Abstract
ClpXP-dependent proteolysis has been implicated in the delayed detection of restriction activity after the acquisition of the genes (hsdR, hsdM, and hsdS) that specify EcoKI and EcoAI, representatives of two families of type I restriction and modification (R-M) systems. Modification, once established, has been assumed to provide adequate protection against a resident restriction system. However, unmodified targets may be generated in the DNA of an hsd+ bacterium as the result of replication errors or recombination-dependent repair. We show that ClpXP-dependent regulation of the endonuclease activity enables bacteria that acquire unmodified chromosomal target sequences to survive. In such bacteria, HsdR, the polypeptide of the R-M complex essential for restriction but not modification, is degraded in the presence of ClpXP. A mutation that blocks only the modification activity of EcoKI, leaving the cell with ≈600 unmodified targets, is not lethal provided that ClpXP is present. Our data support a model in which the HsdR component of a type I restriction endonuclease becomes a substrate for proteolysis after the endonuclease has bound to unmodified target sequences, but before completion of the pathway that would result in DNA breakage.
Within a bacterium that has a classical restriction and modification (R-M) system, the nucleotide sequences that define the targets for attack by the resident restriction endonuclease are concealed by the modification of appropriate bases within them. For some systems this modification is achieved by the methylation of specific adenine residues, and for others it is achieved by methylation of cytosine residues. The restriction endonuclease has the potential to attack DNA from different strains of the same species because foreign DNA generally lacks the protective imprint of the relevant methyltransferase (for reviews see refs. 1 and 2). Restriction of the host cell’s newly synthesized DNA normally is avoided, because the unmethylated strand of each target sequence produced by DNA replication is methylated before the next round of replication. If, however, resident DNA were to acquire unmodified target sequences, would it, like foreign DNA, become a substrate for restriction? In this paper we show that in situations where the modification of the host DNA by a type I R-M system fails, an alternative level of protection impairs the endonuclease activity of the restriction system and the bacteria survive.
A type I R-M system is encoded by three genes: hsdR, hsdM, and hsdS. The three polypeptides, HsdR, HsdM, and HsdS, often designated R, M, and S, assemble to give an enzyme (R2M2S1) that modifies hemimethylated DNA and restricts unmethylated DNA. A smaller complex (M2S1) has only the methyltransferase activity. The S subunit confers target specificity; hence, both complexes and both activities respond to the same nucleotide sequence.
Type I systems of enteric bacteria have been divided into discrete families by tests for cross-hybridization between genes and cross-reactivity with antibodies raised against the archetypal member of each family (3–5). Four families of distantly related systems have been identified (types IA, IB, IC, and ID), and where complementation tests have been done they indicate that enzymes in the same family can interchange subunits, but those from different families cannot (6, 7).
No transcriptional regulation of type I R-M genes has been detected; yet these genes are transferred readily to recipient bacteria devoid of the relevant modification activity (8–10). It is presumed that the cells survive the acquisition of the new R-M system because they become restriction proficient only after the modification activity is established. Experiments in support of this identify a lag of ≈15 generations before the cells become restriction-proficient after the acquisition of hsd genes by conjugation (11). The ClpXP protease was shown to be essential for the effective acquisition of genes specifying type IA and IB systems, and for this reason proteolysis has been implicated in the delayed expression of restriction activity (10).
The acquisition of a new specificity system is not the only situation in which a temporary loss of restriction proficiency has been detected. A well documented example, referred to as restriction alleviation (RA), occurs in response to treatments that damage DNA (12–14). UV light, nalidixic acid, and 2-aminopurine (2-AP) have been shown to induce restriction alleviation. It is possible that the temporary loss of restriction proficiency associated with the establishment of a new specificity is an example of RA. If this is so, ClpXP would be required for the alleviation of restriction in response to DNA damage. We have tested this hypothesis and show ClpXP to be a common requirement for RA in response to the various agents that damage DNA. This led us to identify steps in the molecular pathway that protect bacteria against the potentially lethal effects of restriction after DNA damage in a cell with a resident type I system or after the acquisition of a type I system capable of attacking the resident DNA.
MATERIALS AND METHODS
Bacterial Strains, Phages, Plasmids, and General Microbial Methods.
Bacterial strains are listed in Table 1. Integration-deficient, λhsdcI857 phages were used to transfer hsd alleles to bacterial chromosomes: λNM1367 includes hsdΔRM(F269G)S+; λNM1376, hsdM+S+; λNM1394, hsdM(F269G)S+; and λNM1384, hsdR(A619V) (17). JC9935 was used as the donor of the following derivatives of F′101: F′101–102, hsdKR−M+S+ (11); F′101–301, hsdK− hsdA+ (10); and F′101–103, zjj∷Tn10 hsdKR+Δ(MS)5. F′101–103 was selected after plasmid–chromosome allele exchange, as described for F′101–301 (10). pNK3 was made by transferring the HindIII-SmaI fragment containing hsdR from pBg3 (22) to pACYC184 (23) digested with HindIII and NruI. Media and general methods were as described previously (10).
Table 1.
E. coli K-12 strains
| Strain | Relevant genotype | Source or origin |
|---|---|---|
| C600 | hsdK+ | See ref. 10 |
| 5K | hsdR514 | See ref. 10 |
| CB51 | dam-3 | C. Boyd |
| JC9935 | recA13 | See ref. 10 |
| LE451 | rac-0 recA srl∷Tn10 | Ref. 15 |
| NM477 | Δ(hsdMS)5 | See ref. 10 |
| NM659 | ΔrecA∷cat | This laboratory |
| NM679 | Δ(hsdRMS) | Ref. 16 |
| NM799 | hsdR(A619V) | Refs. 17 and 18 |
| NM802 | ΔhsdR4 | This laboratory |
| SG22007 | ΔclpP∷cat | Ref. 19 |
| SG22080 | ΔclpX∷kan | Ref. 20 |
| SG22129 | ΔclpP∷cat Δ clpX∷kan | S. Gottesman |
| RH6972 | dnaQ∷miniTn10 (mutD) | D. R. F. Leach |
| RS2 | topA10 | Ref. 21 |
| TPC48 | zjj∷Tn10 dnaCts | See ref. 10 |
| NK31 | gyrA96 | Ref. 10 |
| NK167 | hsdK−hsdA+ | Ref. 10 |
| NK300 | rac-0 recA+srl+ | LB451 × P1(C600) |
| NK301 | rac-0 gyrA96 | NK300 × P1(NK31) |
| NK302 | dam | NK301 × P1(CB51) |
| NK303 | ΔclpP | NK301 × P1(SG22007) |
| NK304 | ΔclpX | NK301 × P1(SG22080) |
| NK308 | ΔrecA | NK301 × P1(NM659) |
| NK309 | zjj∷Tn10 dnaCts | NK301 × P1(TPC48) |
| NK310 | hsdR | NK301 × P1(5K) |
| NK311 | Δ(hsdRMS) | NK309 × P1(NM679) |
| NK312 | Δ(hsdRMS) ΔclpX | NK311 × P1(SG22080) |
| NK315 | dam ΔclpX | NK302 × P1(SG22080) |
| NK320 | ΔclpX | NK300 × P1(SG22080) |
| NK323 | ΔclpX ΔrecA | NK304 × P1(NM659) |
| NK324 | Δ(hsdRMS) ΔclpX ΔrecA | NK312 × P1(NM659) |
| NK325 | hsdR ΔclpX | NK310 × P1(SG22080) |
| NK326 | mutD | NK301 × P1(RH6972) |
| NK327 | mutD ΔclpX | NK326 × P1(SG22080) |
| NK329 | topA10 ΔclpP ΔclpX | RS2 × P1(SG22129) |
| NK351 | hsdR(A619V) | NK309 × P1(NM799) |
| NK352 | Δ(hsdMS)5 | NK309 × P1(NM477) |
| NK354 | hsdK−hsdA+ | NK309 × P1(NK167) |
| NK355 | hsdK−hsdA+ ΔclpX | NK354 × P1(SG22080) |
| NK378 | ΔhsdR hsdM(F269G) | NM802 × λNM1367 |
| NK379 | ΔhsdR | NK309 × P1(NM802) |
| NK380 | ΔhsdR ΔclpX | NK379 × P1(SG22080) |
| NK382 | ΔhsdR hsdM(F269G) | NK309 × P1(NK378) |
| NK383 | ΔhsdR hsdM(F269G) ΔrecA | NK382 × P1(NM659) |
| NK384 | ΔhsdR hsdM(F269G) ΔclpX | NK382 × P1(SG22080) |
| NK386 | hsdM(F269G) | NK301 × λNM1394 |
| NK388 | hsdR(A619V)hsdM(F269G) | NK386 × λNM1384 |
Affiliations: C. Boyd, Medical Research Council, Human Genetics Unit, University of Edinburgh; S. Gottesman, National Cancer Institute, Bethesda, MD; D. R. F. Leach, Institute of Cell and Molecular Biology, University of Edinburgh.
Restriction Alleviation.
2-AP (400 μg/ml) was added to midlogarithmic cultures grown at 37°C in LB medium. Intensive aeration was provided before and during the treatment. After 1 h, the cells were washed, resuspended in fresh broth, and tested for restriction. UV-induced RA was measured as described in ref. 24, and RA in response to nalidixic acid was measured as described by (13).
Analysis of Proteins.
Polypeptides were separated by electrophoresis through SDS/polyacrylamide gels (25). Western blots used rabbit antisera against EcoKI or EcoAI and the chemiluminescence detection system (POD) of Boehringer Mannheim.
The stability of proteins was monitored after pulse-labeling with [35S]methionine. Bacteria were grown at 37°C with intensive aeration to an OD600 of 0.2–0.3 in minimal medium supplemented with thiamin and all amino acids except methionine and cysteine. Chloramphenicol (20 μg/ml) maintained the presence of pNK3. Each culture was divided, and 2-AP (400 μg/ml) was added to one aliquot. After 1.5 h, a 1-min pulse of [35S]methionine (25 μCi/ml) was given. Labeling was stopped by diluting each culture with an equal volume of prewarmed LB supplemented with l-methionine (15 μM) or with l-methionine and 2-AP (400 μg/ml). Intensive aeration was maintained, and samples were taken at appropriate intervals. Bacteria were collected by centrifugation, resuspended in SDS sample buffer, and boiled for 5 min, and samples were applied to SDS/polyacrylamide gels for the separation of polypeptides by electrophoresis.
RESULTS
ClpXP Is Necessary for RA.
A simple quantitative test for restriction relies on the fact that most unmodified λ phages are killed when they infect Escherichia coli K-12; the phage genome is a substrate for EcoKI, the resident restriction system. The titer of an unmodified phage lysate (λ.0) on a restricting host relative to that on a nonrestricting derivative is referred to as the efficiency of plating (EOP). Therefore, the inverse of EOP quantifies restriction. RA is detected as a temporary reduction in restriction (hence, an increased EOP) after treatment of genetically restriction-proficient cells with agents that damage DNA.
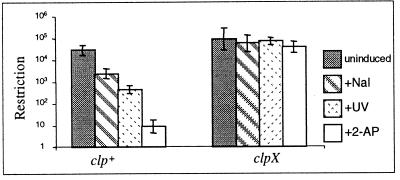
We examined RA for Clp+ and Clp− strains in response to each of three treatments; UV light, nalidixic acid, and 2-AP. For each treatment, ClpX was essential for efficient RA (Fig. 1). A clpP strain was tested for RA in response to 2-AP, and it also was deficient in RA (data not shown). The results support our hypothesis that RA, in response to agents that damage DNA, and the delayed expression of restriction activity after the acquisition of hsd+ genes by an hsd− recipient are both the outcome of a common ClpXP-dependent process. RA for the EcoAI system in response to 2-AP also was shown to be dependent on ClpX (data not shown).
Figure 1.
Restriction of unmodified phage λ by clp+ (NK301, NK300 for nalidixic acid) and clpX (NK304, NK320 for nalidixic acid) bacteria. Only clp+ cells show restriction alleviation.
“Constitutive” RA.
Restriction is alleviated in dam strains (26). It is known that the Dam-methylase identifies the parental DNA strand during mismatch repair, and in dam mutants mismatch repair leads to double-strand breaks (DSBs) (27). This alleviation of restriction in dam strains led us to question whether other mutations that impair the efficiency or fidelity of DNA replication might induce RA. If such a phenotype occurred, would it be dependent on ClpXP? We tested topA, mutD, and dam strains.
Mutants deficient in topoisomerase I, like wild-type cells treated with nalidixic acid, have problems in DNA replication; DSBs may occur when the replication forks stall (28). In contrast, a mutD mutation enhances the error rate of DNA polymerase III (29) and the increased frequency of mismatches may mimic the effect of 2-AP, an analogue of adenine that causes base pair transitions.
Restriction by dam, topA, or mutD strains was at least 100-fold less efficient than restriction by wild-type E. coli K-12 (Fig. 2). If this poor restriction is the result of constitutive expression of RA activated in response to either DNA damage or mismatches, then a mutation in clpX or clpP should restore restriction. Consistent with this prediction, the efficiency of restriction was enhanced by approximately 100-fold in the absence of ClpXP protease (Fig. 2).
Figure 2.
Restriction of unmodified phage λ by dam (NK302), mutD (NK326), and topA (RS2) strains and their clpX derivatives (NK315, NK327, and NK329). It is known that topA strains accumulate compensatory mutations in gyrA or gyrB (21), but the topA10 strain (RS2) is not known to have a compensatory mutation (21), and the topA mutation itself correlates with impaired restriction (G. P. Davies, personal communication).
ClpXP-Deficient, Restriction-Proficient Bacteria Die During Prolonged Exposure to 2-AP.
After prolonged treatment with 2-AP (3–4 h at 400 μg/ml), clp− (NK303 and NK304) but not clp+ (NK301) bacteria become filamentous, a phenotype characteristic of the SOS response. 2-AP does not normally activate the SOS response but, in the absence of ClpXP, it could induce a chain of events that leads to DNA damage. The relevance of a RecA-dependent repair pathway is supported by the observation that recA clpX double mutants (NK323) are supersensitive to 2-AP and do not survive low concentrations (40 μg/ml) of 2-AP in the medium. In contrast, a recA clp+ hsd+ strain (NK308) is no more sensitive to 2-AP than its rec+ counterpart (NK301); recA strains resemble rec+ in their RA response to 2-AP.
Is ClpXP needed in the presence of 2-AP to prevent DNA damage by the resident restriction endonuclease? We made the clpX bacteria deficient in restriction both by deleting the hsd genes (NK312) and by including a mutation in hsdR (NK325), the gene essential for restriction. The restriction-deficient bacteria were not sensitive to 2-AP. Similarly, the hypersensitivity of the recA clpX strain was relieved by inactivation of the endonuclease activity. We suggest that during prolonged treatment with 2-AP, the ClpXP-dependent pathway is essential to prevent EcoKI from causing DNA damage and consequent cell death.
RA Induced by 2-AP Is Associated with a Deficiency of HsdR.
RA is not correlated with a loss of modification activity (14, 30). It could, therefore, be the result of a deficiency in HsdR and the consequent depletion of EcoKI (R2M2S1), but not the modification enzyme (M2S1).
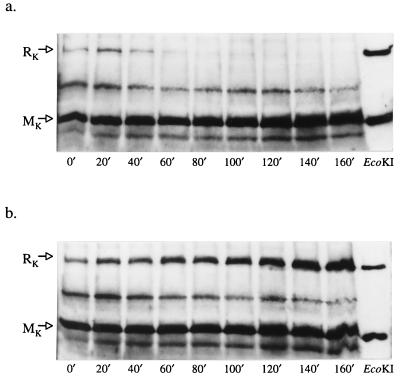
The HsdR and HsdM subunits were monitored by Western blots after the addition of 2-AP to both clp+ and clpX bacteria (Fig. 3). After a lag of 20 min, a reduction in the concentration of HsdR, but not HsdM, was detected. This deficiency of HsdR was found only in clp+ cells in response to 2-AP. RA, therefore, correlated with a ClpX-dependent reduction in the concentration of HsdR, the polypeptide essential for restriction, but not modification.
Figure 3.
Assays for HsdR and HsdM polypeptides after treatment with 2-AP. (a) clp+ bacteria (NK301). (b) clpX bacteria (NK304). In the absence of 2-AP (data not shown), the assays for clp+ and clpX bacteria were indistinguishable from those seen in b. EcoKI polyclonal antibody, used in these Western blots, fails to detect HsdS, but detects HsdR and HsdM and some other E. coli proteins.
HsdR Is Degraded in clp+ Cells Treated with 2-AP.
The very low concentration of HsdR detected in Clp+ cells after a period of growth in the presence of 2-AP (Fig. 3) is consistent with the degradation of HsdR in the presence of ClpXP, but it could be argued that ClpXP in some way affects the synthesis rather than the degradation of HsdR.
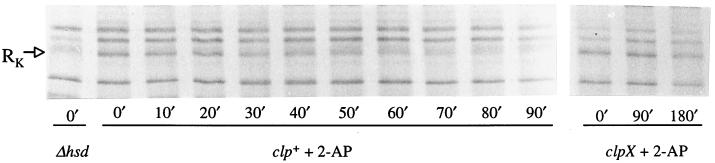
We therefore assayed the stability of HsdR in clp+ and clpX cells in response to treatment with 2-AP. The preferred experiment was to rely on the chromosomal hsdR gene, but the signal generated from a single copy of hsdR was weak compared with those generated by other proteins. Gene dosage was increased by cloning hsdR in pACYC184, a low-copy-number vector. clp+ hsd+ (NK301) and clpX hsd+ (NK304) bacteria transformed with the hsdR+ plasmid (pNK3) were treated with 2-AP for 90 min to allow the establishment of RA before they were pulse-labeled with [35S]methionine. HsdR was unstable in Clp+ but not ClpX− cells after 2-AP treatment (Fig. 4). In the absence of 2-AP (data not shown) the HsdR polypeptide was stable in clp+ and clpX cells for at least 180 min.
Figure 4.
The stability of HsdR in vivo after treatment with 2-AP. Labeled polypeptides separated by electrophoresis through SDS-polyacrylamide gels (6%) were detected by autoradiography. An extract from a strain lacking HsdR (NK311/pACYC184) was analyzed in the first track. Samples from clp+ and clpX bacteria containing pNK3 were taken at the time intervals indicated after pulse labeling.
These results are consistent with 2-AP as the activator of a RA pathway in which HsdR is susceptible to ClpXP-dependent proteolysis.
Functional EcoKI Is Obligatory for the Loss of HsdR That Is Characteristic of RA.
Is active EcoKI necessary to generate the signal that leads to ClpXP-dependent degradation of HsdR? To answer this question we tested whether 2-AP-induced depletion of HsdR occurs in restriction-deficient mutants. One of the mutants tested has a missense mutation in hsdR (NK351), and the other (NK352) has a wild-type hsdR gene, but hsdM and hsdS are deleted so that HsdR cannot form an EcoKI complex.
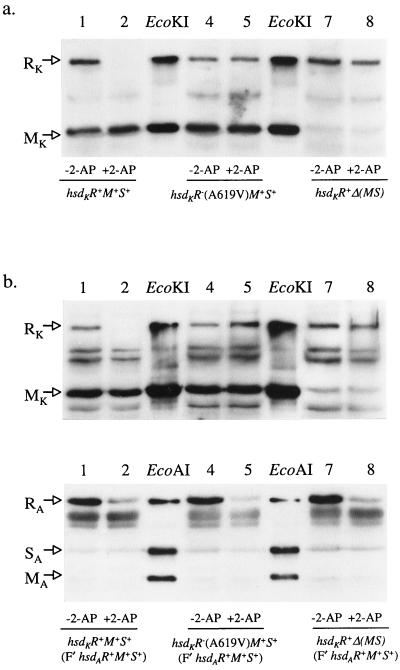
HsdR was not depleted in either mutant in response to 2-AP (Fig. 5a). This finding implies that a functional endonuclease is required for induction of the pathway that leads to degradation of HsdR. If the products of restriction by a type I enzyme are the stimulus for RA, the endonuclease activity of one R-M system should induce RA for a different system. We tested whether a functional type IB system (EcoAI), for which RA is regulated in a ClpXP-dependent manner, induced degradation of the HsdR polypeptide of the inactive type IA system, EcoKI.
Figure 5.
Hsd subunits were monitored, after treatment with 2-AP, using antibodies raised against the relevant R-M complex. HsdR is degraded only when it is a part of a functional complex. (a) Degradation of HsdR is prevented by a missense mutation in hsdR (track 5) or by the absence of HsdM and S (track 8). (b) The presence of functional EcoAI has no effect on the degradation of the HsdR subunit of EcoKI (Upper), even though the HsdR subunit of EcoAI itself is degraded (Lower, lanes 2, 5, and 8). The control tracks for EcoAI contain a mixture of polypeptides in which HsdM and HsdS are present in molar excess to give strong signals with antibody.
We transferred F′hsdAR+M+S+ (F′101–301) to the three strains used in the previous experiment (Fig. 5a): hsdKR+M+S+, hsdKR−M+S+, and hsdKR+Δ(MS). The transconjugants, both untreated and treated with 2-AP, were assayed for EcoAI- and EcoKI-dependent restriction in vivo and for the presence of HsdR polypeptides. 2-AP caused RA of functional R-M systems, and HsdR from any restriction-proficient complex was lost (Fig. 5b). However, for the nonfunctional EcoKI complex, RK remains even in the presence of functional EcoAI. These data require that the stimulus for RA is family-specific and therefore is not simply the product of restriction.
Mutations Predicted to Confer a Restriction-Proficient, Modification-Deficient (r+m−) Phenotype Cause Restriction Alleviation.
It is logical to expect that a mutation conferring an r+m− phenotype would be lethal. We chose to investigate a mutation in hsdM (F269G) that abolishes methyltransferase activity but has no effect on the binding of the cofactor S-adenosylmethionine and therefore is predicted to leave a functional endonuclease (31). This hsdM mutation was transferred from a λhsd phage (λNM1367) to the chromosome of an hsdR strain. The presence of hsdM(F269G) (NK378) was associated with the anticipated m− phenotype. We tested the naive prediction that the acquisition of an F′ with a functional hsdR gene would generate r+m− transconjugants and these would die. However, we found no difference between the survival of the recipients upon acquisition of F′hsdR+ and the survival of recipients receiving the control F′ lacking an hsdR+ allele (Fig. 6). We extended our experiment to include a recA recipient in which DSBs would not be repaired; transfer of the hsdR+ allele still occurred efficiently (Fig. 6).
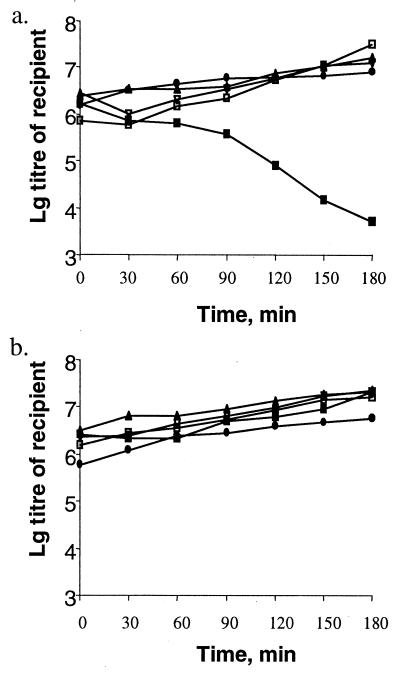
Figure 6.
The survival of hsdR+M−(F269G)S+ cells was assessed after the conjugative transfer of hsdR+ to hsdR−M−S+ recipients. (a) The experiment using F′101–103 (hsdR+). (b) The control experiment with F′101–102 (hsdM+S+). Data are plotted for the following recipients: NK379, hsdR (▴); NK380, hsdR clpX (▿); NK382, hsdRM (□); NK384, hsdRM clpX (■); and NK383, hsdRM recA (●). The data show that hsdR+M−(F269G)S+ cells survive only if the recipient is ClpX+.
The EOP of λ.0 on the hsdR−M−S+ (F′hsdR+) transconjugants was 10−1 in contrast to 5 × 10−4 when the F′hsdR+ was transferred to an hsdR−M+S+ recipient (the EOP of λ.K was 1 in both cases). The low level of restriction by the m− transconjugants is consistent with induction of the RA response. Therefore, the conjugation experiments were extended to include clpX recipients. In the absence of ClpX, transfer of the F′hsdR+ to the hsdR−M−S+ recipient was lethal, consistent with the presence of functional restriction endonuclease (Fig. 6). Our hypothesis predicts that the transconjugant bacteria can survive in the presence of ClpXP because of the activation of the RA pathway. If this suggestion is correct, hsdR+M−S+ bacteria would be deficient in HsdR.
We chose to use chromosomal genes in preference to a plasmid-borne hsdR to test this prediction. We transferred the hsdM(F269G) mutation to the chromosome of NK301, an hsd+ strain. The hsdM recombinants were recognized by their m− phenotype and could not be transduced to give clpX derivatives (data not shown). These derivatives restricted λ.0 with an efficiency indicative of RA (EOP = 10−1). Consistent with the induction of RA, HsdR was missing in the hsdM(F269G) strain that encodes a functional restriction enzyme and present in a derivative with a missense mutation in hsdR (Fig. 7). Importantly, when hsdM(F269G) was replaced with the wild-type allele (see legend to Fig. 7), HsdR was restored. Therefore, the loss of HsdR is a consequence of the hsdM mutation. Our experiments with the modification-deficient mutant show that E. coli has an extraordinary capacity to protect itself against potential DNA damage elicited by a resident type I R-M system.
Figure 7.
The effect of hsdM (F269G) on the level of HsdR. The mutation hsdM (F269G) destroys only the modification activity of EcoKI. The level of HsdR was monitored by Western blots by using antibody against EcoKI. Lanes 1–6 include extracts of strains. Lanes: 1, NK301 (hsd+); 2, NK386 [an hsdM (F269G) derivative of NK301]; 3, an hsd+ derivative of NK386; 4, an hsdR(A619V) derivative of NK386 (NK388) in which alleles of hsd genes were replaced by using λhsd phages that included only hsdMS or hsdR, respectively; 5, NM802 (an hsdR deletion strain); and 6, NK352 (an hsdMS deletion strain).
DISCUSSION
The diagnostic feature of RA is an r− phenotype despite a restriction-proficient genotype (hsd+). The r− phenotype that persists for many generations in a transconjugant after the acquisition of functional hsd genes by an hsd− recipient (11, 32) may be viewed as an example of RA. In this case, the establishment of hsd+ genes in a naive bacterium depends on the ClpXP protease (10). We now have shown that RA in response to a variety of stimuli, including external agents and mutations that affect the fidelity of DNA replication, also requires ClpXP. In two quite different situations the presence of subunits of EcoKI was monitored after the induction of RA. In the first, the bacteria were treated with 2-AP, and in the second, a mutation in hsdM (F269G) was introduced that blocks only the methyltransferase activity of EcoKI (31). In both these examples of ClpXP-dependent RA, a negligible level of HsdR remained. We propose a general pathway for RA in which ClpXP is necessary for the degradation of HsdR and the consequent r− phenotype. According to this scheme, unmodified chromosomal DNA targets would be a signal for the cell to protect its own DNA from restriction. We believe that all the stimuli for RA examined by us rely on the presence of unmodified target sequences.
A particularly severe stimulus is provided by the mutation in hsdM (F269G) that results in a modification-deficient, restriction-proficient EcoKI complex (Fig. 6a). For this mutant to survive, despite an unmodified chromosome, restriction alleviation must be extraordinarily effective. A more common stimulus is DNA damage that elicits RecA-dependent repair. UV irradiation and mutations in dam can cause DSBs (26, 33); nalidixic acid and mutations in topA are likely to generate DSBs by stalling replication. Damage by UV light also leads to lesions in one strand that are repaired postreplicatively (34). RecA-dependent repair relies on homologous recombination. If homologous recombination involves two segments of hemimethylated DNA, the annealing of unmethylated strands or DNA synthesis may generate a localized region of unmethylated DNA. In contrast, both 2-AP and mutD increase the frequency of base pair transitions (29, 35). Some mutations will generate new target sequences, all of which will be unmodified.
Our experiments have shown a ClpXP-dependent loss of HsdR in response to 2-AP. It seems likely that the ClpXP protease itself degrades HsdR, rather than being necessary to maintain or activate another protease. The only protease-deficient mutants found to affect the transmission of the genes encoding EcoKI were clpX and clpP (10). Our experiments also show that HsdR is lost only in cells in which HsdR could produce functional EcoKI. Thus, in the absence of HsdM and HsdS, wild-type HsdR is not degraded; likewise, in the presence of HsdM and HsdS, a missense mutation in hsdR prevents degradation of the nonfunctional polypeptide. The requirement for unmodified targets and functional EcoKI might suggest that DNA breakage initiates the RA response. We argue that DSBs are not involved in the initiation of RA. One reason for doubting this idea is our observation that a recAclp+hsd+ bacterium is no more sensitive to 2-AP than its rec+ counterpart. This finding is not consistent with the creation of DSBs in response to 2-AP. Second, we tested whether active EcoAI, a member of the type IB family of enzymes is sufficient to induce loss of the HsdR subunit of EcoKI in response to treatment with 2-AP. It is not, although it is susceptible to ClpXP-dependent RA. If DSBs are the signal for RA, those made by EcoAI do not provide a signal for degradation of the HsdR subunit of EcoKI. Finally, even in the absence of RecA we readily made strains in which EcoKI is defective in methyltransferase activity (Fig. 6). Because DSBs cannot be repaired in a recA− strain (36), it would appear that in this hsdR+M(F269G) S+ bacterium DSBs are avoided, despite the presence of ≈600 unmodified targets and the coding potential for restriction-proficient, modification-deficient EcoKI. We conclude that ClpXP-dependent degradation of HsdR is able to prevent cutting of the bacterial chromosome. In the absence of ClpXP, however, even rec+ cells fail to survive because EcoKI cuts their chromosomes.
If DSBs are not the stimulus for RA, why does a missense mutation in hsdR prevent degradation of HsdR? The amino acid substitution (A619V) is associated with a defect in the hydrolysis of ATP and probably, therefore, with the ATP-dependent translocation of DNA that precedes the generation of DSBs (18). The missense mutation does not prevent either the binding of EcoKI to its target sequence or the associated ATP-dependent conformational change that is a prerequisite for the restriction pathway (18, 37). Other missense mutations in HsdR also prevent degradation of HsdR (V.A.D. and N.E.M., unpublished observations); therefore, it seems probable that the functional defect, rather than the amino acid substitution per se, determines whether the enzyme is a substrate for ClpXP. We conclude that HsdR is recognized only after the EcoKI complex has embarked on its restriction pathway. It remains to be determined what renders the HsdR subunits susceptible to proteolysis. Nevertheless, the present experiments promote the concept of a remarkably specific control mechanism, effective only once the relevant restriction pathway has been initiated, but able to act before any damage is inflicted on unmodified chromosomal DNA.
The RA response can protect the bacterial chromosome from restriction in the complete absence of modification, but the alleviation is not entirely complete when analyzed by infection with λ.0 (EOP = 10−1). These facts raise two new, but probably related, problems. First, why does phage DNA entering the cell show some susceptibility to restriction whereas the resident bacterial chromosome does not? Second, why do unmodified targets on the chromosome, but not those on incoming phage DNA, stimulate the RA response? At present, it should be borne in mind that the two substrates differ in their location and their association with other proteins.
Our current experiments document the disappearance of HsdR under conditions of RA, and we interpret this as ClpXP-dependent degradation of HsdR. Initial but unsuccessful attempts to detect degradation in vitro used purified HsdR, or EcoKI, as substrate. The in vivo experiments indicate that the substrate is unlikely to be protein alone but, rather, a functional protein–DNA complex.
The role of ClpXP in the disassembly and degradation of the Mu transposase already is known to be complex. MuB apparently protects the MuA–DNA complex from recognition by ClpX and, hence, from disassembly and potential degradation by the protease activity of ClpP (38). These authors suggested “that a protein–complex architecture that uses overlapping sequences for subunit interactions and for targeting a protein for remodeling or destruction provides a useful design for this type of regulation.” By analogy we would suggest that some step in the ATP-dependent DNA translocation by EcoKI leads to the exposure of the target sequence for ClpX.
Our investigation of the relevance of ClpXP to RA has been confined to the type IA and IB families of R-M systems. There is evidence for Dam-mediated RA of a type III system (26). Members of the type IC and ID families are susceptible to RA in response to 2-AP (unpublished results), but transmission of the plasmid-borne type IC hsd genes by conjugation is not dependent on ClpXP (10, 32). Although the assembly pathway of the R2M2S1 complex may provide a lag in the production of the endonuclease after plasmid transfer (39), it would not prevent the cutting of unmodified targets created in cells in which functional endonuclease is already assembled. It is not known whether RA can involve other proteases or other mechanisms, but RA is found for some methylation-dependent restriction systems (24), where DNA damage would not generate target sequences. RA has not been detected for any type II system; rather, RA appears to be characteristic of complex R-M systems.
Our experiments demonstrate that control of the restriction activity of EcoKI is extraordinarily sensitive. It not only copes with the acquisition of hsd genes conferring new specificities and the production of unmodified targets created by repair and mutation, but clp+ cells also survive a mutation that destroys the modification activity of the R-M complex. A similar control system could permit the efficient phase variation of type I R-M systems, a phenomenon recently documented for Mycoplasma pulmonis (40). Molecular mechanisms of the sophisticated interactions that mediate the proteolytic control remain to be determined.
Acknowledgments
We thank our colleagues for their interest throughout this work and, most particularly, Susan Gottesman, David R. F. Leach, Mary O’Neill, Lynn M. Powell, and Frank W. Stahl for their constructive criticism of the manuscript. We are indebted to David Leach for strains and Pavel Janscak for EcoAI. S.M. and V.A.D. thank the Darwin Trust for the award of Scholarships. The research was supported by the Medical Research Council.
ABBREVIATIONS
- R-M
restriction and modification
- RA
restriction alleviation
- 2-AP
2-aminopurine
- DSB
double-strand break
- EOP
efficiency of plating
References
- 1.Barcus V A, Titheradge A J, Murray N E. Genetics. 1995;140:1187–1197. doi: 10.1093/genetics/140.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Redaschi N, Bickle T A. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 773–781. [Google Scholar]
- 3.Murray N E, Gough J A, Suri B, Bickle T A. EMBO J. 1982;1:535–539. doi: 10.1002/j.1460-2075.1982.tb01205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Price C, Pripfl T, Bickle T A. Eur J Biochem. 1987;167:111–115. doi: 10.1111/j.1432-1033.1987.tb13310.x. [DOI] [PubMed] [Google Scholar]
- 5.Titheradge A J, Ternent D, Murray N E. Mol Microbiol. 1996;22:437–447. [PubMed] [Google Scholar]
- 6.Boyer H W, Roulland-Dussoix D. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 7.Fuller-Pace F V, Cowan G M, Murray N E. J Mol Biol. 1985;186:65–75. doi: 10.1016/0022-2836(85)90257-8. [DOI] [PubMed] [Google Scholar]
- 8.Prakash-Cheng A, Chung S S, Ryu J. Mol Gen Genet. 1993;241:491–496. doi: 10.1007/BF00279890. [DOI] [PubMed] [Google Scholar]
- 9.O’Neill M, Chen A, Murray N E. Proc Natl Acad Sci USA. 1997;94:14596–14601. doi: 10.1073/pnas.94.26.14596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makovets S, Titheradge A J, Murray N E. Mol Microbiol. 1998;28:25–35. doi: 10.1046/j.1365-2958.1998.00767.x. [DOI] [PubMed] [Google Scholar]
- 11.Prakash-Cheng A, Ryu J. J Bacteriol. 1993;175:4905–4906. doi: 10.1128/jb.175.15.4905-4906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertani G, Weigle J J. J Bacteriol. 1953;65:113–121. doi: 10.1128/jb.65.2.113-121.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thoms B, Wackernagel W. Mol Gen Genet. 1984;197:297–303. doi: 10.1007/BF00330977. [DOI] [PubMed] [Google Scholar]
- 14.Efimova E P, Delver E P, Belogurov A A. Mol Gen Genet. 1988;214:317–320. doi: 10.1007/BF00337728. [DOI] [PubMed] [Google Scholar]
- 15.Diaz R, Barnsley P, Pritchard R H. Mol Gen Genet. 1979;175:151–157. doi: 10.1007/BF00425531. [DOI] [PubMed] [Google Scholar]
- 16.King G, Murray N E. Mol Microbiol. 1995;16:769–777. doi: 10.1111/j.1365-2958.1995.tb02438.x. [DOI] [PubMed] [Google Scholar]
- 17.Webb J L, King G, Ternent D, Titheradge A J, Murray N E. EMBO J. 1996;15:2003–2009. [PMC free article] [PubMed] [Google Scholar]
- 18.Davies G P, Powell L M, Webb J L, Cooper L P, Murray N E. Nucleic Acids Res. 1998;26:4828–4836. doi: 10.1093/nar/26.21.4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maurizi M R, Clark W P, Katayama Y, Rudikoff S, Pumphrey J, Bowers B, Gottesman S. J Biol Chem. 1990;265:12536–12545. [PubMed] [Google Scholar]
- 20.Gottesman S, Clark W P, de Crecy-Lagard V, Maurizi M R. J Biol Chem. 1993;268:22618–22626. [PubMed] [Google Scholar]
- 21.DiNardo S, Voelkel K A, Sternglanz R, Reynolds A E, Wright A. Cell. 1982;31:43–51. doi: 10.1016/0092-8674(82)90403-2. [DOI] [PubMed] [Google Scholar]
- 22.Sain B, Murray N E. Mol Gen Genet. 1980;180:35–46. doi: 10.1007/BF00267350. [DOI] [PubMed] [Google Scholar]
- 23.Chang A C Y, Cohen S N. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelleher J E, Raleigh E A. J Bacteriol. 1994;176:5888–5896. doi: 10.1128/jb.176.19.5888-5896.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Efimova E P, Delver E P, Belogurov A A. Mol Gen Genet. 1988;214:313–316. doi: 10.1007/BF00337727. [DOI] [PubMed] [Google Scholar]
- 27.Wang T C, Smith K C. J Bacteriol. 1986;165:1023–1025. doi: 10.1128/jb.165.3.1023-1025.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michel B, Ehrlich S D, Uzest M. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Echols H, Lu C, Burgers P M. Proc Natl Acad Sci USA. 1983;80:2189–2192. doi: 10.1073/pnas.80.8.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hiom K J, Sedgwick S G. Mol Gen Genet. 1992;231:265–275. doi: 10.1007/BF00279800. [DOI] [PubMed] [Google Scholar]
- 31.Willcock D F, Dryden D T, Murray N E. EMBO J. 1994;13:3902–3908. doi: 10.1002/j.1460-2075.1994.tb06701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kulik E M, Bickle T A. J Mol Biol. 1996;264:891–906. doi: 10.1006/jmbi.1996.0685. [DOI] [PubMed] [Google Scholar]
- 33.Bonura T, Smith K C. J Bacteriol. 1975;121:511–517. doi: 10.1128/jb.121.2.511-517.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rupp W D. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 2277–2294. [Google Scholar]
- 35.Ronen A. Mutat Res. 1980;75:1–47. doi: 10.1016/0165-1110(80)90026-3. [DOI] [PubMed] [Google Scholar]
- 36.Krasin F, Hutchinson F. J Mol Biol. 1977;116:81–98. doi: 10.1016/0022-2836(77)90120-6. [DOI] [PubMed] [Google Scholar]
- 37.Powell L M, Dryden D T, Murray N E. J Mol Biol. 1998;283:963–976. doi: 10.1006/jmbi.1998.2143. [DOI] [PubMed] [Google Scholar]
- 38.Levchenko I, Yamauchi M, Baker T A. Genes Dev. 1997;11:1561–1572. doi: 10.1101/gad.11.12.1561. [DOI] [PubMed] [Google Scholar]
- 39.Janscak P, Dryden D T, Firman K. Nucleic Acids Res. 1998;26:4439–4445. doi: 10.1093/nar/26.19.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dybvig K, Sitaraman R, French C T. Proc Natl Acad Sci USA. 1998;95:13923–13928. doi: 10.1073/pnas.95.23.13923. [DOI] [PMC free article] [PubMed] [Google Scholar]