Abstract
Nonsyndromic mental retardation is one of the most important unresolved problems in genetic health care. Autosomal forms are far more common than X-linked forms, but, in contrast to the latter, they are still largely unexplored. Here, we report a complex mutation in the ionotropic glutamate receptor 6 gene (GRIK2, also called “GLUR6”) that cosegregates with moderate-to-severe nonsyndromic autosomal recessive mental retardation in a large, consanguineous Iranian family. The predicted gene product lacks the first ligand-binding domain, the adjacent transmembrane domain, and the putative pore loop, suggesting a complete loss of function of the GLUK6 protein, which is supported by electrophysiological data. This finding provides the first proof that GLUK6 is indispensable for higher brain functions in humans, and future studies of this and other ionotropic kainate receptors will shed more light on the pathophysiology of mental retardation.
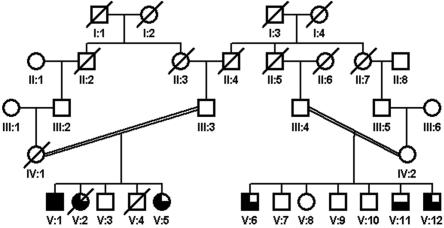
To date, only three genes have been found to be directly linked to nonsyndromic autosomal recessive mental retardation (NS-ARMR): PRSS12 (also called “neurotrypsin” [MIM 606709]), which encodes a trypsin-like serine protease1; CRBN (also called “cereblon” [MIM 609262]),2 which encodes an ATP-dependent Lon protease; and CC2D1A (MIM 610055), which encodes a putative signal transducer that participates in the positive regulation of the I-κB kinase/NF-κB cascade.3 We recently reported eight genomic loci for NS-ARMR (MRT4–MRT11)4 as the result of linkage studies of 78 consanguineous Iranian families. Here, we describe the first of the underlying gene defects, resolving the MRT6 locus [MIM 611092], a 9.98-Mb interval on chromosome 6q16.1-q21 that we found in a family with moderate-to-severe NS-ARMR. The degree of mental retardation (MR) in the affected family members ranged from mild to severe (fig. 1). Patient V:1 had an intelligence quotient (IQ) of 30 at age 50 years. His youngest sister (V:5) had mild MR. Patients V:6 and V:12 also had mild MR, and patient V:11 had an IQ of 55 (moderate MR). The patients did not have neurological problems, congenital malformations, or facial dysmorphisms. Body height, weight, and head circumference were normal in all patients. For patient V:12, we performed a magnetic resonance imaging scan, which revealed no morphological abnormalities. Sample collection and clinical evaluation procedures were performed with the informed consent of the parents and have been described elsewhere.4
Figure 1. .
Family pedigree. Blackened symbols represent patients with severe MR, three-quarters-blackened symbols represent patients with moderate MR, and half-blackened symbols represent patients with mild MR.
The MRT6 locus contains 25 annotated genes; 8 of these (GRIK2 [MIM 138244], CCNC, COQ3, MCHR2, GPR63, KLHL32, POU3F2, and SIM1) were considered to be plausible candidate genes for MR and were selected for mutation screening. We designed PCR primers for the amplification of exonic sequences and splice-site regions, using the Primer3 software.5 PCR amplicons (primer sequences and reaction conditions are available on request) were analyzed by agarose-gel electrophoresis and direct sequencing. In DNA from patients, only a single nonpolymorphic sequence change was detected, apparently a deletion removing exons 7 and 8 of the GRIK2 gene (see fig. 2a for physical coordinates and exon-intron structure), as suggested by our failure to amplify these exons by PCR. GRIK2 encodes GLUK6, a subunit of kainate receptors (KARs) that is highly expressed in the brain. Together with the intensively studied alpha-amino-3-hydroxy-5-methyl-4-isoxazole proprionic acid (AMPA) and N-methyl-d-aspartic acid (NMDA) receptors, KARs belong to the ionotropic receptors that are the targets of glutamate released in excitatory synapses.6,7 Mutation or loss of NMDA and/or AMPA receptors is known to result in severe derangement of neuronal function. Still, extensive analyses of their structure, biophysical properties, and distribution8–10 have so far failed to define a role for ionotropic KARs in higher brain functions. GRIK2 has been associated with autism,11 but this finding could not be substantiated by a recent genomewide linkage study,12 and behavioral studies of Grik2-knockout mice did not yield conclusive results.10 Thus, the involvement of this receptor in cognition has so far remained controversial.
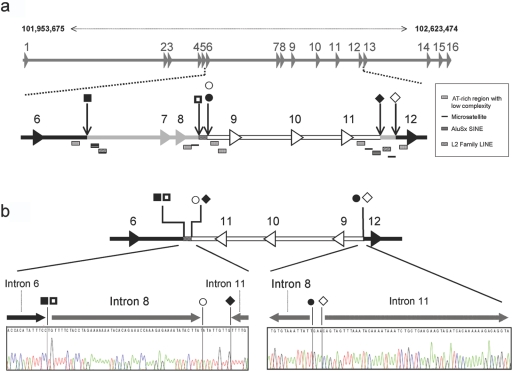
Figure 2. .
Genomic mutation in GRIK2. a, Schematic representation of the GRIK2 wild-type allele. Physical coordinates (top) are according to NCBI build 36.1. Exons are depicted by numbered arrowheads. Black shading indicates unaffected sequence, gray shading indicates deleted regions, and the inversion is white outlined in black. A checkered pattern marks the uninverted sequence fragment 5′ of the deletion of exons 7 and 8. Squares, circles, and diamonds mark the positions of the deletion and inversion borders. Rectangular symbols below the schematic of GRIK2 mark the locations of regions of low sequence complexity or repetitive elements. b, Schematic representation of the GRIK2 mutant allele and the sequences of the 5′ and 3′ genomic junction fragments. LINE = long interspersed nuclear element; SINE = short interspersed nuclear element.
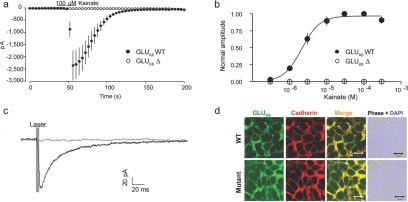
Loss of exons 7 and 8 in GRIK2, as observed in our patients with MR, results in an in-frame deletion of 84 aa between amino acids 317 and 402, close to the first ligand-binding domain (S1) in the extracellular N-terminal region of GLUK6. To study whether this defect was sufficient to impair GLUK6 protein function, we cloned wild-type GLUK6 from human fetal brain RNA (BD Biosciences Clontech), using the pENTR Directional Topo Cloning kit (Invitrogen) in accordance with the manufacturer's instructions. We employed the resulting plasmid for subcloning GRIK2 wild-type cDNA into modified (green fluorescent protein [GFP] replaced with c-Myc tag) pcDNA-DEST47 Gateway vectors (Invitrogen). We then engineered a GLUK6Δ construct lacking exons 7 and 8 by site-directed mutagenesis with the QuikChange Site-Directed Mutagenesis Kit (Stratagene). For a cellular model system, we chose HEK293 cells, because they do not show endogenous GRIK2 expression, and cotransfected them with either plasmid in combination with a green fluorescent transfection marker (1.8 μg cDNA encoding either wild-type GRIK2 or GRIK2Δ plus 0.2 μg pEGFP-N1 vector [BD Biosciences Clontech]). The transfection reagent we used was DreamFect (OZ Biosciences) or Fugene reagent (Roche). For whole-cell patch-clamp recordings, cells were detached with 1 mM EDTA in cold PBS, 24 or 48 h after transfection, were transferred to poly-d-lysin–coated coverslips, and were allowed to settle for at least 4 h. We performed the recordings by using an intracellular solution containing 130 mM CsCl, 9.4 mM NaCl, 1 mM MgCl2, 0.2 mM guanosine triphosphate, 10 mM ethylene glycol tetraacetic acid, and 10 mM HEPES (pH adjusted to 7.3 with CsOH). The cells were superfused with HEPES-buffered solution (140 mM NaCl, 2.4 mM KCl, 10 mM glucose, 10 mM HEPES, 2.5 mM CaCl2, and 1.3 mM MgCl2 [pH adjusted to 7.3 with NaOH]). We treated the cells for 5 min with 200 μg/ml concanavalin A in glucose-free extracellular solution immediately before the recordings, to prevent lectin aggregation. Cells were voltage clamped at −60 mV, by use of an Axoclamp 700A. Recordings were digitized with 5 kHz and were analyzed online by use of IGOR Pro (Wavemetrics). All recordings were performed at room temperature. Our results show that GLUK6Δ completely lost the ability to conduct ions when activated by kainate (fig. 3a) or glutamate (data not shown), independent of agonist dosage (fig. 3b). To exclude the possibility of altered receptor desensitization, we made use of a fast application system13 for performing whole-cell patch-clamp experiments without concanavalin A. For this flash photolysis of glutamate, 40 μM 4-methoxy-7-nitroindolinyl–caged glutamate (Tocris) was perfused continuously on the cells in a volume of 10 ml. A 355-nm UV laser beam (DPSL-355-1W laser [Rapp Optics]) was coupled via a 200-μm quartz fiber optic into a spot illumination adaptor (Rapp Optics), mounted in the epifluorescence path of an Olympus BX51Wl microscope (equipped with a laser beam splitter and a 420-nm-long pass emitter [both from AHF Analysetechnik]) and was focused with a 60× water objective (numerical aperture 0.9 [Olympus]) onto a spot ∼40 μm in diameter. The holding current was constant during washing of the caged glutamate. Again, we observed a complete loss of function of GLUK6Δ compared with the wild-type GLUK6 channels (fig. 3c). We then asked whether this might be caused by the absence of the receptor in the plasma membrane caused by a defect in receptor trafficking. Therefore, we performed GLUK6 staining experiments in transfected HEK293 cells, fixed 48 h after transfection with paraformaldehyde (4%, for 10 min at room temperature) and permeabilized with Triton X-100 in PBS (0.1%, for 5 min at room temperature). The cells were subsequently blocked with normal goat serum (10%, for 1 h at room temperature). For detection of the membrane protein cadherin and GLUK6, we used the antibodies panCadherin (ab6528 [Abcam]) (1:2,000, for 1 h at room temperature) and Anti-GLUR6/7 (05–921 [Upstate Biotechnology]) (1:2,000, for 1 h at room temperature), respectively. They were labeled by the secondary antibodies Alexa Fluor 488 chicken anti-mouse (Invitrogen) (1:1,000, for 1 h at room temperature) and Alexa Fluor 488 chicken anti-rabbit (Invitrogen) (1:1,000, for 1 h at room temperature). Confocal analysis of GLUK6 stainings on a laser-scanning microscope (Zeiss Axiovert 100 M) with a 63× objective, however, clearly showed colocalization of transfected GLUK6Δ with the endogenous membrane protein cadherin in HEK293 cells (fig. 3d). Biotinylation experiments confirmed that these cells had considerable amounts of GLUK6 protein in the plasma membrane (fig. 4).
Figure 3. .
Loss of function of GLUK6Δ. a, Mean currents evoked by 100 μM kainate (for GLUK6 wild type [WT], n=8; for GLUK6Δ, n=9). b, Dose-response curve for kainate, normalized to response in 100 μM kainate. Error bars represent SEM (for GLUK6WT, n=6–8 per reading point; for GLUK6Δ, n=3–9 per reading point). c, Cells expressing GLUK6WT show photolytically evoked glutamate responses without prior incubation in concanavalin A (black line), whereas GLUK6Δ-expressing cells show no currents after fast glutamate application (gray line). Each trace represents the mean of five successive sweeps. d, Immunocytochemistry showing colocalization of both GLUK6 WT and GLUK6Δ with the endogenous plasma membrane protein cadherin. DAPI = 4′,6-diamidino-2-phenylindole.
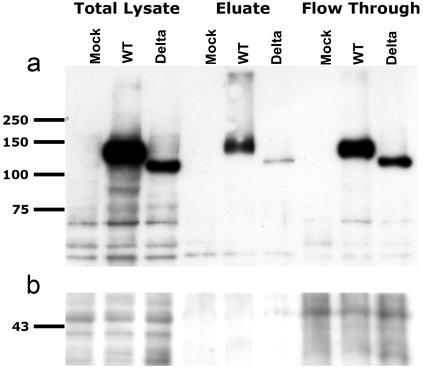
Figure 4. .
GLUK6Δ present on the plasma membrane of transfected HEK293 cells. a, Membrane proteins biotinylated on HEK293 cells overexpressing GLUK6 wild type (WT) or GLUK6Δ (Delta) at 48 h after transfection (Pinpoint [Pierce]). Nontransfected HEK293 cells served as a control (Mock). After cell lysis (Total Lysate samples), biotinylated membrane proteins were bound to a NeutrAvidin affinity column. Nonbiotinylated protein was separated, because it does not bind to the NeutrAvidin affinity column (Flow Through samples). Membrane proteins were eluted with sample buffer (Eluate samples). All fractions were separated on a 10% SDS gel, were blotted, and were probed for GLUK6 (with 05-921 [Upstate Biotechnology] at 1:800). GLUK6 WT and GLUK6Δ are present in the membrane protein fraction (Eluate samples). b, Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) loading control (ab9482 [Abcam]).
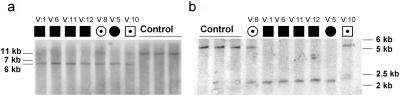
In parallel, we performed PCR experiments with various primer combinations to define the borders of the deletion but failed to amplify a junction fragment. We therefore investigated the 5′ region of the putative deletion in all available family members by Southern-blot analysis. Following standard procedures, we used 10 μg genomic DNA for restriction with BglII and a 32P-labeled probe, directed against a 802-bp sequence upstream of the mutation region (forward primer: 5′-gccattgtgtaagatcatagtgctcagac-3′; reverse primer: 5′-gtatattagaaccactgggaagg-3′). The expected fragment size, corresponding to a GRIK2 allele lacking exons 7 and 8, was 1.5 kb, yet we identified a 6.5-kb band in carriers of the mutant allele (fig. 5a), which pointed to the presence of a more complex genomic rearrangement. We then performed inverse PCR14 on 1 μg of this fragment, isolated with the Qiaquick gel-extraction kit (Qiagen). The fragment was circularized by self-ligation in a 10-μl reaction volume by use of 1.5 U T4 DNA ligase and the reaction buffer provided by the manufacturer. After 16 h of incubation at 12°C, the enzyme was inactivated by increasing the reaction temperature to 65°C for 10 min.
Figure 5. .
Southern blots for patients and heterozygous carriers of the mutation, characterizing the 5′ border (a) (by use of BglII) and the 3′ border (b) (by use of EcoRV) of the mutation.
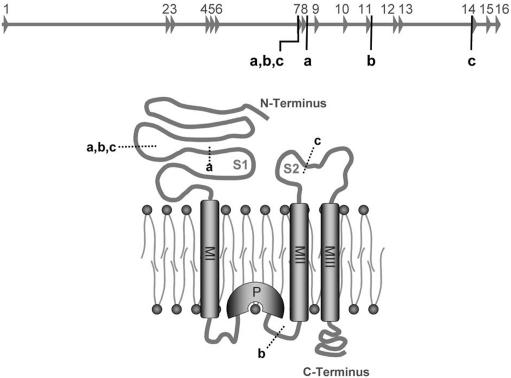
For PCR amplification, we used reverse-oriented primers (forward primer: 5′-catgctcctagaggtgtgcaaatacc-3′; reverse primer: 5′-ctacaatggcaagcgaagtg-3′) located 5′ of the mutation in a region that could be amplified from patient DNA. Cloning and sequencing of the amplicon confirmed the deletion of exons 7 and 8 by uncovering a sequence junction between the remnants of introns 6 and 8. This junction fragment was joined to a sequence fragment belonging to intron 11 in reverse orientation. Through further PCR and sequencing analyses and Southern-blot experiments (fig. 5b), with use of EcoRV for DNA digestion and a 930-bp upstream probe (forward primer: 5′-tggaaattcatagtaaaggtatgtgg-3′; reverse primer: 5′-ctgagtagaaagagagagaaacaatg-3′), we were able to elucidate the full extent of the observed mutation. This is illustrated in figure 2, which shows that, in addition to the ∼120-kb deletion (spanning exons 7 and 8), the mutation comprises an inversion of ∼80 kb, including exons 9, 10, and 11 in combination with a deletion of ∼20 kb of intron 11. At the protein level, this rearrangement can be expected to result in the loss not only of the first ligand-binding domain but also of the adjacent transmembrane domain and the putative pore loop of GLUK6 (fig. 6).
Figure 6. .
Impact of GRIK2 mutation on GLUK6 protein structure. Shown is a schematic representation of the GRIK2 wild-type allele and a cartoon of a GLUK6 monomer. MI–MIII = transmembrane domains; P = pore loop. Letters indicate the start and end of different alterations in the genomic sequence and their corresponding impact on the resulting protein. a = deletion of exons 7 and 8; b = complete genomic rearrangement, as observed in patients with MR; c = region lacking in the putative transcript with a junction between exons 6 and 14.
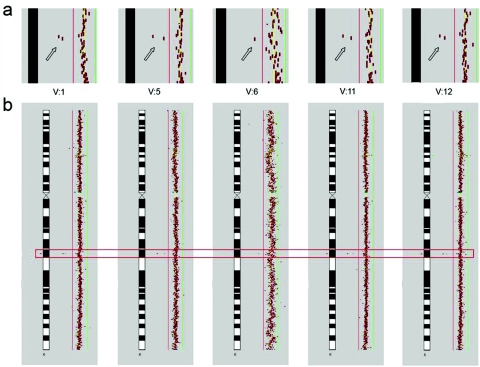
Cosegregation of the mutation with MR in the family was shown by array-based comparative genomic hybridization (array CGH) and Southern-blot experiments (figs. 5 and 7). We also performed Southern blots on a panel of 172 individuals with university education. Neither these nor any of 390 individuals from a random population sample (healthy blood donors) screened by PCR carried the mutation.
Figure 7. .
Array CGH results for patients with MR. Submegabase-resolution array CGH was performed as described elsewhere.15 After normalization by subgrid LOWESS, Cy3:Cy5 intensity ratios of each clone were plotted in a size-dependent manner along the chromosome ideograms, by use of CGHPRO.16 Red and green bars indicate the log2 ratio thresholds of −0.3 (loss) and 0.3 (gain), respectively. A homozygous deletion of 6q16.3 was the only aberration that cosegregated with the disease; it was present in all affected probands and was absent in all unaffected probands tested. Array CGH data shown here have been deposited in NCBI Gene Expression Omnibus (GEO)17 and are accessible through GEO series accession number GSE 7886. a, Close-up of findings for patient 6q16.3, demonstrating a deletion of two BAC clones (RP11-226C08 and RP11-543M18 [arrows]) encompassing GRIK2. (Because of lower hybridization quality for V:6, only one BAC represents the DNA copy-number loss.) b, Overall view of chromosome 6 in the corresponding patients. A red rectangle highlights the region depicted in panel a.
To check for GRIK2 expression, we extracted total RNA from Epstein-Barr virus–transformed lymphoblastoid cell lines of all available patients and healthy siblings by use of the TRIzol reagent (Invitrogen). We isolated mRNA (Dynabeads Oligo (dT)25 [Dynal Biotech]) and generated cDNA by using the SuperScript III Reverse Transcriptase (Invitrogen) together with random hexamers. This cDNA was used to perform PCRs, by use of different combinations of primers from exon 6 with primers from exons 12, 13, and 14. With a combination of primers from exon 6 (5′-gaaaagtggtcgatggaacg-3′) and exon 14 (5′-ggcccacattttgtcatacg-3′), we obtained an aberrant fragment in the cDNA from homozygous and heterozygous carriers of the mutation (data not shown). Compared with the transcript expected from the deletion-inversion, this corresponds to a transcript that translates into a GLUK6 protein, which lacks an additional transmembrane domain (fig. 6). In view of our finding that the deletion of exons 7 and 8 is already sufficient to cause a complete loss of GLUK6 function, we conclude that the patients do not express any GRIK2 gene product with the capacity to form functional ion channels.
GLUK6-containing receptors are present at the presynaptic level,18,19 as well as at the postsynaptic level,20 and the primary involvement of GLUK6 in excitatory synaptic activity suggests that defective GLUK6 might cause cognitive impairment through alterations in local brain circuitry. An attractive hypothesis, in this respect, is that KARs might be involved in the maturation of microcircuits and network formation in brain areas that are important for higher brain functions, such as the hippocampus. In fact, it has been shown that, in mice, the expression level of KAR-subunit mRNAs in the hippocampus is very high during early stages of development, whereas adult animals show lower levels.21,22 Furthermore, it has been found that, in the rodent neonatal hippocampus, KARs are activated by ambient glutamate and thereby regulate spontaneous network activity and functional maturation of hippocampal synapses.23,24
At the cellular level, it is conceivable that defective GLUK6 could also have structural effects. Indeed, GLUK6 has been found to be involved in profilin II–mediated interactions with actin,25,26 through which it might contribute to the stabilization of dendritic spine morphology27 and normal neuritogenesis.28 Further studies of this aspect of GLUK6 function in model organisms will be particularly interesting, because abnormal morphogenesis of dendritic spines is observed in a variety of MR disorders, such as fragile X syndrome (MIM 300624) (for review, see, e.g., the work of Grossmann et al.29). A third avenue to be explored might be the investigation of a putative impact of GLUK6 deficiency on synaptic plasticity. In this context, it is noteworthy that GLUK6 also seems to interact with synapse-associated protein 102,30 for which mutations in the coding gene (DLG3) were found in families with severe X-linked MR.31
By identifying GRIK2 as a likely NS-ARMR gene, we provide strong evidence that functional integrity of this ionotropic glutamate receptor might be a sine qua non for normal human brain function. Given the unclear phenotype of Grik2-deficient mice, this study also indicates that genotype-phenotype studies in man are indispensable for identifying the genes that play a role in complex behavioral traits. Finally, we expect that our results will stimulate research into the role of other KARs in cognition and behavior. GRIK2 is the first gene for NS-ARMR with a known function in the CNS. Its identification is part of an ongoing systematic effort to unravel the molecular causes of recessive MR, which will shed more light on the function of the human brain in health and disease.
Acknowledgments
We express our gratitude to the patients and their families for their cooperation. We thank B. Lipkowitz, M. Schlicht, S. Banihashemi, R. Vazifehmand, M. Kasiri, S. Arjangi, S. Walden, and A. Schönherr for technical assistance. Our work is supported by the Max Planck Innovation Funds, the Deutsche Forschungsgemeinschaft (DFG) Sonderforschungsbereich 577 (to H.H.R.), the Iranian Molecular Medicine Network (to H.N.), other grants from the DFG (Emmy Noether Programm grants SCHM1383/4-1, SFB618, and SFB665), and grants from the German Federal Ministry of Education and Research (Bernstein Center for Computational Neuroscience Berlin) (to D.S.).
Web Resource
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for neurotrypsin, cereblon, CC2D1A, MRT6, GRIK2, and fragile X syndrome)
References
- 1.Molinari F, Rio M, Meskenaite V, Encha-Razavi F, Auge J, Bacq D, Briault S, Vekemans M, Munnich A, Attié-Bitach T, et al (2002) Truncating neurotrypsin mutation in autosomal recessive nonsyndromic mental retardation. Science 298:1779–1781 10.1126/science.1076521 [DOI] [PubMed] [Google Scholar]
- 2.Higgins JJ, Pucilowska J, Lombardi RQ, Rooney JP (2004) A mutation in a novel ATP-dependent Lon protease gene in a kindred with mild mental retardation. Neurology 63:1927–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basel-Vanagaite L, Attia R, Yahav M, Ferland RJ, Anteki L, Walsh CA, Olender T, Straussberg R, Magal N, Taub E, et al (2006) The CC2D1A, a member of a new gene family with C2 domains, is involved in autosomal recessive non-syndromic mental retardation. J Med Genet 43:203–210 10.1136/jmg.2005.035709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Najmabadi H, Motazacker MM, Garshasbi M, Kahrizi K, Tzschach A, Chen W, Behjati F, Hadavi V, Nieh SE, Abedini SS, et al (2006) Homozygosity mapping in consanguineous families reveals extreme heterogeneity of non-syndromic autosomal recessive mental retardation and identifies 8 novel gene loci. Hum Genet 121:43–48 10.1007/s00439-006-0292-0 [DOI] [PubMed] [Google Scholar]
- 5.Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386 [DOI] [PubMed] [Google Scholar]
- 6.Hollmann M, Heinemann S (1994) Cloned glutamate receptors. Annu Rev Neurosci 17:31–108 10.1146/annurev.ne.17.030194.000335 [DOI] [PubMed] [Google Scholar]
- 7.Dingledine R (1991) New wave of non-NMDA excitatory amino acid receptors. Trends Pharmacol Sci 12:360–362 10.1016/0165-6147(91)90602-O [DOI] [PubMed] [Google Scholar]
- 8.Wisden W, Seeburg PH (1993) Mammalian ionotropic glutamate receptors. Curr Opin Neurobiol 3:291–298 10.1016/0959-4388(93)90120-N [DOI] [PubMed] [Google Scholar]
- 9.Lerma J (2003) Roles and rules of kainate receptors in synaptic transmission. Nat Rev Neurosci 4:481–495 10.1038/nrn1118 [DOI] [PubMed] [Google Scholar]
- 10.Mulle C, Sailer A, Perez-Otano I, Dickinson-Anson H, Castillo PE, Bureau I, Maron C, Gage FH, Mann JR, Bettler B, et al (1998) Altered synaptic physiology and reduced susceptibility to kainate-induced seizures in GluR6-deficient mice. Nature 392:601–605 10.1038/33408 [DOI] [PubMed] [Google Scholar]
- 11.Jamain S, Betancur C, Quach H, Philippe A, Fellous M, Giros B, Gillberg C, Leboyer M, Bourgeron T (2002) Linkage and association of the glutamate receptor 6 gene with autism. Mol Psychiatry 7:302–310 10.1038/sj.mp.4000979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, et al (2007) Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet 39:319–328 10.1038/ng1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adesnik H, Nicoll RA, England PM (2005) Photoinactivation of native AMPA receptors reveals their real-time trafficking. Neuron 48:977–985 10.1016/j.neuron.2005.11.030 [DOI] [PubMed] [Google Scholar]
- 14.Ochman H, Gerber AS, Hartl DL (1988) Genetic applications of an inverse polymerase chain reaction. Genetics 120:621–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erdogan F, Chen W, Kirchhoff M, Kalscheuer VM, Hultschig C, Muller I, Schulz R, Menzel C, Bryndorf T, Ropers HH, et al (2006) Impact of low copy repeats on the generation of balanced and unbalanced chromosomal aberrations in mental retardation. Cytogenet Genome Res 115:247–253 10.1159/000095921 [DOI] [PubMed] [Google Scholar]
- 16.Chen W, Erdogan F, Ropers HH, Lenzner S, Ullmann R (2005) CGHPRO—a comprehensive data analysis tool for array CGH. BMC Bioinformatics 6:85 10.1186/1471-2105-6-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett T, Edgar R (2006) Gene Expression Omnibus: microarray data storage, submission, retrieval, and analysis. Methods Enzymol 411:352–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Contractor A, Swanson GT, Sailer A, O’Gorman S, Heinemann SF (2000) Identification of the kainate receptor subunits underlying modulation of excitatory synaptic transmission in the CA3 region of the hippocampus. J Neurosci 20:8269–8278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mulle C, Sailer A, Swanson GT, Brana C, O’Gorman S, Bettler B, Heinemann SF (2000) Subunit composition of kainate receptors in hippocampal interneurons. Neuron 28:475–484 10.1016/S0896-6273(00)00126-4 [DOI] [PubMed] [Google Scholar]
- 20.Bureau I, Dieudonne S, Coussen F, Mulle C (2000) Kainate receptor-mediated synaptic currents in cerebellar Golgi cells are not shaped by diffusion of glutamate. Proc Natl Acad Sci USA 97:6838–6843 10.1073/pnas.97.12.6838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bahn S, Volk B, Wisden W (1994) Kainate receptor gene expression in the developing rat brain. J Neurosci 14:5525–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ritter LM, Vazquez DM, Meador-Woodruff JH (2002) Ontogeny of ionotropic glutamate receptor subunit expression in the rat hippocampus. Brain Res Dev Brain Res 139:227–236 10.1016/S0165-3806(02)00572-2 [DOI] [PubMed] [Google Scholar]
- 23.Lauri SE, Segerstrale M, Vesikansa A, Maingret F, Mulle C, Collingridge GL, Isaac JT, Taira T (2005) Endogenous activation of kainate receptors regulates glutamate release and network activity in the developing hippocampus. J Neurosci 25:4473–4484 10.1523/JNEUROSCI.4050-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lauri SE, Vesikansa A, Segerstrale M, Collingridge GL, Isaac JT, Taira T (2006) Functional maturation of CA1 synapses involves activity-dependent loss of tonic kainate receptor-mediated inhibition of glutamate release. Neuron 50:415–429 10.1016/j.neuron.2006.03.020 [DOI] [PubMed] [Google Scholar]
- 25.Coussen F, Perrais D, Jaskolski F, Sachidhanandam S, Normand E, Bockaert J, Marin P, Mulle C (2005) Co-assembly of two GluR6 kainate receptor splice variants within a functional protein complex. Neuron 47:555–566 10.1016/j.neuron.2005.06.033 [DOI] [PubMed] [Google Scholar]
- 26.Wechsler A, Teichberg VI (1998) Brain spectrin binding to the NMDA receptor is regulated by phosphorylation, calcium and calmodulin. EMBO J 17:3931–3939 10.1093/emboj/17.14.3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ackermann M, Matus A (2003) Activity-induced targeting of profilin and stabilization of dendritic spine morphology. Nat Neurosci 6:1194–1200 10.1038/nn1135 [DOI] [PubMed] [Google Scholar]
- 28.Da Silva JS, Medina M, Zuliani C, Di NA, Witke W, Dotti CG (2003) RhoA/ROCK regulation of neuritogenesis via profilin IIa-mediated control of actin stability. J Cell Biol 162:1267–1279 10.1083/jcb.200304021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grossman AW, Aldridge GM, Weiler IJ, Greenough WT (2006) Local protein synthesis and spine morphogenesis: fragile X syndrome and beyond. J Neurosci 26:7151–7155 10.1523/JNEUROSCI.1790-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia EP, Mehta S, Blair LA, Wells DG, Shang J, Fukushima T, Fallon JR, Garner CC, Marshall J (1998) SAP90 binds and clusters kainate receptors causing incomplete desensitization. Neuron 21:727–739 10.1016/S0896-6273(00)80590-5 [DOI] [PubMed] [Google Scholar]
- 31.Tarpey P, Parnau J, Blow M, Woffendin H, Bignell G, Cox C, Cox J, Davies H, Edkins S, Holden S, et al (2004) Mutations in the DLG3 gene cause nonsyndromic X-linked mental retardation. Am J Hum Genet 75:318–324 [DOI] [PMC free article] [PubMed] [Google Scholar]