Abstract
Mvwf1 is a cis-regulatory mutation previously identified in the RIIIS/J mouse strain that causes a unique tissue-specific switch in the expression of an N-acetylgalactosaminyltransferase, B4GALNT2, from intestinal epithelium to vascular endothelium. Vascular B4galnt2 expression results in aberrant glycosylation of von Willebrand Factor (VWF) and accelerated VWF clearance from plasma. We now report that 13 inbred mouse strains share the Mvwf1 tissue-specific switch and low VWF phenotype, including five wild-derived strains. Genomic sequencing identified a highly conserved 97-kb Mvwf1 haplotype block shared by these strains that encompasses a 30-kb region of high nucleotide sequence divergence from C57BL6/J flanking B4galnt2 exon 1. The analysis of a series of bacterial artificial chromosome (BAC) transgenes containing B4galnt2 derived from the RIIIS/J or C57BL6/J inbred mouse strains demonstrates that the corresponding sequences are sufficient to confer the vessel (RIIIS/J) or intestine (C57BL6/J)-specific expression patterns. Taken together, our data suggest that the region responsible for the Mvwf1 regulatory switch lies within an approximately 30-kb genomic interval upstream of the B4galnt2 gene. The observation that Mvwf1 is present in multiple wild-derived strains suggests that this locus may be retained in wild mouse populations due to positive selection. Similar selective pressures could contribute to the high prevalence of von Willebrand disease in humans.
Electronic supplementary material
The online version of this article (doi:10.1007/s00335-007-9079-4) contains supplementary material, which is available to authorized users.
Introduction
The RIIIS/J inbred mouse strain carries a spontaneous gain-of-function mutation that specifically switches the expression of β1,4-N-acetylgalactosaminyltransferase, B4GALNT2 (previously referred to as GALGT2), from the intestinal epithelial pattern seen in most mice to a vascular endothelial cell-specific pattern. Endothelial cell expression of B4GALNT2 leads to aberrant post-translational modification of endothelial-derived von Willebrand Factor (VWF) with subterminal GalNAc residues, resulting in accelerated clearance and low circulating VWF levels. This unique regulatory mutation was termed Mvwf1, for Modifier of VWF 1 (Mohlke et al. 1999).
Glycosyltransferases serve a critical role in post-translational modification of proteins and are generally either spatially or temporally restricted in their expression programs (Lowe and Marth 2003). Up to 1% of mammalian genes are involved in glycosylation, and 16 human congenital disorders of coagulation have been described, several of which affect coagulation (Haltiwanger and Lowe 2004; Marquardt and Denecke 2003). The physiologic function of the murine B4galnt2 gene and its human ortholog, B4GALNT2, are unknown (Montiel et al. 2003), and B4galnt2 knockout animals are viable and have no discernable phenotype under laboratory conditions (Lowe and Marth 2003). Several spontaneous mutations altering the tissue specificity of a given gene’s expression have been reported, generally resulting in either a change in developmental timing (Crossley et al. 1992; Cunningham and Jane 1996) or a shift from a spatially restricted to a more generalized pattern of expression (Bedell et al. 1995; Duhl et al. 1994; Duttlinger et al. 1993). To our knowledge, Mvwf1 is the only reported example of a regulatory mutation resulting in a switch in gene expression program from one tissue-specific pattern to another restricted tissue-specific pattern.
Dolichos biflorus (DBA) lectin detects terminal nonreducing GalNAc residues such as those generated by B4galnt2. We previously demonstrated that DBA lectin detects the Mvwf1 switch in the B4galnt2 gene expression program from intestine to vessel (Mohlke et al. 1999). Ponder and Wilkinson (1983) surveyed DBA lectin staining patterns in ten inbred mouse strains, including RIII/Ro, an ancestor of the RIIIS/J inbred mouse strain. They described two distinct staining patterns, the first an intestinal epithelial-specific pattern present in eight of the strains surveyed (including C57BL/6) and the second a vascular endothelial-specific pattern in the inbred strains DDK and RIII/Ro. Ponder et al. (1985) subsequently expanded the DBA lectin survey and detected the RIII/Ro pattern in 3 of 29 strains.
We now report the analysis of sequences surrounding the B4galnt2 gene from mice exhibiting the vascular endothelial-specific or gastrointestinal epithelial-specific B4galnt2 expression programs. Our data suggest that the region responsible for the unique Mvwf1 regulatory mutation likely lies within a 30-kb interval upstream of the B4galnt2 structural gene. We identify a number of unrelated inbred mouse strains, including several wild-derived strains, that carry the same Mvwf1 allele, suggesting that this locus may be under positive selection.
Materials and methods
Animals
C57BL6/J, CASA/RkJ, LEWES/EiJ, PERA/EiJ, PERC/EiJ, RIIIS/J, RF/J, Sf/CamEiJ, SWR/J, and WSB/EiJ males 5-8 weeks old were obtained from The Jackson Laboratory (Bar Harbor, ME). DDK mice were a gift from Dr. C. Sapienza (Temple University). Idaho (Id) outbred wild-derived mice (Miller et al. 2002) were provided by Dr. R. Miller (University of Michigan). All protocols employed were approved by the University Committee on Use and Care of Animals (UCUCA) at the University of Michigan.
Preparation of biological samples
DNA was either obtained directly from The Jackson Laboratory or prepared from frozen tissue as previously described (Nichols et al. 1994). Total RNA was prepared as previously described (Mohlke et al. 1999). Platelet-poor plasma was collected as previously described (Nichols et al. 1994) and stored at -80°C until analysis. Formalin-fixed tissues were paraffin embedded by the Tissue Core of the University of Michigan Comprehensive Cancer Center. Frozen tissues from six wild-derived inbred strains (Supplementary Table 1) were purchased from RIKEN BioResources Center (Japan). Formalin-fixed tissues from the inbred wild-derived mouse strain WSA were provided by Dr. Michael Potter (NIH). Formalin-fixed, paraffin-embedded whole bowels from the inbred mouse strains MA, STS, and GR were provided by Dr. Marco Breuer (The Netherlands Cancer Institute).
RIIIS/J genomic sequencing
Primers were designed to generate overlapping amplicons averaging 800–900 bp in length from the C57BL6/J mouse chromosome 11 build 35, which is publicly available at NCBI, using Primer3 (Rozen and Skaletsky 2000). PCR products were purified as previously described (Mohlke et al. 1999) and sequencing of both strands was performed at the University of Michigan DNA Sequencing Core. An RIIIS/J consensus sequence for each amplicon was assembled to create an RIIIS/J contig using Seqman Lasergene software (DNASTAR, Inc.). Large polymorphisms were confirmed by long-range PCR (Expand Long Template PCR System, Roche) per the manufacturer’s instructions or Southern blot as previously described (Bahou et al. 1988). The RIIIS/J genomic sequence assembly has been deposited at NCBI (GenBank accession number EF372924).
Genotyping
Screening of a panel of DNA from multiple inbred mouse strains (Supplementary Table 1) was performed using RIIIS/J polymorphisms 5 kb and 10 kb upstream of B4galnt2 exon 1 by genomic PCR (primer sets 1 and 2 in Supplementary Table 2) using PfuTurboHotstart (Stratagene) for amplicons smaller than 3 kb or Expand Long Template PCR System (Roche) for amplicons larger than 3 kb per the manufacturer’s instructions.
For a haplotype analysis, genomic sequencing was performed as described above (primer sets 1, 2, and 29–53 in Supplementary Table 2). Unrooted parsimony trees were generated for individual amplicons using PAUP v4.0 beta (bootstrap analysis, heuristic method, n = 1000 replicates) and used to define groups with similar sequences. A parsimony tree, distance tree, and neighbor-joining tree were similarly generated using ordered concatenated sequences with PAUP v4.0 beta.
Analysis for the Mvwf1 tissue-specific switch
DBA lectin staining was performed on formalin-fixed, paraffin-embedded tissues as previously described (Mohlke et al. 1999) using HRP-DBA lectin (H-Y Labs, Inc.). Plasma VWF levels were determined by ELISA in triplicate as previously described (Mohlke et al. 1998). Aliquots of pooled C57BL6/J plasma were used to generate a standard curve. VWF values for at least three individual animals were averaged to ascertain each strain’s VWF level.
Bacterial artificial chromosomes (BACs)
Three overlapping C57BL6/J BACs containing the B4galnt2 gene (Fig. 1A) were obtained. The C57BL/6J BAC clone RP23-271O13 (B6 BAC-1, Fig. 1A) was identified by screening the RPCI-23 library segment 2 filters (Research Genetics) with a probe generated by PCR with primer set 25 (Supplementary Table 2) spanning exons 1 and 2 of the B4galnt2 cDNA and 446 bp of 5′ upstream genomic sequence. The RP24-158F18 and RP24-247D8 BACs (B6 BAC-2 and B6 BAC-3 respectively, Fig. 1A) were identified to contain B4galnt2 by genomic sequence alignments with the C57BL6/J mouse chromosome 11 build 35 (available at http://www.ncbi.nlm.nih.gov/). All three BACs were obtained from the BACPAC Resources Center (Oakland, CA). The integrity of the BAC inserts was confirmed by PCR for the presence of the previously described sequence tagged sites (STSs) (Mohlke et al. 1998) that were predicted to lie within the regions of the BACs and by HindIII, EcoRI, and SpeI restriction digests.
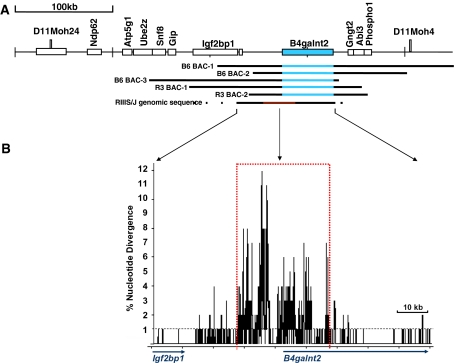
Fig. 1.
Schematic of the Mvwf1 genomic region, BACs, and RIIIS/J sequence. A Schematic of the nonrecombinant interval between D11Moh24 and D11Moh4 (Mohlke et al. 1996); the B4galnt2 gene is shown in blue (distances are referenced to C57BL6/J). Genomic regions spanned by the inserts of the C57BL6/J (B6) and the RIIIS/J (R3) BAC transgenes are shown; blue regions identify the location of the B4galnt2 structural gene within the BAC insert. The homologous genomic regions sequenced in RIIIS/J are indicated, with the region of high sequence divergence in red. B Plot of single nucleotide differences between RIIIS/J and C57BL6/J; the B4galnt2 structural gene is annotated below the x axis. Larger genomic differences were deleted, resulting in an approximately 10-kb smaller interval in this analysis than the C57BL6/J genomic distance. The average sequence difference between inbred strains is 1%, indicated by the dashed black line. An approximately 30-kb region of high sequence divergence (dotted red box) flanks B4galnt2 exon 1
An RIIIS/J BAC library was commissioned from BioS&T (Montreal, Quebec). The library was screened for BAC clones likely to contain the B4galnt2 gene with a probe positioned 2 kb upstream of B4galnt2 (primer set 28 in Supplementary Table 2). Two positive clones, C8 and H1 (R3 BAC-1 and R3 BAC-2 respectively, Fig. 1A), with unique insert sizes were identified using pulsed-field gel electrophoresis. End sequencing and alignment to C57BL6/J mouse chromosome 11 build 35 predicted both BACs to contain the entire B4galnt2 gene (Fig. 1A). The integrity of the BAC inserts was confirmed by HindIII restriction digests, genomic PCR of 16 amplicons spanning the predicted genomic inserts (primer sets 8–23 in Supplementary Table 2), and Southern blots as previously described (Bahou et al. 1988).
Transgenic animals
BACs were purified using the NucleoBond BAC Maxi kit (Clontech). Transgenic mice were generated at the University of Michigan Transgenic Animal Model Core. Purified DNA was microinjected into fertilized eggs obtained from the mouse strains indicated below and pronuclear microinjection was performed as described (Nagy et al. 2007). The B6 BAC-1 was injected into (B4galnt2 knockout × SWR/J)F1 oocytes. The B6 BAC-2 and B6 BAC-3 were injected into SWR/J oocytes. R3 BAC-1 and R3 BAC-2 were injected into (C57BL/6 × SJL)F2 oocytes. Founder animals were backcrossed to a B4galnt2-deficient background using a PCR-based genotyping strategy (all primers are in Supplementary Table 2) to identify the BAC vector (primer set 5), the B4galnt2 null allele (primer set 6), and a 118-bp insertion present 5 kb upstream of B4galnt2 on the SWR/J (Mvwf1) B4galnt2 allele (primer set 7).
Results
RIIIS/J genomic sequence and structure
RIIIS/J genomic sequence was obtained spanning a region homologous to 105 kb in C57BL6/J (annotated in Fig. 1) containing the entire B4galnt2 structural gene, upstream intergenic region, and extending into the upstream gene Igf2bp1. An approximately 30-kb region of greater than 2–3% single nucleotide divergence from C57BL6/J was identified that flanks B4galnt2 exon 1 (indicated in red in Fig. 1). In addition to single nucleotide differences (SNPs), this region also contains size variation in simple and complex tandem repeats as well as larger insertions and deletions. The net effect of these larger genomic changes decreases the size of the RIIIS/J upstream intergenic interval by 10 kb compared to C57BL6/J.
Survey of mouse strains for the Mvwf1 allele and tissue-specific switch
Ponder and Wilkinson (1983) previously described a switch in DBA lectin staining pattern from bowel to vessel similar to Mvwf1 in several inbred mouse strains, including the RIII strain from which RIIIS/J is descended. Thus, Mvwf1 may be present in inbred mouse strains other than RIIIS/J. DNA from 59 inbred strains (Supplementary Table 1) was screened for the Mvwf1 allele using two size polymorphisms 5 and 10 kb upstream of B4galnt2 exon 1, including all available descendants of the strains described by Ponder et al. (1985). Thirteen unrelated (Beck et al. 2000) inbred strains carrying the Mvwf1 allele were identified (Table 1), including five independent wild-derived strains. The presence of the Mvwf1 allele in wild-derived strains suggests that Mvwf1 may also be present in wild mouse populations. Alternatively, accidental genetic contamination, which has been documented to occur between conventional inbred strains (Naggert et al. 1995), could explain distribution of a single rare mutant allele among multiple laboratory strains. To test this hypothesis, DNA from recently established wild-derived outbred mouse colonies maintained by Dr. R. Miller at the University of Michigan Miller et al. 2002) was screened for the Mvwf1 allele. Mvwf1 polymorphisms were found in individuals from two distinct wild-derived M. m. domesticus colonies, implying that the Mvwf1 allele is present in the wild mouse populations from which these mice were derived.
Table 1.
Mouse strain phenotypes
| Strain ID# | Strain name | DBA lectin staining pattern | Plasma VWF + /-1 SD (arbitrary units) | 2.7-kb del (15 kb upstream) | 6.4-kb del (10 kb upstream) | 120-bp ins (5 kb upstream) |
|---|---|---|---|---|---|---|
| 1 | 129x1/SvJ | bowel+, vessel- | nd | nd | wt | wt |
| 2 | A/J | bowel+, vessel- | nd | nd | wt | wt |
| 9 | C57BL/6J | bowel+, vessel- | nd | wt | wt | wt |
| 15 | CASA/RkJa | bowel+, vessel- | 18.87 + /−2.37 | nd | nr | wt |
| 24 | DDK | bowel-, vessel+ | 1.47 + /−0.80 | nd | del | ins |
| 26 | GR | bowel-, vessel+ | nd | nd | nd | ins |
| 28 | Id12bb | bowel+, vessel- | nd | nd | nd | wt |
| 29 | Ih12bb | bowel-, vessel+ | nd | nd | nd | ins |
| 32 | KK/H1J | bowel-, vessel+ | 6.26 + /−0.74 | del | del | ins |
| 33 | LEWES/EiJa | bowel-, vessel+ | 3.65 + /−0.32 | del | del | ins |
| 35 | MA | bowel-, vessel+ | nd | nd | nd | ins |
| 37 | MOLF/EiJa | bowel+, vessel- | 17.81 + /−0.80 | wt | nr | wt |
| 46 | PERA/EiJa | bowel-, vessel+ | 1.70 + /−0.61 | del | del | ins |
| 47 | PERC/EiJa | bowel-, vessel+ | 11.72 + /−1.67 | del | del | ins |
| 51 | RF/J | bowel-, vessel+ | 3.53 + /−1.00 | del | del | ins |
| 52 | RIIIS/J | bowel-, vessel+ | 2.36 + /−0.18 | del | del | ins |
| 53 | SF/CamEiJa | bowel-, vessel+ | 2.48 + /−0.22 | del | del | ins |
| 60 | SWR/J | bowel-, vessel+ | nd | del | del | ins |
| 63 | WSAa | bowel-, vessel+ | nd | del | nd | ins |
| 64 | WSB/EiJa | bowel-, vessel- | 22.35 + /−1.52 | wt | del | ins* |
Data for mouse strains for small-bowel DBA lectin staining pattern, plasma VWF level, and corresponding results of the screen by PCR for RIIIS/J size polymorphisms: wt = wild-type (similar to C57BL6/J); ins* = intermediate PCR size with Mvwf1 SNPs; del = deletion, ins = insertion; nd = not determined; nr = no results
aWild-derived inbred mouse strains
bFrom recently derived outbred M. m. domesticus colonies
To confirm that the presence of the Mvwf1 allele, as detected by the presence of two RIIIS/J polymorphisms described above, corresponds to the Mvwf1 tissue-specific switch, DBA lectin staining was performed on bowels from the 13 strains identified above and plasma VWF levels were determined for the nine available strains maintained at The Jackson Laboratory. DBA lectin detects B4galnt2-specific subterminal GalNAc residues, as demonstrated by the absence of DBA lectin staining in any tissues from B4galnt2 knockout animals. All 13 Mvwf1 strains exhibited a vessel(+), bowel(–) DBA lectin staining pattern identical to RIIIS/J (Table 1). A homozygote for the Mvwf1 allele from Dr. Miller’s wild-derived colony also exhibited the RIIIS/J lectin staining pattern. A survey of plasma VWF antigen levels from nine Mvwf1 strains and three non-Mvwf1 wild-derived strains (Table 1) found that the Mvwf1 strains have low VWF levels relative to the three non-Mvwf1 wild-derived strains, consistent with a decrease in VWF level due to vascular endothelial B4galnt2 expression, as previously demonstrated for RIIIS/J (Mohlke et al. 1996).
Characterization of a highly conserved Mvwf1 haplotype block
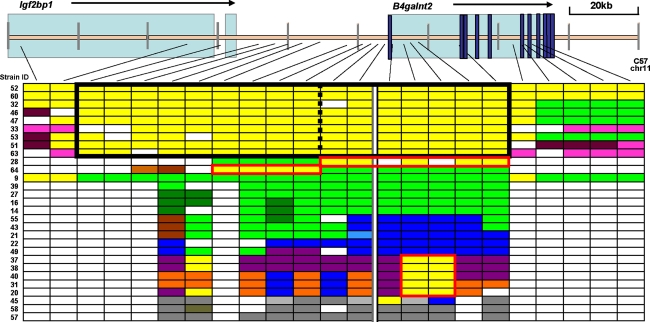
Genomic sequencing from 13 regions with an average amplicon length of approximately 800 bp was performed over a 55-kb genomic region encompassing the approximately 30-kb region of high sequence divergence (see Supplementary Table 3 for a summary of the number of SNPs at each amplicon). Nine Mvwf1 strains and wild-derived inbred strains from the M. musculus clade (Tucker 2007), M. spretus, and M. spicilegus were included in this analysis. All nine of the Mvwf1 strains surveyed were found to share a unique, highly conserved common haplotype block spanning the entire region that was distinct from all other strains tested (represented in Fig. 2; amplicons with Mvwf1 SNPs are yellow; the haplotype block is outlined by the solid black box). Refinement of the haplotype data effort by sampling sequence from ten additional regions (5 upstream and 5 downstream) determined that the shared Mvwf1 haplotype block is approximately 97kb in length and encompasses the entire upstream intergenic region and most of the B4galnt2 structural gene. Seven of 20 non-Mvwf1 mouse strains surveyed were found to share two or more contiguous SNP blocks identical to the Mvwf1 haplotype (highlighted in the red boxes in Fig. 2), suggesting recombined alleles partially derived from the Mvwf1 founder allele. To determine if these shared regions affect B4galnt2 tissue-specific expression, DBA lectin staining was performed on bowel from strains representative of each candidate recombined haplotype pattern (Table 1). MOLF/EiJ (Strain ID #37) and Id12b (Strain ID #28) both exhibit a wild-type gut(+), vessel(−) pattern, suggesting that the genomic region responsible for the Mvwf1 tissue-specific switch in B4galnt2 gene expression program lies upstream of the B4galnt2 structural gene and proximal promoter region (boundary indicated in Fig. 2 by the black dashed line within the Mvwf1 haplotype block). DBA lectin staining was absent in both bowel and vessel in WSB/EiJ (Strain ID #64), suggesting the presence of an independent B4galtnt2 loss-of-function mutation on this allele.
Fig. 2.
Mvwf1 haplotype block. A highly conserved Mvwf1 haplotype block (yellow) is shared by the Mvwf1 inbred strains and distinct from other wild-derived inbred strains representative of the M. m. musculus clade, M. spretus, and M. spicilegus. Strain ID numbers are indicated on the left (see Supplementary Table 1). Colors were assigned to similar sequences based upon PAUP phylogeny trees; missing data are white. The B4galnt2 exon 1 boundary is highlighted by the thick white vertical line. The Mvwf1 haplotype block is outlined in black. Shorter segments in seven additional strains (28, 64, 37, 38, 40, 31, 20) have two or more contiguous amplicons with sequence identity to the Mvwf1 strains (outlined in the red boxes)
BAC transgenes confer tissue-specific B4galnt2 expression
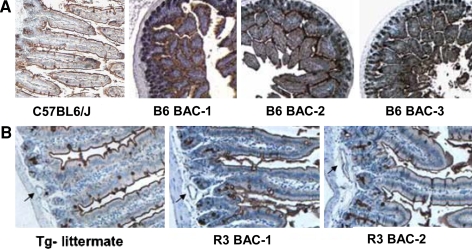
A strain-specific overlapping BAC transgene strategy (Fig. 1A) with inserts derived from either the C57BL6/J (intestine+) mouse strain or the RIIIS/J (vessel+) mouse strain was used to identify a minimal genomic interval that contains intestine- and/or vessel-specific regulatory elements. All three C57BL6/J BAC transgenes restored an intestinal DBA lectin staining pattern in B4galnt2-deficient animals (Fig. 3), suggesting that the critical regulatory elements required for intestinal epithelial-specific expression are contained within the 84-kb overlapping region shared by the three BACs. Both RIIIS/J BAC transgenes restored a vascular expression pattern (Fig. 3), suggesting that the critical regulatory elements necessary for the vascular endothelial-specific expression program are contained within the 108-kb overlapping genomic region shared by these two BACs. Taken together, these data suggest that a master regulatory region may be localized to the genomic interval shared by these five BACs.
Fig. 3.
BAC transgenic bowel DBA lectin staining. DBA lectin staining of small bowel; positive lectin staining is brown. A Lectin-stained bowel from C57BL6/J and F1 transgenic animals generated with each of the C57BL6/J BACs (vessels are positive from the endogenous B4galnt2 gene; F2 transgenic B4galnt2-/- animals exhibit staining in bowel but not vessels, while B4galnt2-/- animals exhibit no staining in any tissue, data not shown). B Lectin-stained bowel from transgenic animals generated with the two RIIIS/J BACs and a transgene negative (Tg-) littermate. The endogenous gene confers intestinal staining in all animals. Arrows indicate serosal blood vessels
Discussion
The inbred mouse strain RIIIS/J exhibits a tissue-specific switch in the expression of β1,4-N-acetylgalactosaminyltransferase, or B4GALNT2, from the intestinal epithelial pattern seen in most mice to a vascular endothelial cell-specific pattern (Mohlke et al. 1999). Examples of regulatory mutations in other genes have been previously reported, which change either temporal or spatial gene expression programs. Although large genomic rearrangements such as those resulting from chromosome translocations can also result in tissue-specific switches in gene expression programs by moving genes from their native genomic context to another differentially regulated locus, previous work (Mohlke et al. 1999) and our current data exclude such a large genomic rearrangement as a mechanism for the Mvwf1 switch. In humans, diseases such as hereditary persistence of fetal hemoglobin (Cunningham and Jane 1996) can result from mutations that alter the timing of gene expression during development, although tissue specificity generally remains unchanged. Spontaneous mutations have been reported in mice that specifically decrease gene expression in the “normal” tissue while upregulating gene expression in other tissues, including examples such as the W-sash allele affecting c-kit (Duttlinger et al. 1993) and the Steel-contrasted allele of the melanocyte growth factor gene (Bedell et al. 1995). However, in these cases the gene expression pattern has been changed from a restricted tissue-specific pattern to a more ubiquitous pattern of gene expression. In contrast, the Mvwf1 regulatory mutation results in a unique switch in gene expression program from one tissue-specific pattern to another entirely distinct tissue-specific expression program.
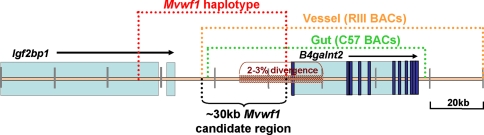
Previous work demonstrated that Mvwf1 results from a cis-acting mutation (Mohlke et al. 1999) genetically localized to a large 264-kb genomic interval surrounding the B4galnt2 gene and containing nine flanking genes (Mohlke et al. 1996). Although the use of alternative tissue-specific promoters has been described as a mechanism for regulating tissue-specific gene expression (Morishita et al. 1995), 5′-RACE analysis appears to exclude this explanation for the Mvwf1 regulatory switch. Taken together, the BAC expression data and multiple-strain analyses reported here (summarized in Fig. 4) refine the candidate interval for the Mvwf1 regulatory mutation from the previous 264-kb genomic segment to a 30-kb intergenic region upstream of the B4galnt2 structural gene. These results position the candidate region for the Mvwf1 regulatory mutation well outside of the B4galnt2 structural gene and proximal promoter region and suggest that the Mvwf1 mutation alters the function of one or more enhancer and/or repressor elements acting over a large genomic distance.
Fig. 4.
Summary of the Mvwf1 candidate region. The genomic regions defined by the BAC transgenic experiments (RIIIS/J BACs in orange, C57BL6/J BACs in green), haplotype block analysis (red), and RIIIS/J genomic region of high sequence divergence (maroon hatch marks) are shown. The deduced 30-kb Mvwf1 candidate interval is indicated in black
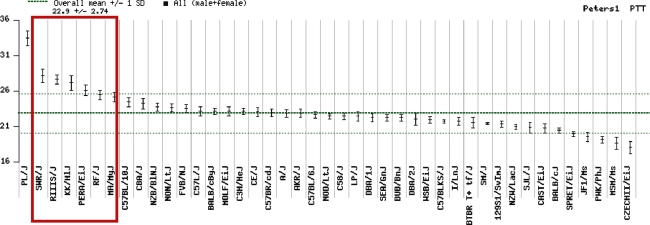
The genomic sequence within this region contains an approximately 30-kb region of strikingly high sequence divergence (>2–3%) in RIIIS/J compared to C57BL6/J that flanks B4galnt2 exon 1. This degree of sequence divergence is 3- to 30-fold greater than the sequence difference expected between modern laboratory strains (Wade et al. 2002). This 30-kb segment lies within a conserved 97-kb haplotype block shared by all 13 inbred mouse strains shown to exhibit the Mvwf1 switch in DBA lectin staining pattern from bowel to vessel first described by Ponder and Wilkinson (1983). Our data identify the cause of the Mvwf1 tissue-specific switch in the B4galnt2 gene expression program (and the resulting low circulating VWF levels) as a single, highly conserved Mvwf1 founder allele that is common among inbred laboratory mouse strains. Six of these Mvwf1 strains were included in a survey of coagulation parameters as part of the Mouse Phenome Database project (Peters and Barker 2006), accounting for six of the seven longest activated partial thromboplastin times (aPTTs) reported in the database (Fig. 5). Thus, Mvwf1 is a common allele among inbred mouse strains and the major cause of a prolonged aPTT in laboratory mice.
Fig. 5.
Mouse Phenome Database: aPTT data. The six Mvwf1 strains in the Mouse Phenome Database (Peters and Barker 2006) are indicated by the red box
The identification of Mvwf1 in multiple conventional inbred strains suggested that this allele may have been present in the Mus musculus clade from which the conventional inbred mouse strains were derived. Indeed, several of the Mvwf1 strains reported here are wild-derived mouse strains that were created and maintained independent from conventional inbred laboratory mouse strains, supporting the persistence of Mvwf1 specifically in the wild M. m. domesticus populations from which these wild-derived strains are descended (LEWES/EiJ was derived from founders trapped in Delaware, PERA/EiJ and PERC/EiJ from Peru, Sf/CamEiJ from California, and WSA from Maryland). The identification of Mvwf1 in two independent and recently derived outbred wild mouse colonies from Idaho suggests that the Mvwf1 allele continues to be maintained in contemporary wild mouse populations.
The existence of such a highly divergent, polymorphic genomic segment within a single species is quite unusual and raises interesting questions about the genetic origin of the Mvwf1 allele. Although there are differences in the sizes of tandem repeats within this region between the Mvwf1 strains, Mvwf1 single nucleotide differences are nearly perfectly conserved. The conservation of single nucleotide differences is also shared by the wild-derived Mvwf1 mouse strains. The existence of this highly conserved region within M. m. domesticus could be the result of selection, of suppression of recombination, or of a founder event. Indeed, the presence of a genomic region of such high nucleotide divergence is suggestive of balancing selection at or near this locus (Charlesworth 2006). Alternatively, this divergent segment could have arisen as the result of an introgression event from a closely related species such as other members of the genus Mus (Lundrigan et al. 2002). There is evidence that fragments of genomic DNA can be exchanged between mammalian species, including M. spretus, which is sympatric with M. m. domesticus (Hardies et al. 2000; Orth et al. 2002). A recently introduced foreign allele would be expected to have had less time to accrue sequence changes and could account for the large size of the Mvwf1 haplotype block. If the Mvwf1 divergent region is the result of an introgression event, then the species from which this allele was originally derived might also exhibit the phenotype conferred by the Mvwf1 regulatory mutation.
Regardless of its origin, the presence of the highly conserved Mvwf1 haplotype block among multiple, independent, wild-derived inbred strains, the diversity in geographic location and temporal spacing of capture of the original wild-derived strain founder mice, and the presence of the Mvwf1 allele in recently derived outbred mouse colonies are all consistent with the maintenance of the Mvwf1 allele as a natural variant in wild M. m. domesticus populations. Taken together, these data suggest the possibility that the Mvwf1 phenotype may confer a fitness advantage, resulting in selective pressure that accounts for the prevalence of the Mvwf1 allele in wild mouse populations. Even if additional evidence for natural selection can be obtained, the underlying mechanism may be difficult to precisely define. A direct effect of reduced plasma VWF (perhaps via decreased thrombotic risk) is one possibility. However, the responsible protein could also be any of the large number of other endothelial protein products (in addition to VWF) that are likely to be altered by Mvwf1. It is also possible that loss of GI epithelial B4galnt2 expression, rather than gain of endothelial expression, confers a fitness advantage in wild mouse populations. If the underlying explanation is indeed reduced plasma VWF, then similar positive selective pressures may have contributed to the high prevalence of von Willebrand disease in multiple mammalian species, including humans.
Electronic supplementary material
Acknowledgments
The authors acknowledge Dr. Wanda Filipiak, Galina Gavrilina, Dr. Thom Saunders, and Dr. Maggie Van Keuren for preparation of transgenic mice at the Transgenic Animal Model Core of the University of Michigan’s Biomedical Research Core Facilities. This work was also supported by the Tissue Core of the University of Michigan Comprehensive Cancer Center. Transgenic Core and Tissue Core support was provided by The University of Michigan Cancer Center (NIH grant number CA46592), The University of Michigan Center for Organogenesis, and The University of Michigan Multipurpose Arthritis Center (NIH grant number AR20557). Many thanks to the following for generously providing mouse tissue samples, DNA, and/or mice of interest: Dr. Thomas Glaser at the University of Michigan (eight of the inbred strains used in the initial screen), Dr. Richard Miller and Jessica Sewald at the University of Michigan (Idaho wild-derived outbred mice), Dr. C. Sapienza at Temple University (DDK), Dr. Michael Potter at the National Institutes of Health (WSA), and Dr. Marco Breuer at the Netherlands Cancer Institute (MA, STS, GR). This work was supported by NIH grant 1 F32 HL78185-01 (J.J.), a National Hemophilia Foundation Judith Graham Pool Fellowship (J.J.), American Heart Association Fellow-to-Faculty Award #0575033N (J.J), National Science Foundation grant DEB 0212667 (P.T.), and NIH grant 4 R37-HL 036963 (D.G.). D.G. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s00335-007-9079-4) contains supplementary material, which is available to authorized users.
Contributor Information
Jill M. Johnsen, Email: jjohnsen@med.umich.edu
David Ginsburg, Email: ginsburg@umich.edu.
References
- Bahou WF, Bowie EJ, Fass DN, Ginsburg D (1988) Molecular genetic analysis of porcine von Willebrand disease: tight linkage to the von Willebrand factor locus. Blood 72:308–313 [PubMed]
- Beck JA, Lloyd S, Hafezparast M, Lennon-Pierce M, Eppig JT et al. (2000) Genealogies of mouse inbred strains. Nat Genet 24:23–25 [DOI] [PubMed]
- Bedell MA, Brannan CI, Evans EP, Copeland NG, Jenkins NA et al. (1995) DNA rearrangements located over 100 kb 5′ of the Steel (Sl)-coding region in Steel-panda and Steel-contrasted mice deregulate Sl expression and cause female sterility by disrupting ovarian follicle development. Genes Dev 9:455–470 [DOI] [PubMed]
- Charlesworth D (2006) Balancing selection and its effects on sequences in nearby genome regions. PLoS Genet 2:e64 [DOI] [PMC free article] [PubMed]
- Crossley M, Ludwig M, Stowell KM, De Vos P, Olek K et al. (1992) Recovery from hemophilia B Leyden: an androgen-responsive element in the factor IX promoter. Science 257:377–379 [DOI] [PubMed]
- Cunningham JM, Jane SM (1996) Hemoglobin switching and fetal hemoglobin reactivation. Semin Hematol 33:9–23 [PubMed]
- Duhl DM, Vrieling H, Miller KA, Wolff GL, Barsh GS (1994) Neomorphic agouti mutations in obese yellow mice. Nat Genet 8:59–65 [DOI] [PubMed]
- Duttlinger R, Manova K, Chu TY, Gyssler C, Zelenetz AD et al. (1993) W-sash affects positive and negative elements controlling c-kit expression: ectopic c-kit expression at sites of kit-ligand expression affects melanogenesis. Development 118:705–717 [DOI] [PubMed]
- Haltiwanger RS, Lowe JB (2004) Role of glycosylation in development. Annu Rev Biochem 73:491–537 [DOI] [PubMed]
- Hardies SC, Wang L, Zhou L, Zhao Y, Casavant NC et al. (2000) LINE-1 (L1) lineages in the mouse. Mol Biol Evol 17:616–628 [DOI] [PubMed]
- Lowe JB, Marth JD (2003) A genetic approach to Mammalian glycan function. Annu Rev Biochem 72:643–691 [DOI] [PubMed]
- Lundrigan BL, Jansa SA, Tucker PK (2002) Phylogenetic relationships in the genus mus, based on paternally, maternally, and biparentally inherited characters. Syst Biol 51:410–431 [DOI] [PubMed]
- Marquardt T, Denecke J (2003) Congenital disorders of glycosylation: review of their molecular bases, clinical presentations and specific therapies. Eur J Pediatr 162:359–379 [DOI] [PubMed]
- Miller RA, Harper JM, Dysko RC, Durkee SJ, Austad SN (2002) Longer life spans and delayed maturation in wild-derived mice. Exp Biol Med (Maywood) 227:500–508 [DOI] [PubMed]
- Mohlke KL, Nichols WC, Westrick RJ, Novak EK, Cooney KA et al. (1996) A novel modifier gene for plasma von Willebrand factor level maps to distal mouse chromosome 11. Proc Natl Acad Sci USA 93:15352–15357 [DOI] [PMC free article] [PubMed]
- Mohlke KL, Purkayastha AA, Westrick RJ, Ginsburg D (1998) Comparative mapping of distal murine chromosome 11 and human 17q21.3 in a region containing a modifying locus for murine plasma von Willebrand factor level. Genomics 54:19–30 [DOI] [PubMed]
- Mohlke KL, Purkayastha AA, Westrick RJ, Smith PL, Petryniak B et al. (1999) Mvwf, a dominant modifier of murine von Willebrand factor, results from altered lineage-specific expression of a glycosyltransferase. Cell 96:111–120 [DOI] [PubMed]
- Montiel MD, Krzewinski-Recchi MA, Delannoy P, Harduin-Lepers A (2003) Molecular cloning, gene organization and expression of the human UDP-GalNAc:Neu5Acalpha2–3Galbeta-R beta1,4-N-acetylgalactosaminyltransferase responsible for the biosynthesis of the blood group Sda/Cad antigen: evidence for an unusual extended cytoplasmic domain. Biochem J 373:369–379 [DOI] [PMC free article] [PubMed]
- Morishita K, Johnson DE, Williams LT (1995) A novel promoter for vascular endothelial growth factor receptor (flt-1) that confers endothelial-specific gene expression. J Biol Chem 270:27948–27953 [DOI] [PubMed]
- Naggert JK, Mu JL, Frankel W, Bailey DW, Paigen B (1995) Genomic analysis of the C57BL/Ks mouse strain. Mamm Genome 6:131–133 [DOI] [PubMed]
- Nagy A, Gerstenstein M, Vintersten K, Behringer R (2007) Manipulating the Mouse Embryo: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press)
- Nichols WC, Cooney KA, Mohlke KL, Ballew JD, Yang A et al. (1994) von Willebrand disease in the RIIIS/J mouse is caused by a defect outside of the von Willebrand factor gene. Blood 83:3225–3231 [PubMed]
- Orth A, Belkhir K, Britton-Davidian J, Boursot P, Benazzou T et al. (2002) [Natural hybridization between 2 sympatric species of mice, Mus musculus domesticus L. and Mus spretus Lataste]. C R Biol 325:89–97 [DOI] [PubMed]
- Peters LL, Barker JE (2006) MPD:6225, 6226, 6227. Mouse Phenome Database website, The Jackson Laboratory, Bar Harbor, ME, USA. Available at http://www.jax.org/phenome [accessed 30 May 2006]
- Ponder BA, Wilkinson MM (1983) Organ-related differences in binding of dolichos-biflorus agglutinin to vascular endothelium. Dev Biol 96:535–541 [DOI] [PubMed]
- Ponder BA, Festing MF, Wilkinson MM (1985) An allelic difference determines reciprocal patterns of expression of binding-sites for dolichos-biflorus lectin in inbred strains of mice. J Embryol Exp Morphol 87:229–239 [PubMed]
- Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds), Bioinformatics Methods and Protocols: Methods in Molecular Biology. (Totowa, NJ: Humana Press), pp. 365–386 [DOI] [PubMed]
- Tucker PK (2007) Systematics of the genus Mus. In: Fox J, Barthold S, Davisson MT, Newcomer C, Quimby F et al. (eds), Mouse in Biomedical Research, 2nd ed. (Boston: Elsevier Press), pp 13–23
- Wade CM, Kulbokas EJ III, Kirby AW, Zody MC, Mullikin JC et al. (2002) The mosaic structure of variation in the laboratory mouse genome. Nature 420:574–578 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.