Abstract
Objective
Sickle cell anemia is a genetic blood disease resulting from production of mutant β-globin (βS) and has severe clinical consequences. It is known that a higher cellular γ-globin level, e.g., higher ratio of cellular γ-globin to βS-globin (γ/βS ratio), inhibits sickle hemoglobin (HbS) polymerization tendency. Hence, therapeutic treatment of sickle cell anemia has been focused on introducing γ-globin gene into red blood cells to increase the cellular γ/βS ratio. Here, we have introduced ribozymes and small interfering RNAs (siRNAs) against βS-globin mRNA into blood cells as a means to increase the γ/βS ratio.
Methods
Single and multi-ribozymes against βS-globin mRNA have been tested in vitro and in human erythroleukemia K562βS cells that stably express exogenous βS-globin gene. Primary human hematopoietic progenitor cells were also transfected with multi-ribozyme and the γ/(γ+β) ratio determined and compared with cells transfected with long hairpin β-globin cDNA and synthetic siRNA genes.
Results
We have found that the multi-ribozyme zb21A containing two ribozyme units effectively reduces βS-globin mRNA both in vitro and in K562βS cells. The γ-globin mRNA to βS-globin mRNA ratio in the multi-ribozyme transfected cells is about a factor of 2 more than that in the control cells. We have also found that the γ/(γ+β) ratio in the transfected hematopoietic progenitor cells is increased by more than 2-fold in cells treated with multi-ribozyme zb21A or siRNA ib5.
Conclusion
Our results suggest that introducing multi-ribozymes or siRNAs into red blood cells are comparable in their effectiveness to increase the ratio of cellular γ-globin mRNA to β- or βS-globin mRNA, providing possible strategies to increase the effectiveness of γ-globin gene transfer as gene therapy for treatment of patients with sickle cell anemia.
Sickle cell anemia is an inheritable blood disease caused by a single nucleotide mutation in the human β-globin gene (codon 6 GAG→GTG). Individuals homozygous for this mutant β-globin gene, designated as βS-globin, have severe clinical symptoms, because the abnormal hemoglobin, sickle cell hemoglobin [HbS (β6 Glu→Val)], produced by this gene can polymerize under low oxygen tension, change red blood cell rheology and shape (sickle), and result in vasoocclusive crisis, infarction, and organ damage [1]. It is known that in some patients with milder sickle cell anemia symptoms, there are higher fetal hemoglobin [HbF (α2γ2)] levels in their red blood cells (RBCs) [2,3]. Increased HbF facilitates the formation of the asymmetric hybrid of HbF and HbS that cannot be incorporated into the HbS polymer and reduces the overall tendency of HbS polymerization as well as the sickling propensity of the blood cells [4,5]. Hydroxyurea can increase HbF to therapeutically significant levels in sickle cell patients, although only half of the adults enrolled in a large efficacy trial were responsive and children appear to be more responsive than adults [6]. Hence, increasing the level of HbF in RBCs and the ratio of HbF to HbS, or of γ-globin to βS-globin (γ/βS), becomes an attractive consideration of gene therapy for the treatment of sickle cell anemia. During recent years, research on gene therapy for sickle cell anemia has focused on high-level expression of a transferred human γ-globin gene (or an anti-sickling mutant β-globin gene), with limited success being reported in murine models and human progenitor cells by using engineered retroviral or lentiviral vectors [7-13]. In contrast, such a strategy has shown more promise for β-thalassemia, another severe hemoglobin disorder [14-16].
An alternate way of increasing the γ/βS ratio in RBCs is to reduce the expression of endogenous βS-globin. For anti-sickling treatment, this may be more effective because the harmful effect of HbS polymerization is greatly dependent on HbS concentration, thus even a relatively small decrease in HbS concentration may greatly reduce the tendency of HbS polymerization and sickling [17-21]. An additional challenge in therapeutic strategies to increase HbF is that HbF is not uniformally distributed resulting in a subpopulation of cells in which HbF cannot be detected [22]. The most desired outcome is to increase % HbF in all red cells [23].
Ribozymes and interference RNA (RNAi) are useful tools for the suppression of specific gene expression by reducing the amount of corresponding mRNA. Ribozymes are a group of catalytic RNAs that react with RNA molecules in a sequence-specific manner [24-26]. Specifically designed ribozymes have been used as potential therapeutic agents for the suppression of many harmful viral and cellular mRNAs [27-29]. For suppressing the βS-globin gene, a trans-splicing ribozyme that converts βS-globin mRNA into γ-globin-like mRNA has been reported [30], and a single hammerhead ribozyme against βS-globin mRNA linked with a luciferase reporter has been tested in transgenic mouse [31]. In our laboratories, a multi-ribozyme against α-globin mRNA has been found to be effective in reducing α-globin mRNA levels in K562 cells [32]. RNAi is an evolutionarily conserved cellular pathway that involves generation of active small interfering RNA (siRNA) from long double stranded mRNA through RNaseIII endonuclease Dicer, and the 21-23 nt siRNA incorporates into a RNA-induced silencing complex (RISC), and then triggers degradation of complementary homologous mRNA [33-35]. In regards to the suppression of βS-globin, significant reduction of mouse adult β-globin mRNA by siRNA has been reported in MEL cells [36].
Since hemoglobin production is primarily under transcription control, in this proof-of-concept study specially designed ribozymes and multi-ribozymes against βS-globin mRNA have been tested in a cell free system and in K562βS cells with significant increase in cellular γ/βS ratio. The multi-ribozymes were also assayed in primary human hematopoietic progenitor cells and found to increase the γ/(γ+β) expression ratio. Comparisons with other strategies to target β-globin mRNA using specific long hairpin β-globin RNA and siRNAs against β-globin mRNA showed similar increases of the γ/(γ+β) ratio for the multi-ribozyme and siRNAs.
Methods
Ribozyme and siRNA oligonucleotides
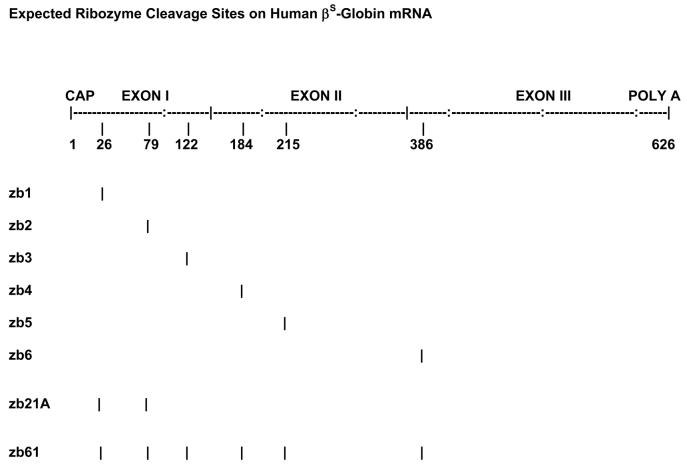
Six genes of single ribozymes zb1, zb2, zb3, zb4, zb5, and zb6 against human βS-globin mRNA were synthesized with a 22-bp core sequence, as zb1, TGTTTGAGGTTGCTAGTGctgatgagtccgtgaggacgaaACACAGTTGTGTCAGAAG; zb2, ACAGGGCAGTAACGGCActgatgagtccgtgaggacgaaACTTCTCCTCAGGAGTCA; zb3, GCCCAGGGCCGCCACCACCctgatgagtccgtgaggacgaaACTTCATCCACTTCACC; zb4, AGATCCCCAAAGctgatgagtccgtgaggacgaaACTCAAAGAACCTC; zb5, TTAGGGTTGCCCATctgatgagtccgtgaggacgaaACAGCATCAGGA; and zb6, TGGGCCAGCACACActgatgagtccgtgaggacgaaACCAGCCGTTGCC; and their presumed cleavage sites on human βS-globin mRNA are shown in Fig. 1. In order to facilitate ligation in later steps, restriction site pairs Kpn I/Xho I, Xho I/Cla I, Cla I/EcoR I, EcoR I/BamH I, BamH I/ Eag I, and Eag I/ Sac I were added to both ends of the synthetic oligonucleotides zb1-zb6, respectively.
Figure 1.
Expected ribozyme cleavage sites.
Three siRNA genes were synthesized as hairpin structures with a 9-bp loop, as ib2, GGTGAACGTGGATGAAGTTGGTttcgaaagaACCAACTTCATCCACGTTCACC; ib4, GGTGAAGGCTCATGGCAAGAAAttcgaaagaTTTCTTGCCATGAGCCTTCACC; and ib5, GAAAGTGCTCGGTGCCTTTAGTGATttcgaaagaATCACTAAAGGCACCGAGCACTTTC corresponding to β-globin mRNA 104-125, 230-251, and 248-272, respectively.
The oligonucleotides were synthesized as sense and antisense chains using Applied Biosystems 308 DNA Synthesizer (Applied Biosystems, Foster City, CA) and annealed for ligation.
Plasmids for producing ribozymes and mRNA substrates in vitro
Six annealed ribozyme oligonucleotides were inserted into pBluescript II (Stratagene, La Jolla, CA) to form plasmids pKSzb1, pKSzb2, pKSzb3, pKSzb4, pKSzb5, and pKSzb6. The 0.47-kb NgoM IV/Xho I fragment containing the ribozyme zb2 from pKSzb2 was inserted into the 2.59-kb NgoM IV/Xho I vector immediately ahead of the ribozyme zb1 gene to form plasmid pKSzb21, which contained the multi-ribozyme gene zb21 which consisted of tandemly arranged ribozyme genes zb2 and zb1, linked by a Xho I site. In a similar manner, the other four ribozyme genes, zb6, zb5, zb4, and zb3, were stepwise added on pKSzb21, and finally plasmid pKSzb61 which contained multi-ribozyme zb61, composed of tandemly arranged ribozyme genes, zb6, zb5, zb4, zb3, zb2, and zb1, was constructed. In addition, plasmid pKSzb21A containing multi-ribozyme zb21A was constructed from pKSzb21 by replacing the Xho I site between the zb2 and zb1 sequences with an antisense βS-globin mRNA fragment (GGTGCACCATGCATGGTGTC). All these single and multi-ribozyme genes in plasmids described above were located between the T7 and T3 promoter and derived from plasmid pBluescript II.
Plasmid pKShac’ containing the T7 promoter directed human α-globin cDNA was described previously [32]. Plasmids pKShbSc’ and pKShgc’ containing the T7 promoter directed human βS-globin cDNA and γ-globin cDNA were constructed by respectively placing human β-globin cDNA fragment from pHE7 and Aγ-globin cDNA fragment from pHE9 [37] into pBluescript II after the T7 promoter. A β6 Glu→Val mutation [19] was introduced into β-globin cDNA to produce human βS-globin cDNA in pKShbSc’.
Plasmids for ribozyme expression in K562 cells
The human βS-globin gene fragment (exon 1-IVS1-exon 2-IVS2-exon 3) excluding the promoter was engineered into an NcoI/Pst1 fragment (1268 bp) and inserted downstream of the human cytomegalovirus (CMV) immediate early promoter in the pSP64 vector (Promega, Madison, IL) to make a pCMV-βS construct. The neomycin resistant gene in plasmid pCMV-βS was replaced by a hygromycin resistant gene, and the 0.49-kb Hind III/BamH I fragment of its βS-globin gene was replaced by single and multi-ribozyme genes zb1-zb6, zb21A, and zb61 to form plasmids pHC-zb1-pHC-zb6, pHC-zb21A, and pHC-zb6, in which the corresponding ribozyme and multi-ribozyme genes were directed by the CMV promoter. A similar plasmid with no ribozyme gene insertion was constructed as pHC-0. In addition, a 1.68-kb luciferase gene fragment from pGL3-control (Promega) was inserted into plasmid pCMV-βS after codons 18 and 56 of the βS-globin gene. The resultant plasmids pNC-βS18:luc and pNC-βS56:luc, respectively, contained a βS-globin N-terminal 1-18 and 1-56 amino acid coding sequence fused with a complete luciferase gene directed by the CMV promoter.
Plasmids for ribozyme, long hairpin β-globin mRNA and siRNA expression in progenitor cells
Plasmid pHLG-zb21A containing the multi-ribozyme zb21A gene directed by a shortened human locus control region (LCR) of the β-globin gene cluster and γ-globin promoter was constructed by modifying plasmid pKLCRa2-Zc [32] in following manner: (i) HS1 fraction of LCR was deleted; (ii) α2-globin promoter was replaced by Gγ-globin promoter (-728 - -1); (iii) Zc gene was replaced by multi-ribozyme gene zb21A; and (iv) neomycin resistant gene was replaced by hygromycin resistant gene from pTKhyg (BD Biosciences, Palo Alto, CA). The control plasmid pHLG-0 was constructed by deleting the zb21A gene of pHLG-zb21A. Plasmids pCA-0 and pCA-zb21A are similar to pHC-0 and pHC-zb21A, except the hygromycin resistant gene was deleted, and the LCR-Gγ globin promoter was replaced by CMV promoter.
The long hairpin β-globin mRNA expression plasmid pHC-Fbi1 was constructed from pHC-zb21A, in which the zb21A portion was replaced by a hairpin sequence corresponding to β-globin cDNA 1-133, with a 26-bp loop.
siRNA expression plasmids, pEU-ib2, pEU-ib4, and pEU-ib5, were constructed by inserting corresponding siRNA gene ib2, ib4, and ib5 (see Ribozyme and siRNA Oligonucleotides) into plasmid pSilencer2.0-U6 (Ambion, Austin, TX) between the U6 promoter and TTTTTGGAA sequence [38] and a 0.40-kb enhancer sequence of CMV immediate early promoter (27-431 in pCMVβ, BD Biosciences, Palo Alto, CA) was placed before the U6 promoter [39].
In-vitro transcription and ribozyme cleavage assay
32P-labeled human α-, βS-, and γ-globin mRNAs were respectively prepared by in-vitro transcription using MAXIscript T7 kit (Ambion) with [α-32P]-UTP as the labeled substrate (800 Ci/mmole, Amersham Pharmacia Biotech, Piscataway, NJ) with Spe I linearized pKShac’ and Xho I linearized pKShbSc’ and pKShgc’ as templates. Unlabeled single and multi-ribozyme RNA zb1-zb6, zb21A, and zb61 were prepared using the same kit with Acc65 I linearized pKSzb1-pKSzb6, pKSzb21A and pKSzb61 as templates. Antisense zb21A RNA was synthesized using a MAXIscript T3 kit (Ambion) with Hind III linearized pKSzb21A as template. All in-vitro transcription products were purified with a RNeasy mini kit (Qiagen, Valencia, CA) and their concentrations were estimated by the total isotope incorporation or by UV absorption.
The in-vitro ribozyme cleavage reaction has been described previously [32], except that we used incubation times of 20 and 60 min.
Cell culture and transfection
K562βS cells are derived from human erythroleukemia cells K562 by transfection of the human βS-globin expression plasmid pCMV-βS. For stable integration into K562 cells, the vector contained a neomycin resistant gene cassette and was transfected into K562 cells, and stable clones were selected using G418 (Invitrogen Corp., Carlsbad, CA). Stable expression of βS-globin was confirmed by RT-PCR and sequencing of the reaction product to confirm the presence of the βS-globin mutation. The expression level of βS-globin mRNA in K562βS cells is 391±0.9 attomole/μg total RNA.
The conditions of K562βS cell culture, stable transfection, and separation of single cell colonies have been described previously [32]. Plasmid DNAs used in transfection were prepared using QIAwell 8 plasmid kits (Qiagen). The cells were cultured in RPMI media with no added antibiotics right before transfection.
K562 cells were used in the transient transfection experiment, and LipofectAMINE 2000 (Invitrogen) was used as the transfection reagent. The transient transfection procedure followed the manufacturer’s protocol. Stable transfection and single-cell colony culture have been described previously [32], except we used hygromycin (200 μg /ml) as the selection reagent.
Primer extension assay
Total RNA was prepared from transfected cells or single-cell colonies using RNeasy mini kits (Qiagen). Total mRNA was prepared from total RNA using GenElute mRNA Miniprep Kit (Sigma, St Louis, MO).
Relative amounts of cellular βS-globin mRNA, ribozyme RNA, and other mRNAs in the K562βS cells were determined by primer extension assay. 5′-32P-labeled oligonucleotide primers complementary to βS-globin mRNA, α-globin mRNA, γ-globin mRNA, β-actin mRNA, and ribozyme RNAs, respectively (see Supplemental Data, Table 1S) were prepared with [γ-32P]-ATP (>5000Ci/mmole, Amersham Pharmacia Biotech) and T4 polynucleotide kinase (New England Biolabs, Beverly MA) reaction, and were purified with a QIAquick nucleotide removal kit (Qiagen). We added 0.1 - 0.5 pmole of 32P-primers to reaction mixtures containing total RNA sample, 1 M NaCl, 0.2 M tris-HCl (pH 7.0), and 0.1 M EDTA in a total volume of 100 μl. The mixtures were incubated at 70°C for 3 min and then at 55°C for 1 hr. Then, 0.1 volume of 3 M NaOAc and 2 volumes of ethanol were added, and the mixtures were placed at -20°C for at least 1 hr. The annealed 32P-primer-RNA products were precipitated, washed and then dissolved in 11 μl H2O. We then added 4 μl 5× first stranded buffer containing 15 mM MgCl2 (Invitrogen), 1 μl dNTP (10 mM), 1 μl DTT (0.1 M), 1 μl RNsin (39 units, Promega), and 2 μl MMLV reverse transcriptase (400 units, Invitrogen). The extension reaction proceeded at 42°C for 1 hr. To stop the reaction, we added 20 μl loading buffer II (Ambion), the mixtures were heated at 95°C for 3 min., placed in 0°C bath for 3 min, and then loaded on a 5% polyacrylamide gel with 8 M urea. After electrophoresis, the radioactive bands corresponding to different mRNAs or ribozyme RNAs were located by autoradiography, and the radioactivity of each band was determined by direct counting of the bands that had been cut from the gel using a Beckman LS 7000 scintillation counter. The relative amount of βS-globin mRNA, ribozyme RNA, and other mRNAs was calculated from the radioactivity data using β-actin mRNA as an internal standard. In order to simplify the calculation, 32P-primers for the different RNA tests were labeled with the same specific activity, except for βS-globin mRNA. The γ/βS ratio was directly calculated by the ratio of relative amount of γ-globin mRNA and βS-globin mRNA.
βS-globin mRNA and other RNAs were also determined by primer extension using total mRNA samples. In such tests, because both βS-globin mRNA and the expressed ribozyme existed in the samples, excess degradation of βS-globin mRNA by the co-existed ribozyme may happen. We have found that such degradation can be completely inhibited by adding 0.1 μM antisense-ribozyme RNA into the annealing mixture before primer extension (results not shown). The antisense zb21A RNA can be easily prepared by in-vitro transcription with T3 promoter using pKSzb21A as template.
βS-Globin:luciferase fusion protein assay
βS-globin18:luciferase and βS-globin56:luciferase fusion proteins produced in K562 cells transfected with pNC-βS18:luc and pNC-βS56:luc were directly detected in transient transfection cell lysates using luminometer (Perkin-Elmer Victor2 1420 luminometer, Wellesley, MA) with Bright-Glo reagent (Promega) following the manufacturer’s protocol. The cell lysate transfected by luciferase expression plasmid pGL3-control was used as a transfection efficiency control.
Hematopoietic progenitor cell cultures
Blood was obtained from consenting normal volunteers from the NIH Department of Transfusion Medicine and hematopoietic progenitor cells were harvested and grown in liquid culture [40]. Mononuclear cells were isolated by centrifugation on Ficoll-Hypaque (BioWhittaker, Walkersville, MD). Cells were cultured in α-minimal essential medium supplemented with 10% fetal bovine serum (FBS) (both from Gibco, Grand Island, NY), 10% conditioned medium from bladder carcinoma 5637 cultures, 1.5 mM glutamine (Biofluids, Rockville, MD), 1 μg/ml cyclosporin A (Sigma Chemical Co., St. Louis, MO), and antibiotics. After 5-7 days, non-adherent cells were washed twice with Dulbecco’s phosphate buffered saline without Ca+2 and Mg+2, and transferred to erythropoietin containing medium which consisted of α-minimal essential medium supplemented with 30% FBS (both from Gibco), 1% deionized bovine serum albumin, 10-6 M dexamethasone, 10-5 M β-mercapthoethanol, 0.3 mg/ml human hollo-transferrin (all from Sigma Chemical Co.), 10 ng/ml human recombinant stem cell factor (PeproTech, Rocky Hill, NJ), 1 U/ml human recombinant erythropoietin (EPO) (Amgen Inc., Thousand Oaks, CA), and antibiotics. Cultures were incubated at 37°C in an atmosphere of 5% CO2 and 100% humidity in standard incubators. On day 1 of culture with EPO stimulation, cells were transfection by electroporation with plasmids containing either a multi-ribozyme gene, a long hairpin β-globin cDNA gene, or a siRNA gene. Electroporation was performed with the Amaxa electroporation system (Amaxa, Gaithersburg, MD) using their human CD34 cell nucleofector kit, following the included protocol.
Cells were harvested and mRNA analyzed on day 12 of culture following EPO stimulation. First-strand cDNA was synthesized from 1 mg of total RNA using MuLV reverse transcriptase (RT) and oligo-d(T)16 (Applied Biosystems, Foster City, CA). Quantitative real-time RT-PCR was used to determine the level of mRNA expression with gene-specific primers and fluorescent labeled Taqman probes on a 7700 Sequence Detector (Applied Biosystems) [41]. Probes were designed to span exon junctions in order to prevent the amplification of any contaminating genomic DNA and were fluorescently labeled with FAM (6-carboxy-fluorescein) as the 5′-fluorescent reporter and TAMERA (6-carboxy-tetramethyl-rhodamine) as the 3′ end quencher. Probes and primers were generated using Primer Express (Applied Biosystems). PCR reaction conditions were 50°C for 2 min, 95°C for 4 min and 40 cycles of 95°C (melting temperature) for 15 sec and 60°C (annealing-extension temperature) for 1 min. At low amplification, threshold cycle number (Ct) is directly proportional to the amount of corresponding specific mRNA. Standard curves were created using serial dilutions of plasmids containing the cDNA of interest. Human β-actin was used to normalize all results.
Results
In-vitro cleavage of βS-globin mRNA by ribozymes
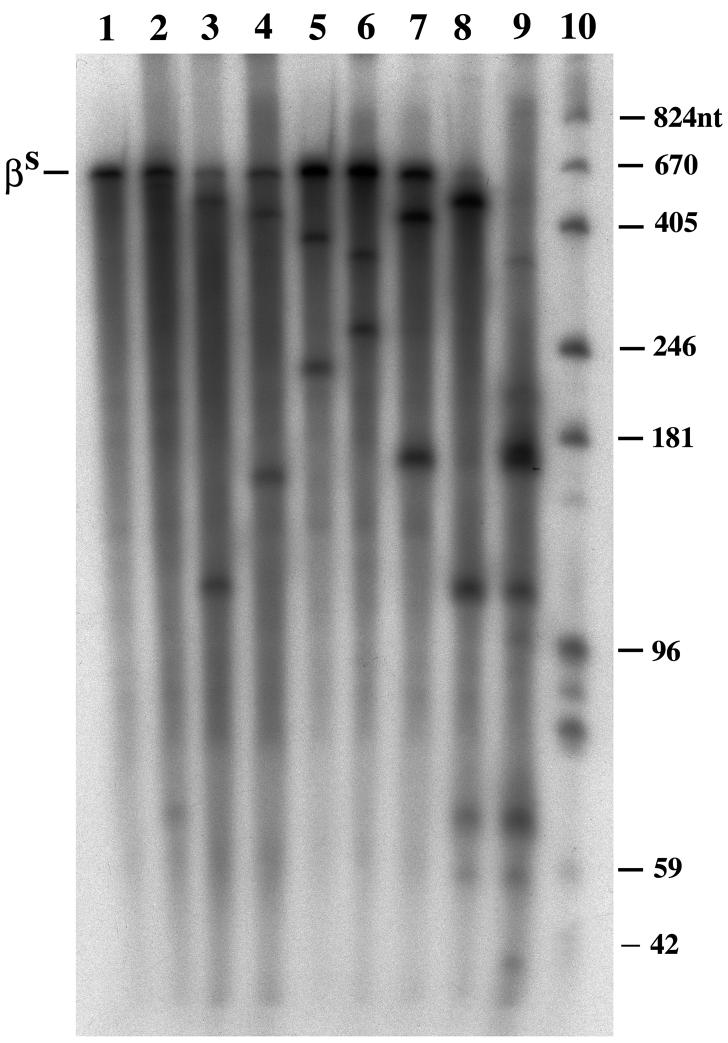
Six single ribozymes zb1-zb6 and two multi-ribozymes zb21A and zb61 were tested for their ability to cleave synthetic βS-globin mRNA in vitro. At a 4:1 molar ratio of ribozyme to substrate, the ribozymes were incubated with 32P-βS-globin mRNA at 37°C for 20 and 60 min. As showed in Fig. 2, the cleavage of βS-globin mRNA was observed in all ribozyme reactions, but with differing degrees of cleavage. Considerable cleavage of βS-globin mRNA was seen in the zb2 and zb3 reactions, but complete cleavage of βS-globin mRNA was only seen in the multi-ribozyme reactions. Both zb21A and zb61 cleaved βS-globin mRNA completely within 20 min, while no single ribozyme did this even at 60 min. These results indicate that the multi-ribozymes are more effective than the single ribozymes used in these experiments in the cleavage of βS-globin mRNA.
Figure 2.
Cleavage of 32P-βS-globin mRNA by ribozymes in vitro. Sample lanes: lane 1, no ribozyme; lane 2, zb1; lane 3, zb2; lane 4, zb3; lane 5, zb4; lane 6, zb5; lane 7, zb6; lane 8, zb21A; lane 9, zb61 and lane 10, RNA size marker. Substrate: βS, 32P-βS-globin mRNA. Reactions were performed at 37°C for 60 min.
Specificity of βS-globin ribozymes
To test the substrate specificity of these ribozymes, [32P]-α-globin mRNA was used as the substrate and incubated with ribozymes zb2, zb21A, and zb61 at 37°C for 60 min. and no cleavage in α-globin mRNA is found while βS-globin mRNA is well cleaved. Similarly, interaction of the βS-globin ribozymes with γ-globin mRNA was assessed using [32P]-γ-globin mRNA as the substrate and incubated with ribozymes zb2, zb21A, and zb61 at 37°C for 60 min. In contrast to the cleavage of βS-globin mRNA, no cleavage in γ-globin mRNA is observed (see Supplemental Data, Figs. 1S and 2S).
Reduction of βS-globin mRNA level in K562βS cells by ribozymes
In order to assess the efficacy of these ribozymes in living cells, single and multi-ribozyme expression plasmids pHC-zb1-pHC-zb6, pHC-zb21A, and pHC-zb61 were transfected into K562βS cells. Cells transfected with plasmid pHC-0 was used as the control. The ribozyme expression level in each transfection mixture was also monitored, and only the samples in which ribozyme expression fell in the range of 1.7 to 1.9 times the level for β-actin mRNA were used to make comparison. The results showed that there was reduction of βS-globin mRNA in 4 of the 7 transfected cell mixtures (Fig. 3). The greatest reduction was found in cells transfected with plasmids containing the multi-ribozyme zb21A, and a lesser degree of βS-globin reduction was observed in cells transfected with multi-ribozyme zb61, and with single ribozymes zb1 and zb2. Single ribozymes zb3, zb4, and zb5 were not very effective in K562βS cells. No results were obtained for zb6 because its expression was too low following transfection into K562βS cells.
Figure 3.
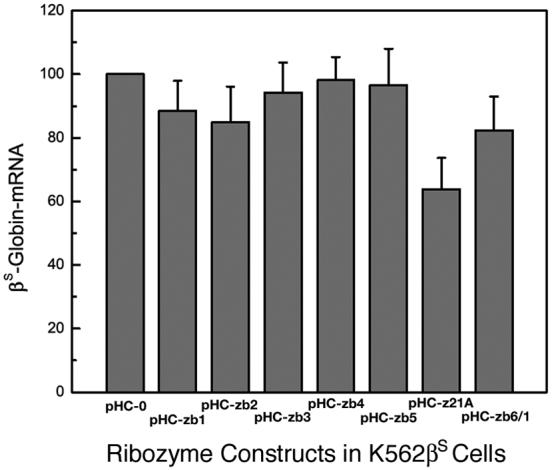
Reduction of βS-globin mRNA by ribozymes in K562βS cells. K562βS cells were transfected with different ribozyme constructs and total RNA extracted. The amounts of βS-globin mRNA, β-actin mRNA, and ribozyme RNA were determined by primer extension assays and normalized to β-actin mRNA. The βS-globin mRNA value obtained for K562βS cells transfected with pHC-0 (control plasmid) is set at 100%. RNA samples from cells transfected with (lanes 1-8, from left): pHC-0; pHC-zb1; pHC-zb2; pHC-zb3; pHC-zb4; pHC-zb5; pHC-zb21A; and pHC-zb61.
Increase in the ratio of cellular γ-globin mRNA to βS-globin mRNA in K562βS cells by multi-ribozyme zb21A
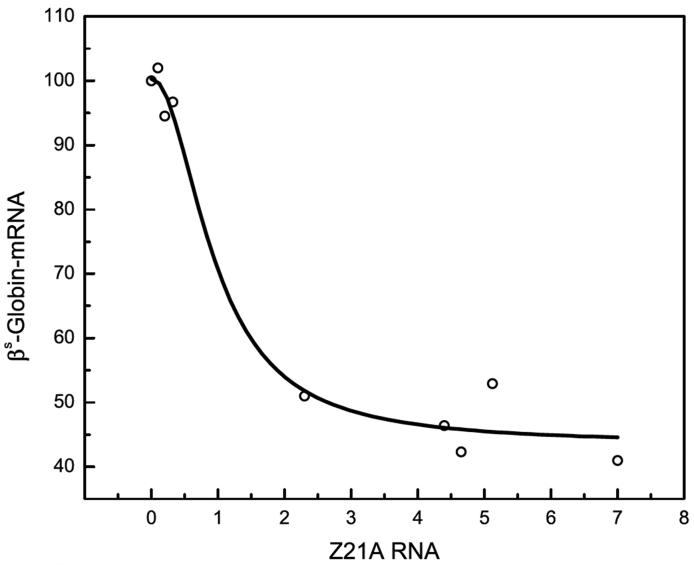
Ribozyme expression in a transfection cell mixture can vary among individual cells [32]. Thus, in order to determine the relationship between βS-globin mRNA reduction and multi-ribozyme zb21A expression level, single cell colonies were selected from K562βS cells stably transfected with pHC-zb21A. Total mRNA was prepared from different clonal cell lines and the relative amounts of βS-globin mRNA and of multi-ribozyme zb21A were determined. We have found that in clonal lines expressing low levels of ribozyme, there was little or no βS-globin mRNA reduction. While, significant βS-globin mRNA reduction was observed in clonal lines with higher ribozyme expression (Fig. 4).
Figure 4.
Relation between the reduction of βS-globin mRNA and the expression of multi-ribozyme zb21A in single cell colony cultures. K562βS cells transfected with pHC-zb21A and single colony cultures were used for test. The relative cellular βS-globin mRNA level was explained in Fig. 5. The multi-ribozyme expression level was presented as the ratio of multi-ribozyme zb21A RNA to β-actin mRNA.
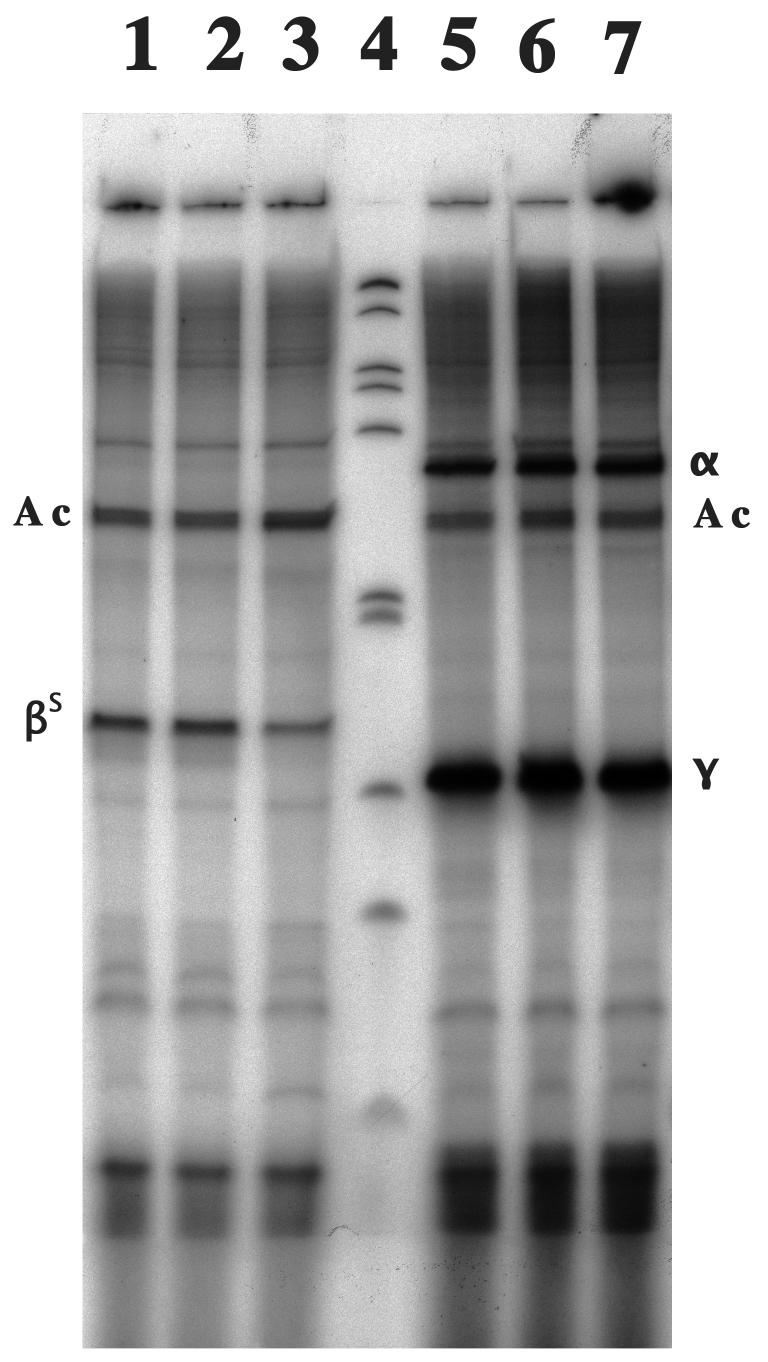
The effective reduction in βS-globin expression was compared with the expression of α- and γ-globin expression. The mean reduction of βS-globin mRNA in single cell colonies with high zb21A expression was 53.6% ± 9.7%. The ratio of γ-globin mRNA to βS-globin mRNA in these cells was 1.7 - 2.0 times higher than that in control cells (Fig. 5).
Figure 5.
Cellular α-globin mRNA, βS-globin mRNA and γ-globin mRNA in K562βS cells transfected with pHC-zb21A. Primer extension assay results: lanes 1 and 5: no plasmid; lanes 2 and 6: pHC-0 (control plasmid); lane 3 and 7: pHC-zb21A (multi-ribozyme zb21A); lane 4, DNA size marker. 32P-primer: lane 1-3: E-at3, E-b5; lanes 5-7: E-at3, E-a1, and E-g1. Primer extension products: α, α-globin mRNA; βS, βS-globin mRNA; γ, γ-globin mRNA; and Ac, β-actin mRNA.
Reduction of βS-globin:luciferase fusion protein in single K562 cell colonies
As another measure of zb21A ribozyme activity, βS-globin:luciferase fusion genes were constructed linking the luciferase reporter gene downstream of codons 18 and 56 of βS-globin to make pNC-βS18:luc and pNC-βS56:luc. These plasmids were used as reporters of βS-globin gene expression in K562 cells. Plasmids pNC-βS18:luc and pNC-βS56:luc were transfected into K562 cells with or without stable high zb21A expression, and transient expression of the luciferase reporter gene was detected (Table 1). A 56.9% reduction of βS-globin18:luciferase was found in K562 cells harboring plasmid pHC-zb21A compared with K562 cells transfected with plasmid pHC-0. A similar result was found in βS-globin56:luciferase detection. These data were consistent with the reduction seen for βS-globin mRNA earlier.
Table 1.
βS:luc activity in K562 cells transfected with multi-ribozyme gene
| Report gene Reduction plasmid | Luciferase activity (×105 units) in | Relative | Corrected | ||
|---|---|---|---|---|---|
| K562/pHC-0 (control) | K562/pHC-zb21A (multi-ribozyme) | luciferase activity* | relative activity** | of βS:luc expression | |
| pGL3-control | 2763 | 2428 | 0.879*** | ||
| pNC-βS18:luc | 1702 | 645 | 0.379 | 0.431 | 56.9% |
| pNC-βS56:luc | 381 | 149 | 0.390 | 0.444 | 55.6% |
Relative luciferase activity is the luciferase activity in K562/pHC-zb21A related to that in control cells.
Corrected relative activity is the relative luciferase activity corrected with transfection efficiency.
This value means the transfection efficiency of K562/pHC-z21A cells related to control cells.
Increase in the ratio of cellular γ-globin mRNA to β-globin mRNA in human hematopoietic progenitor cells by multi-ribozyme, long hairpin mRNA, and siRNA
Primary human hematopoietic progenitor cells stimulated with EPO were used to test the ability to reduce β-globin expression during erythroid differentiation using the multi-ribozyme zb21A that proved to be effective in K562 cells. The changes of the γ/(γ+β) ratio are shown in Table 2. The greatest increase in the γ/(γ+β) ratio of 2.3-fold relative to control was observed in cells transfected with plasmids containing the zb21A gene directed by the LCR-γ-globin promoter (pHLG-zb21A). This is due in part to a decrease in β-globin mRNA of 0.52 ± 0.06-fold and increase in γ-globin mRNA of 1.95 ± 0.2-fold compared to control cultures. In contrast, control plasmid pHLG-0 had only a modest effect on the γ/(γ+β) ratio. A significant increase in the γ/(γ+β) ratio of 2.0-fold relative to control was observed with pCA-zb21A that contained multi-ribozyme zb21A gene directed by CMV promoter, although this increase was not as great as that for pHLG-zb21A that incorporated the erythroid specific LCR-γ-globin promoter. Changes in globin gene expression with the pCA-zb21A plasmid were a to 0.59 ± 0.01-fold decrease in β-globin mRNA and a 1.68 ± 0.19-fold increase in γ-globin mRNA compared to control cultures.
Table 2.
The γ/(γ+β) ratio in progenitor cells transfected with multi-ribozyme gene
| Samples | Experiment 1 | Experiment 2 | Mean | ||
|---|---|---|---|---|---|
| γ/(γ+β)* | relative γ/(γ+β) | γ/(γ+β) | relative γ/(γ+β) | relative γ/(γ+β)** | |
| Control/Mock | 0.26 | 1.00 | 0.20 | 1.00 | 1.00 |
| pHLG-0 | 0.39 | 1.48 | 0.32 | 1.57 | 1.53 |
| pLHG-zb21A | 0.58 | 2.23 | 0.49 | 2.40 | 2.32 |
| pCA-0 | 0.33 | 1.26 | 0.29 | 1.42 | 1.34 |
| pCA-zb21A | 0.49 | 1.86 | 0.44 | 2.15 | 2.01 |
β-globin mRNA and γ-globin mRNA are calculated relative to β-actin mRNA.
Mean relative γ/(γ+β) is the mean value from Experiments 1 and 2.
The ability of long hairpin mRNA designed from the β-globin sequence to affect β-globin expression during erythroid differentiation was also determined. A long hairpin mRNA expression construct corresponding to the β-globin mRNA sequence from 1 to 133 with a 26 bp loop (pHC-Fbi1) was assayed directly in the primary hematopoietic progenitor cell cultures stimulated with erythropoietin and β- and γ-globin expression determined at the completion of the 12-day culture period. Introduction of this construct into differentiating erythroid progenitor cells did not increase in the γ/(γ+β) ratio significantly greater than the control plasmid pHLG-0 (Tables 2 and 3), although there was a reduction in β-globin expression by 0.7± 0.2-fold and a detectable increase in γ-globin expression compared to control cultures.
Table 3.
The γ/(γ+β) ratio in progenitor cells transfected with long hairpin β-globin cDNA and siRNA genes
| Samples | Experiment 1 | Experiment 2 | Mean | ||
|---|---|---|---|---|---|
| γ/(γ+β)* | relative γ/(γ+β) | γ/(γ+β) | relative γ/(γ+β) | relative γ/(γ+β)** | |
| Control/Mock | 0.24 | 1.00 | 0.08 | 1.00 | 1.00 |
| pHC-Fbi1 | 0.40 | 1.67 | 0.09 | 1.13 | 1.40 |
| pEU-ib2 | 0.41 | 1.71 | 0.20 | 2.16 | 1.94 |
| pEU-ib4 | 0.48 | 1.99 | 0.15 | 1.63 | 1.81 |
| pEU-ib5 | 0.50 | 2.05 | 0.21 | 2.33 | 2.19 |
β-globin mRNA and γ-globin mRNA are calculated relative to β-actin mRNA.
Mean relative γ/(γ+β) is the mean value from Experiments 1 and 2.
Constructs were also designed to express siRNA directed against β-globin. The siRNA constructs were designed to target nucleotides the β-globin transcript region from 104 to 125 (ib2), from 230 to 251 (ib4), and from 248 to 272 (ib5). These constructs were directly tested in the primary hematopoietic progenitor cell cultures undergoing erythroid differentiation. The γ/(γ+β) ratio data are summarized in Table 3. The three siRNA constructs increased the γ/(γ+β) ratio with the most effective being siRNA ib5 with an increase of 2.19 ± 0.2-fold compared to control, resulting from a decrease in β-globin mRNA by 0.56 ± 0.30-fold and an increase in γ-globin mRNA by 1.7 ± 1.0-fold compared to control cultures.
Discussion
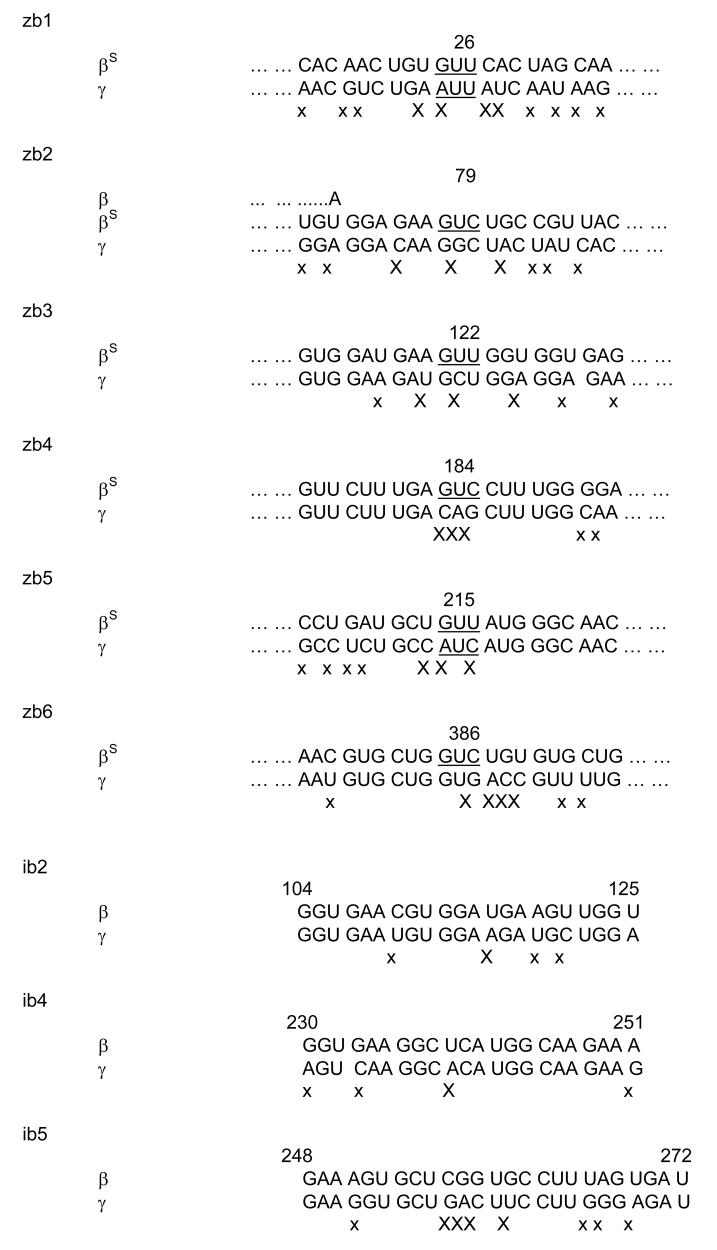
The high degree (70%) of sequence homology between β-globin mRNA and γ-globin mRNA necessitates assessing the specificity of ribozyme and siRNA constructs directed against β-globin for cleavage of β-globin mRNA in the presence of γ-globin mRNA. In this work, six sites in βS-globin mRNA were selected for ribozyme design against βS-globin mRNA (Fig. 6) to ensure at least one mismatch was located in the cleavage site of the ribozyme [42]. Our results have shown that all of those ribozymes attacked βS-globin mRNA or β-globin mRNA rather than γ-globin mRNA (Figs. 2S and 5). In addition, our results show that the ribozyme zb21A designed to cleave βS-globin mRNA also targeted normal β-globin mRNA (Table 2), indicating that the ribozyme does not discriminate a single nucleotide difference at a site away from the cleavage site. Similarly, three sequences of β-globin mRNA were used to design siRNAs against β-globin mRNA (Fig. 6) with at least one mismatch between the β-globin and γ-globin sequences located in the middle region of the siRNA [43]. We observed that siRNAs decreased β-globin mRNA and not γ-globin mRNA demonstrating specificity of these siRNA.
Figure 6.
Ribozyme target sequences and siRNA related sequences in βS-globin mRNA and related sequences in γ-globin mRNA. Underlined: ribozyme target sequence; X and x: base(s) that differ between β-globin and βS-globin mRNA and corresponding γ-globin mRNA. X was the mismatch near the cleavage site of ribozyme or in the middle region of the siRNA. Note the difference between β- and βS-globin mRNA is showed in zb2. The number is according to β-globin mRNA or cDNA.
Choosing the target cleavage site and designing the corresponding ribozyme are mostly empirical. In our previous study [32] and in this work, we have found that multi-ribozymes, e.g., a ribozyme containing more than one target/catalytic sequences, even with only two ribozyme sequences (such as zb21A), is more effective than single ribozymes. The coordinate action of two ribozymes has also been reported in trans-splicing ribozymes [44]. The higher efficacy of multi-ribozymes may be due to the fact that they bind the target mRNA in many locations simultaneously and change the mRNA conformation in such a manner that the mRNA becomes more available for cleavage in some sites. In culture, the CMV promoter was able to drive multi-ribozyme expression in K562βS cells to reduce βS-globin. However, we have observed that the construct that employed the erythroid specific LCR enhancer linked to the γ-globin promoter to drive the multi-ribozyme gene expression was more effective than the construct in which the CMV promoter was used to drive the multi-ribozyme gene expression in reducing β-globin gene expression (data not shown) and increasing the γ/(γ+β) ratio in primary hematopoietic progenitor cells (Table 2).
Long double stranded RNA has been regarded as the precursors of cellular small RNAs. However, for most mammalian cells, direct introduction of long double stranded RNA may trigger interferon regulated non-specific RNA destruction and protein synthesis arrest except for embryo cells in which non-specific antiviral responses are not prevalent [45]. Here, we have found that the introduction of a long hairpin β-globin mRNA expression in human hematopoietic progenitor cells has only a minimal effect on the γ/(γ+β) ratio (Table 3) and with no apparent toxic effect. This result implies that like typical embryonic cells, differentiating human hematopoietic progenitor cells may not have a specific antiviral mechanism for non-specific RNA destruction.
As with the ribozymes, little is known about the relationship between efficacy and individual siRNA sequence. As showed in Table 3, we find that siRNA ib5 is more effective than ib2 and ib4 in increasing the γ/(γ+β) ratio in hematopoietic progenitor cells. In order to obtain the most effective siRNA construct, a systematic search of more siRNA sequences is required.
The increase of γ/(γ+β) ratio in EPO stimulated primary human hematopoietic progenitor cells by introducing multi-ribozyme, long hairpin β-globin mRNA, or siRNA results from both the decrease of β-globin mRNA and the increase of γ-globin mRNA (data not shown). The former is the predicted result of ribozyme and siRNA, and the latter was somewhat unexpected. The reduction of β-globin with a compensatory increase of γ-globin production suggests a possible competitive regulatory control mechanism for globin gene expression beyond that described between the γ-globin and β-globin promoters [46]. While both the CMV promoter and erythroid specific LCR-γ-globin promoter driving multi-ribozyme expression are able to increase the overall γ/(γ+β) ratio, the erythroid specific promoter construct is more effective. The increased effectiveness of the promoter derived from portions of the β-like globin cluster is consistent with other studies using an analogous gene transfer approach for sickle cell anemia [31].
It has been shown that a single ribozyme with expression driven by a human β-globin promoter linked to an LCR derived enhancer is able to reduce βS-globin production in erythroid cells in vivo in a transgenic mouse model [31]. Toward increasing the efficiency of the ribozyme technology for decreasing β-globin expression, we have found that the effectiveness of the multi-ribozyme zb21A is greater than single ribozymes and comparable to the siRNA ib5, the siRNA with the highest activity in reducing β-globin mRNA. The γ/(γ+β) ratio can be increased 2-fold or more by introducing multi-ribozyme zb21A or siRNA ib5 into primary human hematopoietic progenitor cells stimulated to undergo erythroid differentiation (Tables 2 and 3). The combination of targeting βS-globin expression and increasing the γ/(γ+βS) ratio may be very useful in reducing the tendency of HbS polymerization and cell sickling in sickle blood cells. The potential of using high-efficiency gene transfer techniques in hematopoietic cells, such as the lentiviral vector to obtain long stable gene suppression, has been suggested using a combination of ribozyme and short hairpin RNA strategies against HIV-1 infection [47]. Because HbF is not uniformally distributed, increasing the abundance of HbF in a large proportion of red cells has been an important goal in gene transfer approaches to treat sickle cell anemia. Toward this end, targeting βS-globin gene expression by introduction a multi-ribozyme, for example, combined with a gene transfer technique to increase γ-globin gene expression may be more beneficial in increasing the γ/(γ+β) ratio than either strategy alone. The polymerization potential of a HbS mixture with 20% or more HbF at 32-34 g/dl approaches that for sickle trait [48]. Reaching this level of HbF in all sickle erythrocytes is a useful therapeutic goal as it would reduce the overall polymerization potential to that associated with the clinically benign sickle trait.
Supplementary Material
Cleavage of 32P-α-globin mRNA and 32P-βS-globin mRNA by ribozymes in vitro. Sample lanes: lanes 1-4, 32P-α-globin mRNA; lanes 5-8, 32P-βS-globin mRNA. Ribozyme added: lanes 1 and 5, none; lanes 2 and 6, zb2; lanes 3 and 7, zb21A; lanes 4 and 8, zb61. Substrates: α, 32P-α-globin mRNA; βS, 32P-βS-globin mRNA.
Cleavage of 32P-βS-globin mRNA and 32P-γ-globin mRNA by ribozymes in vitro. Sample lanes (from left): lanes 1-4, 32P-βS-globin mRNA; lanes 5-8, 32P-γ-globin mRNA; lane 9, RNA size marker. Ribozyme added to lanes 1 and 5, none; lanes 2 and 6, zb2; lanes 3 and 7, zb21A; and lanes 4 and 8, zb61. Substrates: βS, 32P-βS-globin mRNA; γ, 32P-γ-globin mRNA.
Acknowledgments
This work is supported by research grant from the National Institutes of Health (R01HL-024525).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bunn HF. Pathogenesis and treatment of sickle cell disease. N. Engl. J. Med. 1997;337:762–769. doi: 10.1056/NEJM199709113371107. [DOI] [PubMed] [Google Scholar]
- 2.Perrine RP, Pembrey ME, John ME, Perrine S, Shoup F. Natural history of sickle cell anemia in Saudi Arabs. A study of 270 subjects. An. Intern. Med. 1978;88:1–6. doi: 10.7326/0003-4819-88-1-1. [DOI] [PubMed] [Google Scholar]
- 3.Powars DR, Weiss JN, Chan LS, Schroeder WA. Is there a threshold level of fetal hemoglobin that ameliorates morbidity in sickle cell anemia? Blood. 1984;63:921–926. [PubMed] [Google Scholar]
- 4.Bookchin RM, Nagel RL, Balazs T. Role of hybrid tetramer formation in gelatin of hemoglobin S. Nature. 1975;256:667–668. doi: 10.1038/256667a0. [DOI] [PubMed] [Google Scholar]
- 5.Sunshine HR, Hofrichter J, Eaton WA. Gelation of sickle cell hemoglobin in mixture with normal adult and fetal hemoglobins. J. Mol. Biol. 1979;133:435–467. doi: 10.1016/0022-2836(79)90402-9. [DOI] [PubMed] [Google Scholar]
- 6.Steinberg MH. Pathophysiologically based drug treatment of sickle cell disease. Trends Pharmacol. Sci. 2006;27:204–210. doi: 10.1016/j.tips.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 7.Sabatino DE, Seidel NE, Aviles-Mendoza GJ, et al. Long-term Expression of γ-globin mRNA in mouse erythrocytes from retrovirus vectors containing the human γ-globin gene fused to the ankyrin-1 promoter. Proc. Natl. Acad. Sci. USA. 2000;97:13294–13299. doi: 10.1073/pnas.230453097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.May C, Rivella S, Callegari J, et al. Therapeutic hemoglobin synthesis in beta-thalassaemic mice expressing lentivirus-encoded human beta-globin. Nature. 2000;406:82–86. doi: 10.1038/35017565. [DOI] [PubMed] [Google Scholar]
- 9.Pawliuk R, Westerman KA, Fabry ME, et al. Correction of sickle cell disease in transgenic mouse models by gene therapy. Science. 2001;294:2368–2371. doi: 10.1126/science.1065806. [DOI] [PubMed] [Google Scholar]
- 10.Person DA, Hargrove PW, Allay ER, Hanawa H, Nienhuis AW. The degree of phenotypic correction of murine β-thalassemia intermedia following lentiviral-mediated transfer of a human γ-globin gene is influenced by chromosomal position effects and vector copy number. Blood. 2003;101:2175–2183. doi: 10.1182/blood-2002-07-2211. [DOI] [PubMed] [Google Scholar]
- 11.Ward M, Sattler R, Grossman IR, et al. A stable murine-based RD114 retroviral packaging line efficiently transduces human hematopoietic cells. Mol. Ther. 2003;8:804–812. doi: 10.1016/j.ymthe.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Levasseur DN, Ryan TM, Pawlik K, Townes TM. Correction of a mouse model of sickle cell disease: lentiviral/antisickling beta-globin gene transduction of unmobilized, purified hematopoietic stem cells. Blood. 2003;102:4312–4319. doi: 10.1182/blood-2003-04-1251. [DOI] [PubMed] [Google Scholar]
- 13.Imren S, Fabry ME, Westerman KA, et al. High-level beta-globin expression and preferred intragenic integration after lentiviral transduction of human cord blood stem cells. J. Clin. Invest. 2004;114:953–962. doi: 10.1172/JCI21838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rivella S, May C, Chadburn A, Riviere I, Sadelain M. A novel murine model of Cooley anemia and its rescue by lentiviral-mediated human beta-globin gene transfer. Blood. 2003;101:2932–2939. doi: 10.1182/blood-2002-10-3305. [DOI] [PubMed] [Google Scholar]
- 15.Hanawa H, Hargrove PW, Kepes S, Srivastava DK, Nienhuis AW, Persons DA. Extended beta-globin locus control region elements promote consistent therapeutic expression of a gamma-globin lentiviral vector in murine beta-thalassemia. Blood. 2004;104:2281–2290. doi: 10.1182/blood-2004-03-0863. [DOI] [PubMed] [Google Scholar]
- 16.Puthenveetil G, Scholes J, Carbonell D, et al. Successful correction of the human beta-thalassemia major phenotype using a lentiviral vector. Blood. 2004;104:3445–3453. doi: 10.1182/blood-2004-04-1427. [DOI] [PubMed] [Google Scholar]
- 17.Noguchi CT. Intercellular polymer formation in sickle cell syndromes: theoretical and experimental analysis of AS, SC and SS cells. Biophys. J. 1984;45:1153–1158. [Google Scholar]
- 18.Nagel RL. Sickle cell anemia is a multigene disease: sickle painful crises, a case in point. Am. J. Hematol. 1993;42:96–101. doi: 10.1002/ajh.2830420119. [DOI] [PubMed] [Google Scholar]
- 19.Ho C, Willis BF, Shen TJ, et al. Roles of α114 and β87 amino acid residues in the polymerization of hemoglobin S: Implications of gene therapy. J. Mol. Biol. 1996;263:475–485. doi: 10.1006/jmbi.1996.0590. [DOI] [PubMed] [Google Scholar]
- 20.Christoph GW, Hofrichter J, Eaton WA.Understanding the shape of sickled red cells Biophys. J 2005881371–6. Epub 2004 Nov 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rotter M, Aprelev A, Adachi K, Ferrone FA. Molecular crowding limits the role of fetal hemoglobin in therapy for sickle cell disease. J. Mol. Biol. 2005;347:1015–1023. doi: 10.1016/j.jmb.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Steinberg MH, Lu ZH, Barton FB, Terrin ML, Charache S, Dover GJ. Fetal hemoglobin in sickle cell anemia: determinants of response to hydroxyurea. Multicenter Study of Hydroxyurea. Blood. 1997;89:1078–1088. [PubMed] [Google Scholar]
- 23.Chang YP, Maier-Redelsperger M, Smith KD, et al. The relative importance of the X-linked FCP locus and beta-globin haplotypes in determining haemoglobin F levels: a study of SS patients hemozygous for beta S haplotypes. Br. J. Haematol. 1997;96:806–814. doi: 10.1046/j.1365-2141.1997.d01-2094.x. [DOI] [PubMed] [Google Scholar]
- 24.Cech TR, Zaug AJ, Grabowski PJ. In vitro splicing of the ribosomal RNA precursor of Tetrahemena: Involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell. 1981;27:487–496. doi: 10.1016/0092-8674(81)90390-1. [DOI] [PubMed] [Google Scholar]
- 25.Forster AC, Symons RH. Self-cleavage of virusoid RNA is performed by the proposed 55-nucleotide active site. Cell. 1987;50:9–16. doi: 10.1016/0092-8674(87)90657-x. [DOI] [PubMed] [Google Scholar]
- 26.Chowrira BM, Burke JM. Binding and cleavage of nucleic acids by “hairpin” ribozyme. Biochemistry. 1991;30:8518–8522. doi: 10.1021/bi00099a003. [DOI] [PubMed] [Google Scholar]
- 27.Sioud M. Application of performed hammerhead ribozymes in the gene therapy of cancer (review) Int. J. Mol. Med. 1999;4:381–4. doi: 10.3892/ijmm.3.4.381. [DOI] [PubMed] [Google Scholar]
- 28.Gaughan DJ, Whitehead AS. Function and biological applications of catalytic nucleic acids. Biochim. Biophys. Acta. 1999;1445:1–20. doi: 10.1016/s0167-4781(99)00021-4. [DOI] [PubMed] [Google Scholar]
- 29.Amado RG, Mitsuyasu RT, Rosenblatt JD, et al. Anti-human immunodeficiency virus hematopoietic progenitor cell-delivered ribozyme in a phase I study: myeloid and lymphoid reconstitution in human immunodeficiency virus type-1-infected patients. Hum. Gene Ther. 2004;15:251–262. doi: 10.1089/104303404322886101. [DOI] [PubMed] [Google Scholar]
- 30.Lan N, Howrey RP, Lee S-W, Smith CA, Sullenger BA. Ribozyme-mediated repair of sickle β-globin mRNAs in erythrocyte precursors. Science. 1998;280:1593–1596. doi: 10.1126/science.280.5369.1593. [DOI] [PubMed] [Google Scholar]
- 31.Alami R, Gilman JG, Feng YQ, et al. Anti βS-ribozyme reduces βS mRNA levels in transgenic mice: Potential application to the gene therapy of sickle cell anemia. Blood Cells Mol. Dis. 1999;25:110–19. doi: 10.1006/bcmd.1999.0235. [DOI] [PubMed] [Google Scholar]
- 32.Shen TJ, Ikonomi P, Smith R, Noguchi CT, Ho C. Multi-ribozyme targeting of human α-globin gene expression. Blood Cells Mol. Dis. 1999;25:361–373. doi: 10.1006/bcmd.1999.0266. [DOI] [PubMed] [Google Scholar]
- 33.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CD. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1988;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 34.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 35.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 36.Zhao N, Zu Z-X, Liu C-M, Dong W-J, Liu D-P, Liang C-C. Knockdown of mouse adult β-globin gene expression in MEL cells by retrovirus vector-mediated RNA interference. Mol. Biotech. 2004;28:195–199. doi: 10.1385/MB:28:3:195. [DOI] [PubMed] [Google Scholar]
- 37.Shen TJ, Ho NT, Zou M, et al. Production of human normal adult and fetal hemoglobins in Escherichia coli. Protein Eng. 1997;10:1085–1097. doi: 10.1093/protein/10.9.1085. [DOI] [PubMed] [Google Scholar]
- 38.Sui G, Soohoo C, Affar EB, et al. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia XG, Zhou H, Ding H, Affar E, Shi Y, Xu Z. An enhanced U6 promoter for synthesis of short hairpin RNA. Nucleic Acids Res. 2003;31:e100. doi: 10.1093/nar/gng098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fibach E. Techniques for studying stimulation of fetal hemoglobin production in human erythroid cultures. Hemoglobin. 1998;22:445–458. doi: 10.3109/03630269809071542. [DOI] [PubMed] [Google Scholar]
- 41.Yu X, Shacka JJ, Eells JB, et al. Erythropoietin receptor signaling is required for normal brain development. Development. 2002;129:505–516. doi: 10.1242/dev.129.2.505. [DOI] [PubMed] [Google Scholar]
- 42.Werner M, Uhlenbeck OC. The effect of base mismatches in the substrate recognition helices of hammerhead ribozymes on binding and catalysis. Nucleic Acids Res. 1995;23:2092–2096. doi: 10.1093/nar/23.12.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller VM, Gouvion CM, Davidson BL, Paulson HL. Targeting Alzheimer’s disease genes with RNA interference: an efficient strategy for silencing mutant alleles. Nucleic Acids Res. 2004;32:661–668. doi: 10.1093/nar/gkh208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lan N, Rooney BL, Lee SW, Howrey RP, Smith CA, Sullenger BA. Enhancing RNA repair efficiency by combining trans-splicing ribozymes that recognize different accessible sites on a target RNA. Mol. Ther. 2000;2:245–255. doi: 10.1006/mthe.2000.0125. [DOI] [PubMed] [Google Scholar]
- 45.Billy E, Brondani V, Zhang H, Muller U, Filipowicz W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl. Acad. Sci. USA. 2001;98:14428–14433. doi: 10.1073/pnas.261562698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jane SM, Ney PA, Vanin EF, Gumucio DL, Nienhuis AW. Identification of a stage selector element in the human gamma-globin gene promoter that fosters preferential interaction with the 5′ HS2 enhancer when in competition with the beta-promoter. EMBO J. 1992;11:2961–2969. doi: 10.1002/j.1460-2075.1992.tb05366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li MJ, Kim J, Li S, et al. Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol. Ther. 2005;12:900–909. doi: 10.1016/j.ymthe.2005.07.524. [DOI] [PubMed] [Google Scholar]
- 48.Noguchi CT, Rodgers GP, Serjeant G, Schechter AN. Levels of fetal hemoglobin necessary for treatment of sickle cell disease. New England J. Med. 1988;318:96–99. doi: 10.1056/NEJM198801143180207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cleavage of 32P-α-globin mRNA and 32P-βS-globin mRNA by ribozymes in vitro. Sample lanes: lanes 1-4, 32P-α-globin mRNA; lanes 5-8, 32P-βS-globin mRNA. Ribozyme added: lanes 1 and 5, none; lanes 2 and 6, zb2; lanes 3 and 7, zb21A; lanes 4 and 8, zb61. Substrates: α, 32P-α-globin mRNA; βS, 32P-βS-globin mRNA.
Cleavage of 32P-βS-globin mRNA and 32P-γ-globin mRNA by ribozymes in vitro. Sample lanes (from left): lanes 1-4, 32P-βS-globin mRNA; lanes 5-8, 32P-γ-globin mRNA; lane 9, RNA size marker. Ribozyme added to lanes 1 and 5, none; lanes 2 and 6, zb2; lanes 3 and 7, zb21A; and lanes 4 and 8, zb61. Substrates: βS, 32P-βS-globin mRNA; γ, 32P-γ-globin mRNA.