected for proliferation in rich medium, that the AP− strain could grow as efficiently as the AP+ strain in peptone soy broth was surprising and suggests that the AP− strain ATCC 28753 may possess additional secreted peptidases not present in the AP+ strain (see the Discussion). In the presence of pre-elafin, the rapid proliferation of both strains in Mueller-Hinton medium was barely affected. In contrast, growth in peptone soy broth was modestly reduced at later time points for the AP− strain and was significantly slowed down for the AP+ strain, thereby confirming the correlation between AP inhibition and reduced growth in complex medium.
FIG. 4.
Antimicrobial assays of P. aeruginosa strains possessing or lacking AP activity preincubated with pre-elafin, cementoin, and TLCK. The effect of the various inhibitors on the percent CFU of the indicated P. aeruginosa strains (AP+, ATCC 33348; AP−, ATCC 27853) was performed as described in Fig. 1. The error bars represent standard deviation of the mean determined from three independent experiments performed in duplicate.
FIG. 5.
Growth curves of P. aeruginosa strains ATCC 27853 (AP−) and ATCC 33348 (AP+) in rich (Mueller-Hinton) and complex (peptone-soy broth) media in the presence or absence of 8 μM pre-elafin (PE). Overnight P. aeruginosa precultures were diluted to 0.1 A600/ml, and growth in the indicated media in the presence or absence of pre-elafin was monitored spectroscopically at 600 nm. The error bars represent standard deviation of the mean determined from two independent experiments.
Pre-elafin-derived peptides interact strongly with P. aeruginosa and to negatively charged artificial membranes.
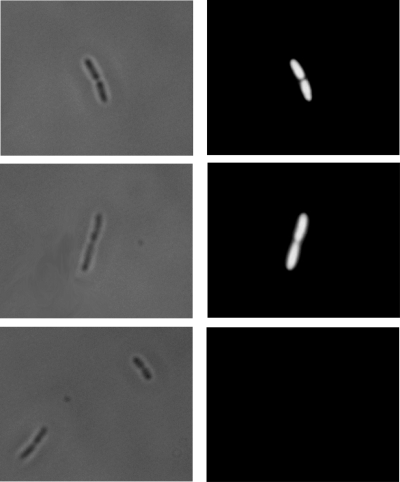
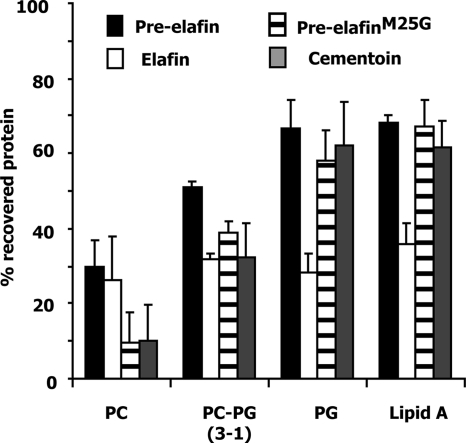
Binding of pre-elafin and derived peptides to the lipopolysaccharide (LPS) structure of gram-negative bacteria was recently reported (21). In vivo, this suggests that the outer membrane of gram-negative bacteria may attract pre-elafin and/or derived peptides and help concentrate these antimicrobials at sites of infection. As evidenced by the intensity of fluorescence (Fig. 6), when P. aeruginosa strains possessing or not possessing AP activity were incubated in the presence of fluorescein-labeled pre-elafin, both cell surfaces were shown to be heavily decorated by the labeled peptide. In contrast, no fluorescence was observed when the cells were incubated with free fluorescein under the same experimental conditions. As a first step toward elucidating what governs the interaction between pre-elafin and bacterial membranes, we performed binding experiments with the elafin derived peptides and artificial membranes varying in lipid composition. Lipid A is the lipid anchor portion of LPS, and liposomes composed of this lipid were used to model the outer membranes of gram-negative bacteria. Microbial membranes, in contrast to mammalian plasma membranes, are enriched into negatively charged phospholipids, and liposomes composed exclusively of phosphatidylcholine (neutral), phosphatidylglycerol (negatively charged), or phosphatidylcholine-phosphatidylglycerol in a 3:1 ratio were also assayed for binding. As shown in Fig. 7, >60% of pre-elafin, pre-elafinM25G, and cementoin were recovered in the membrane fraction after ultracentrifugation when preincubated with negatively charged liposomes (phosphatidylglycerol and lipid A). This percentage decreased to ∼30 when incubated with only slightly charged liposomes (phosphatidylcholine-phosphatidylglycerol [3:1]) and, for the cementoin peptide, only minimal interaction (<10%) was noted with neutral (phosphatidylcholine) liposomes. In contrast, although elafin interacted less strongly with artificial membranes (∼ 30%), the binding was not influenced by the phospholipid composition of artificial membranes. These data therefore suggest that binding of the cementoin peptide to microbial membranes is mediated by ionic interactions, whereas that of elafin appears to be governed primarily by hydrophobic interaction.
FIG. 6.
Binding of pre-elafin to bacteria. Bright-field (left column) and fluorescence (right column) images of the same field were acquired after P. aeruginosa strains ATCC 33348 (top) and ATCC 27853 (middle) were incubated in the presence of fluorescein-labeled pre-elafin. As a control (bottom), strain ATCC 33348 was first incubated with free fluorescein prior to processing for microscopic examination.
FIG. 7.
Binding experiments of pre-elafin, pre-elafinM25G, and pre-elafin domains to artificial membranes varying in lipid composition. PC, phosphatidylcholine; PG, phosphatidylglycerol. The error bars represent the standard deviation of the mean determined from three independent experiments.
DISCUSSION
In agreement with Simpson et al. (31), we confirmed here that both the cementoin and the elafin domains contribute to the antibacterial activity of pre-elafin against P. aeruginosa. However, while earlier studies suggested that the antipeptidase function of elafin is not involved in bacterial growth (22, 31), our data clearly indicate that this domain selectively inhibits a secreted P. aeruginosa serine-peptidase. Furthermore, several lines of evidence suggest that the main target of elafin inhibition is AP. First, growth in peptone soy broth of a P. aeruginosa strain (ATCC 27853) lacking AP activity (25) is only modestly inhibited by pre-elafin, and this growth-inhibiting effect is entirely due to the cementoin domain alone. Second, pre-elafin-directed growth inhibition of a P. aeruginosa strain possessing AP activity (ATCC 33348) correlates with its ability to inhibit Chromozym PL hydrolysis, an AP-specific substrate (3). Third, among the secreted P. aeruginosa peptidases, TLCK specifically inhibits AP (3), and we showed here that pre-elafin and TLCK similarly affect growth of an AP-proficient P. aeruginosa strain. Fourth, consistent with the known role of pre-elafin as an inhibitor of selected members of the serine-peptidase family, AP is currently the only known P. aeruginosa-secreted peptidase belonging to this family (3). Finally, and importantly, we demonstrated that pre-elafin mutants with attenuated inhibitory activity toward previously identified peptidase targets either slightly or poorly inhibit Chromozym PL hydrolysis, and the extent of inhibition correlates with a reduced bacterial proliferation in complex medium.
It was shown that P. aeruginosa strains possessing AP activity are more virulent than AP-deficient strains in an animal model of corneal infection (9, 25, 35). AP is also postulated to play a major role in the pathogenesis of acute and chronic lung infections caused by P. aeruginosa (18, 32, 39). That P. aeruginosa growth is compromised by elafin-directed inhibition of this secreted peptidase therefore constitutes a novel and important finding. Although no standard MICs could be determined with up to 150 μg of pre-elafin/ml (data not shown), the growth-inhibiting property of the elafin domain in complex medium in vitro is certainly relevant for its action in vivo given the scarcity of nutrients available for P. aeruginosa in its ecological niche. It is intriguing that strain ATCC 27853, which lacks AP activity (25), grew as efficiently as the AP-proficient strain ATCC 33348. The reason for this finding is unknown but is possibly linked to the expression of a secreted peptidase, distinct from the enzyme activities assayed in the present study, which would be present in strain ATCC 27853 but absent from strain ATCC 33348 (AP+). A putative candidate could be the recently characterized P. aeruginosa small peptidase. This enzyme was purified from a strain (PA103) originally described as devoid of hydrolytic activity and was found to contribute to P. aeruginosa corneal virulence (19). However, further studies are needed to clarify this issue.
Pre-elafin and derived peptides were recently shown to bind the smooth and rough forms of bacterial LPS, possibly through ionic interactions with lipid A (21). Using artificial membranes composed of lipid A, we confirmed here that pre-elafin and its separate domains associate with the lipid core of LPS, the cementoin domain more readily than the elafin domain. Interestingly, our data further indicated that the polycationic cementoin domain (+4 net charge) has a net preference for binding to negatively charged membranes. In contrast, the elafin domain, despite possessing a +3 net charge, binds to both neutral and negatively charged membranes with similar affinity, suggesting hydrophobic interaction. Hence, like most linear antimicrobial peptides (34), the antimicrobial property of the cementoin peptide appears to rely on its cationic nature, and both structural and biophysical studies are currently under way to elucidate its precise mode of action. Its ability to bind the negatively charged microbial membranes may, additionally, help target and concentrate the antipeptidase domain of pre-elafin at the bacterial cell surface to facilitate the rapid and efficient inhibition of bacterial peptidases just upon their release.
Several WAP-motif-containing proteins from various origins were shown to possess antimicrobial activity. These include human SLPI (13), pre-elafin (22, 31), Eppin (42), mouse SWAM1 and SWAM2 (12), snake Omwaprin (12), and equine eNAP2 (6). The mode of action of these peptides is largely unknown, although direct evidences that Eppin and Omwaprin bind and disrupt bacterial membrane on selected gram-negative (Eppin) and gram-positive (Omwaprin) bacteria were recently reported (12, 42). Based on their inability to inhibit neutrophil-secreted peptides, which are targets for SLPI and pre-elafin, many WAP-motif proteins were proposed not to possess antipeptidase function. However, the implication of a putative antipeptidase function directed toward bacterial peptidases, as demonstrated here for pre-elafin and earlier for the equine WAP-containing protein eNAP2 (7), certainly warrants reexamination for other WAP-motif proteins. eNAP2, which has no inhibitory function against neutrophil secreted peptidases, was shown to selectively inhibit microbial subtilisin A and proteinase K. In this context, the reported broad-spectrum antimicrobial activity of pre-elafin and other WAP-motif proteins may thus result from several unique features of the molecule, some providing specificity and others not. The recent study reported by Zaiou et al. (43) with the human cathelicidin precursor (hCAP18/LL-37) provides a good example that a single polypeptide may carry several structural properties, each one contributing an antibacterial function. Apart from the well-characterized polycationic C-terminal moiety (LL-37) of hCAP-18, the conserved N-terminal cathelin domain of the precursor molecule acts as a cysteine-peptidase inhibitor and possesses antimicrobial activity. Interestingly, the antibacterial activity of the two peptides exhibited distinct specificities, cathelin being active against some gram-negative and gram-negative species, but not P. aeruginosa, and LL-37 exclusively against gram-negative bacteria.
P. aeruginosa chronic infections progressively lead to lung tissue damage caused, largely, by the sustained inflammation status and the associated release of neutrophil peptidases such as neutrophil elastase and myeloblastin. In a widely used animal model of chronic P. aeruginosa infection the administration of two potent inhibitors of neutrophil elastase (monocyte neutrophil elastase inhibitor and α1-peptidase inhibitor) was successful in suppressing P. aeruginosa proliferation and in attenuating tissue injury (4, 41). Since neither of these inhibitors possesses antimicrobial activity in vitro, it was concluded that inhibition of neutrophil elastase helps decrease the extent of inflammation and protects against the degradation of innate defense molecules, which in turn, facilitates bacterial clearance. Because pre-elafin is also a potent inhibitor of neutrophil peptidases and with demonstrated antimicrobial activities against P. aeruginosa, future studies should be directed to evaluate its efficiency in animal models of chronic P. aeruginosa infections.
Acknowledgments
We thank Caroline Duchaine (Université Laval) and Joseph Lam (University of Guelph) for bacterial strains and Roger Lévesque and François Sanschagrin (Université Laval) for their help with the pseudomonal peptidase assays.
This study was supported by grants from the Bayer/Talecris/CBS/Hema-Quebec Partnership Fund and the Réseau de Recherche en Santé Respiratoire du FRSQ (Y.B.). A.B. acknowledges a studentship from the Centre de Recherche sur la Fonction, la Structure et l'Ingénierie des Protéines (Université Laval).
Footnotes
Published ahead of print on 19 November 2007.
REFERENCES
- 1.Bingle, L., T. D. Tetley, and C. D. Bingle. 2001. Cytokine-mediated induction of the human elafin gene in pulmonary epithelial cells is regulated by nuclear factor-κB. Am. J. Respir. Cell Mol. Biol. 25:84-91. [DOI] [PubMed] [Google Scholar]
- 2.Bourbonnais, Y., C. Larouche, and G. M. Tremblay. 2000. Production of full-length human pre-elafin, an elastase specific inhibitor, from yeast requires the absence of a functional yapsin 1 (Yps1p) endoprotease. Protein Expr. Purif. 20:485-491. [DOI] [PubMed] [Google Scholar]
- 3.Caballero, A. R., J. M. Moreau, L. S. Engel, M. E. Marquart, J. M. Hill, and R. J. O'Callaghan. 2001. Pseudomonas aeruginosa protease IV enzyme assays and comparison to other Pseudomonas proteases. Anal. Biochem. 290:330-337. [DOI] [PubMed] [Google Scholar]
- 4.Cantin, A. M., and D. E. Woods. 1999. Aerosolized prolastin suppresses bacterial proliferation in a model of chronic Pseudomonas aeruginosa lung infection. Am. J. Respir. Crit. Care Med. 160:1130-1135. [DOI] [PubMed] [Google Scholar]
- 5.Chernish, R. N., and S. D. Aaron. 2003. Approach to resistant gram-negative bacterial pulmonary infections in patients with cystic fibrosis. Curr. Opin. Pulm. Med. 9:509-515. [DOI] [PubMed] [Google Scholar]
- 6.Couto, M. A., S. S. Harwig, J. S. Cullor, J. P. Hughes, and R. I. Lehrer. 1992. eNAP-2, a novel cysteine-rich bactericidal peptide from equine leukocytes. Infect. Immun. 60:5042-5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Couto, M. A., S. S. Harwig, and R. I. Lehrer. 1993. Selective inhibition of microbial serine proteases by eNAP-2, an antimicrobial peptide from equine neutrophils. Infect. Immun. 61:2991-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doucet, A., D. Bouchard, M. F. Janelle, A. Bellemare, S. Gagne, G. M. Tremblay, and Y. Bourbonnais. 2007. Characterization of human pre-elafin mutants: full antipeptidase activity is essential to preserve lung tissue integrity in experimental emphysema. Biochem. J. 405:455-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engel, L. S., J. A. Hobden, J. M. Moreau, M. C. Callegan, J. M. Hill, and R. J. O'Callaghan. 1997. Pseudomonas deficient in protease IV has significantly reduced corneal virulence. Investig. Ophthalmol. Vis. Sci. 38:1535-1542. [PubMed] [Google Scholar]
- 10.Erwin, A. L., and D. R. VanDevanter. 2002. The Pseudomonas aeruginosa genome: how do we use it to develop strategies for the treatment of patients with cystic fibrosis and Pseudomonas infections? Curr. Opin. Pulm. Med. 8:547-551. [DOI] [PubMed] [Google Scholar]
- 11.Fitch, P. M., A. Roghanian, S. E. Howie, and J. M. Sallenave. 2006. Human neutrophil elastase inhibitors in innate and adaptive immunity. Biochem. Soc. Trans. 34:279-282. [DOI] [PubMed] [Google Scholar]
- 12.Hagiwara, K., T. Kikuchi, Y. Endo, Huqun, K. Usui, M. Takahashi, N. Shibata, T. Kusakabe, H. Xin, S. Hoshi, M. Miki, N. Inooka, Y. Tokue, and T. Nukiwa. 2003. Mouse SWAM1 and SWAM2 are antibacterial proteins composed of a single whey acidic protein motif. J. Immunol. 170:1973-1979. [DOI] [PubMed] [Google Scholar]
- 13.Hiemstra, P. S., R. J. Maassen, J. Stolk, R. Heinzel-Wieland, G. J. Steffens, and J. H. Dijkman. 1996. Antibacterial activity of antileukoprotease. Infect. Immun. 64:4520-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 15.Kipnis, E., T. Sawa, and J. Wiener-Kronish. 2006. Targeting mechanisms of Pseudomonas aeruginosa pathogenesis. Med. Mal. Infect. 36:78-91. [DOI] [PubMed] [Google Scholar]
- 16.Kunkel, T. A. 1985. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82:488-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livermore, D. M., and N. Woodford. 2006. The beta-lactamase threat in Enterobacteriaceae, Pseudomonas, and Acinetobacter. Trends Microbiol. 14:413-420. [DOI] [PubMed] [Google Scholar]
- 18.Malloy, J. L., R. A. Veldhuizen, B. A. Thibodeaux, J. O'Callaghan, R., and J. R. Wright. 2004. Pseudomonas aeruginosa protease IV degrades surfactant proteins and inhibits surfactant host defense and biophysical functions. Am. J. Physiol. Lung Cell. Mol. Physiol. 288:L409-L415. [DOI] [PubMed] [Google Scholar]
- 19.Marquart, M. E., A. R. Caballero, M. Chomnawang, B. A. Thibodeaux, S. S. Twining, and R. J. O'Callaghan. 2005. Identification of a novel secreted protease from Pseudomonas aeruginosa that causes corneal erosions. Investig. Ophthalmol. Vis. Sci. 46:3761-3768. [DOI] [PubMed] [Google Scholar]
- 20.Matsumoto, K. 2004. Role of bacterial proteases in pseudomonal and serratial keratitis. Biol. Chem. 385:1007-1016. [DOI] [PubMed] [Google Scholar]
- 21.McMichael, J. W., A. Roghanian, L. Jiang, R. Ramage, and J. M. Sallenave. 2005. The antimicrobial antiproteinase elafin binds to lipopolysaccharide and modulates macrophage responses. Am. J. Respir. Cell Mol. Biol. 32:443-452. [DOI] [PubMed] [Google Scholar]
- 22.Meyer-Hoffert, U., N. Wichmann, L. Schwichtenberg, P. C. White, and O. Wiedow. 2003. Supernatants of Pseudomonas aeruginosa induce the Pseudomonas-specific antibiotic elafin in human keratinocytes. Exp. Dermatol. 12:418-425. [DOI] [PubMed] [Google Scholar]
- 23.Mihaila, A., and G. M. Tremblay. 2001. Human alveolar macrophages express elafin and secretory leukocyte protease inhibitor. Z. Naturforsch. C 56:291-297. [DOI] [PubMed] [Google Scholar]
- 24.Molhuizen, H. O., and J. Schalkwijk. 1995. Structural, biochemical, and cell biological aspects of the serine proteinase inhibitor SKALP/elafin/ESI. Biol. Chem. 376:1-7. [DOI] [PubMed] [Google Scholar]
- 25.O'Callaghan, R. J., L. S. Engel, J. A. Hobden, M. C. Callegan, L. C. Green, and J. M. Hill. 1996. Pseudomonas keratitis: the role of an uncharacterized exoprotein, protease IV, in corneal virulence. Investig. Ophthalmol. Vis. Sci. 37:534-543. [PubMed] [Google Scholar]
- 26.Richards, M. J., J. R. Edwards, D. H. Culver, and R. P. Gaynes. 1999. Nosocomial infections in medical intensive care units in the United States. Crit. Care Med. 27:887-892. [DOI] [PubMed] [Google Scholar]
- 27.Sadikot, R. T., T. S. Blackwell, J. W. Christman, and A. S. Prince. 2005. Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit. Care Med. 171:1209-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 29.Schalkwijk, J., A. Chang, P. Janssen, G. J. De Jongh, and P. D. Mier. 1990. Skin-derived antileucoproteases (SKALPs): characterization of two new elastase inhibitors from psoriatic epidermis. Br. J. Dermatol. 122:631-641. [DOI] [PubMed] [Google Scholar]
- 30.Schalkwijk, J., O. Wiedow, and S. Hirose. 1999. The trappin gene family: proteins defined by an N-terminal transglutaminase substrate domain and a C-terminal four-disulphide core. Biochem. J. 340:569-577. [PMC free article] [PubMed] [Google Scholar]
- 31.Simpson, A. J., A. I. Maxwell, J. R. Govan, C. Haslett, and J. M. Sallenave. 1999. Elafin (elastase-specific inhibitor) has anti-microbial activity against gram-positive and gram-negative respiratory pathogens. FEBS Lett. 452:309-313. [DOI] [PubMed] [Google Scholar]
- 32.Smith, L., B. Rose, P. Tingpej, H. Zhu, T. Conibear, J. Manos, P. Bye, M. Elkins, M. Willcox, S. Bell, C. Wainwright, and C. Harbour. 2006. Protease IV production in Pseudomonas aeruginosa from the lungs of adults with cystic fibrosis. J. Med. Microbiol. 55:1641-1644. [DOI] [PubMed] [Google Scholar]
- 33.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 34.Toke, O. 2005. Antimicrobial peptides: new candidates in the fight against bacterial infections. Biopolymers 80:717-735. [DOI] [PubMed] [Google Scholar]
- 35.Traidej, M., A. R. Caballero, M. E. Marquart, B. A. Thibodeaux, and R. J. O'Callaghan. 2003. Molecular analysis of Pseudomonas aeruginosa protease IV expressed in Pseudomonas putida. Investig. Ophthalmol. Vis. Sci. 44:190-196. [DOI] [PubMed] [Google Scholar]
- 36.Tremblay, G. M., M. F. Janelle, and Y. Bourbonnais. 2003. Anti-inflammatory activity of neutrophil elastase inhibitors. Curr. Opin. Investig. Drugs 4:556-565. [PubMed] [Google Scholar]
- 37.Vacheron, M. J., E. Clottes, C. Chautard, and C. Vial. 1997. Mitochondrial creatine kinase interaction with phospholipid vesicles. Arch. Biochem. Biophys. 344:316-324. [DOI] [PubMed] [Google Scholar]
- 38.Wiedow, O., J. M. Schroder, H. Gregory, J. A. Young, and E. Christophers. 1990. Elafin: an elastase-specific inhibitor of human skin: purification, characterization, and complete amino acid sequence. J. Biol. Chem. 265:14791-14795. [PubMed] [Google Scholar]
- 39.Wilderman, P. J., A. I. Vasil, Z. Johnson, M. J. Wilson, H. E. Cunliffe, I. L. Lamont, and M. L. Vasil. 2001. Characterization of an endoprotease (PrpL) encoded by a PvdS-regulated gene in Pseudomonas aeruginosa. Infect. Immun. 69:5385-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams, S. E., T. I. Brown, A. Roghanian, and J. M. Sallenave. 2006. SLPI and elafin: one glove, many fingers. Clin. Sci. 110:21-35. [DOI] [PubMed] [Google Scholar]
- 41.Woods, D. E., A. Cantin, J. Cooley, D. M. Kenney, and E. Remold-O'Donnell. 2005. Aerosol treatment with MNEI suppresses bacterial proliferation in a model of chronic Pseudomonas aeruginosa lung infection. Pediatr. Pulmonol. 39:141-149. [DOI] [PubMed] [Google Scholar]
- 42.Yenugu, S., R. T. Richardson, P. Sivashanmugam, Z. Wang, G. O'Rand, M., F. S. French, and S. H. Hall. 2004. Antimicrobial activity of human EPPIN, an androgen-regulated, sperm-bound protein with a whey acidic protein motif. Biol. Reprod. 71:1484-1490. [DOI] [PubMed] [Google Scholar]
- 43.Zaiou, M., V. Nizet, and R. L. Gallo. 2003. Antimicrobial and protease inhibitory functions of the human cathelicidin (hCAP18/LL-37) prosequence. J. Investig. Dermatol. 120:810-816. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, L., J. Parente, S. M. Harris, D. E. Woods, R. E. Hancock, and T. J. Falla. 2005. Antimicrobial peptide therapeutics for cystic fibrosis. Antimicrob. Agents Chemother. 49:2921-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]