Abstract
The transmembrane aspartate receptor of bacterial chemotaxis regulates an associated kinase protein in response to both attractant binding to the receptor periplasmic domain and covalent modification of four adaptation sites on the receptor cytoplasmic domain. The existence of at least 16 covalent modification states raises the question of how many stable signaling conformations exist. In the simplest case, the receptor could have just two stable conformations (“on” and “off”) yielding the two-state behavior of a toggle-switch. Alternatively, covalent modification could incrementally shift the receptor between many more than two stable conformations, thereby allowing the receptor to function as a rheostatic switch. An important distinction between these models is that the observed functional parameters of a toggle-switch receptor could strongly covary as covalent modification shifts the equilibrium between the on- and off-states, due to population-weighted averaging of the intrinsic on- and off-state parameters. By contrast, covalent modification of a rheostatic receptor would create new conformational states with completely independent parameters. To resolve the toggle-switch and rheostat models, the present study has generated all 16 homogeneous covalent modification states of the receptor adaptation sites, and has compared their effects on the attractant affinity and kinase activity of the reconstituted receptor–kinase signaling complex. This approach reveals that receptor covalent modification modulates both attractant affinity and kinase activity up to 100-fold, respectively. The regulatory effects of individual adaptation sites are not perfectly additive, indicating synergistic interactions between sites. The three adaptation sites at positions 295, 302, and 309 are more important than the site at position 491 in regulating attractant affinity and kinase activity, thereby explaining the previously observed dominance of the former three sites in in vivo studies. The most notable finding is that covalent modification of the adaptation sites alters the receptor attractant affinity and the receptor-regulated kinase activity in a highly correlated fashion, strongly supporting the toggle-switch model. Similarly, certain mutations that drive the receptor into the kinase activating state are found to have correlated effects on attractant affinity. Together these results provide strong evidence that chemotaxis receptors possess just two stable signaling conformations and that the equilibrium between these pure on- and off-states is modulated by both attractant binding and covalent adaptation. It follows that the attractant and adaptation signals drive the same conformational change between the two settings of a toggle. An approach that quantifies the fractional occupancy of the on- and off-states is illustrated.
Keywords: bacterial chemotaxis, two-component signaling pathway, CheA, adaptation site, transmembrane signal
INTRODUCTION
In motile bacteria, chemotaxis toward or away from attractants or repellents is mediated by the chemotaxis signaling pathway in which transmembrane receptors serve to modulate the activity of an associated histidine kinase (Parkinson 1993; Blair 1995; Falke et al. 1997; Grebe and Stock 1998; Armitage 1999; Bren and Eisenbach 2000). This bacterial chemotaxis pathway is representative of a large class of homologous receptor-regulated phosphorelay pathways ubiquitous in prokaryotes and lower eukaryotes (Swanson and Simon 1994; Wurgler-Murphy and Saito 1997). The aspartate receptor of Escherichia coli and Salmonella typhimurium is an extensively studied example of the transmembrane receptors that typically regulate such pathways. The aspartate receptor and its closest prokaryotic relatives initiate taxis to a range of different stimuli including chemicals, heat, osmotic pressure, and light (Bibikov et al. 1997; Falke et al. 1997; Nishiyama et al. 1999; Perazzona and Spudich 1999). The cytoplasmic domains of these taxis receptors are highly conserved, suggesting that they share similar molecular signaling mechanisms (Le Moual and Kirkland 1996), as confirmed by the construction of functional chimeric proteins formed by fusing different domains from closely related chemoreceptors (Krikos et al. 1985; Slocum et al. 1987; Tatsuno et al. 1994; Weerasuriya et al. 1998). Furthermore, active chimeric proteins have been generated by fusing chemoreceptor domains with receptor domains from more distantly related prokaryotic receptors or even unrelated eukaryotic receptors, indicating that a large group of receptors may share similar mechanisms of kinase regulation (Moe et al. 1989; Utsumi et al. 1989; Baumgartner et al. 1994).
The structure of the chemotaxis receptors has been extensively characterized by biochemical and crystallographic studies (Falke and Kim 2000; Falke and Hazelbauer 2001). A schematic view of the receptor is shown in Fig. 1. The aspartate chemotaxis receptor is a homodimer that binds aspartate in the periplasmic domain and propagates a signal across the membrane bilayer and through the cytoplasmic domain to the histidine kinase. The smallest unit of receptor structure is the homodimer, which is stable both in the presence or absence of ligand (Milligan and Koshland 1988). There is strong negative cooperativity between the two ligand binding sites present in the homodimer, effectively limiting the aspartate receptor to one ligand bound per dimer (Milburn et al. 1991; Biemann and Koshland 1994; Danielson et al. 1994; Kolodziej et al. 1996). The receptor is primarily helical in structure, consisting of well-defined regions of 4-helix bundle architecture (Milburn et al. 1991; Pakula and Simon 1992; Chervitz and Falke 1995; Chervitz et al. 1995; Bass and Falke 1999; Kim et al. 1999). Independent lines of evidence indicate that the transmembrane signal is carried by a 1–2-Å piston-type displacement of one of the four transmembrane helices, termed the signaling helix, toward the cytoplasm upon attractant binding (Chervitz and Falke 1996; Falke and Hazelbauer 2001). However, the mechanism by which the conformational signal is transmitted through the cytoplasmic domain remains unknown, except that it subtly rearranges the packing of the cytoplasmic helices (Bass and Falke 1999). The cytoplasmic 4-helix bundle provides a structural scaffold for the formation of a stable ternary signaling complex consisting of the receptor, the coupling protein CheW, and the histidine kinase CheA (Gegner et al. 1992; Schuster et al. 1993). In this ternary complex, the receptor stimulates CheA autophosphorylation activity, which subsequently results in the transfer of the phosphate group to the response regulator CheY (Borkovich et al. 1989; Ninfa et al. 1991). Although a great deal of structural information has been obtained for most individual components of the chemotaxis system (for reviews see Djordjevic and Stock 1998; Falke and Kim 2000), the overall structure of the assembled ternary complex remains unknown. Further, the receptor dimers and other pathway components assemble into patches of poorly characterized structure in the cell (Maddock and Shapiro 1993; Lybarger and Maddock 2001). Such higher order interactions between receptor dimers are known to play an important role in receptor adaptation, and could have other significant functions (Le Moual et al. 1997; Li et al. 1997; Bornhorst and Falke 2000; Li and Weis 2000).
Figure 1.
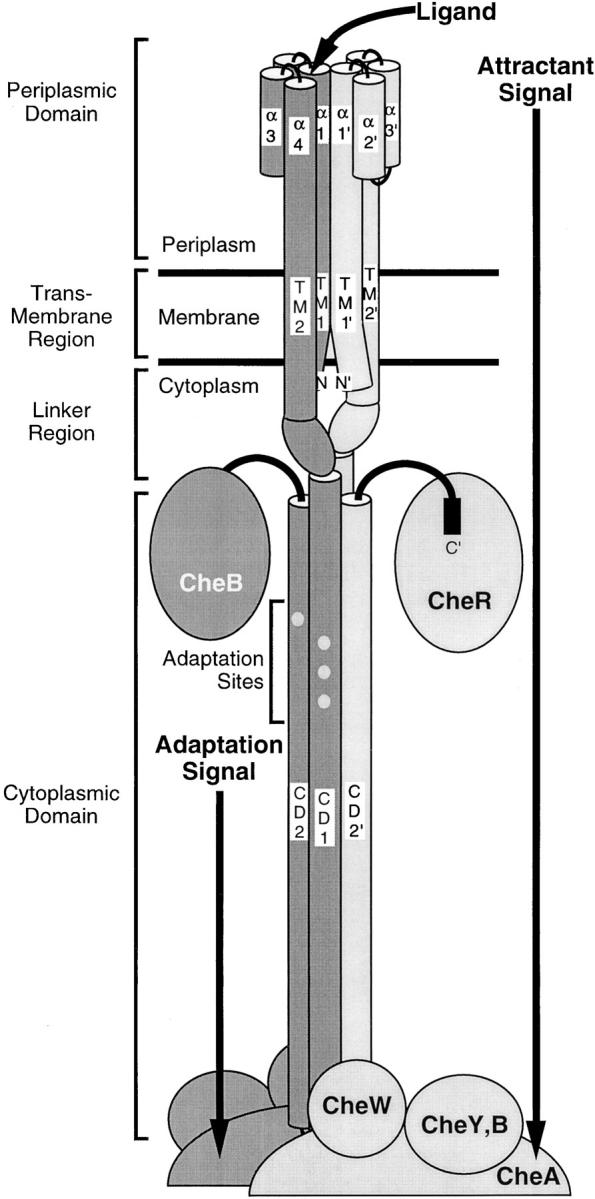
Schematic model of the full-length homodimeric membrane bound receptor in the associated signaling complex (Falke and Hazelbauer 2001). Cylinders represent helical domains determined by crystallographic or cysteine and disulfide scanning methods. The two 60-kD receptor subunits are differentially shaded. The receptor provides a scaffold for the formation of a large super-molecular signaling complex. Proposed docking sites for the methyltransferase CheR, methylesterase CheB, histidine kinase CheA, coupling protein CheW and motor response regulator CheY are shown. The core ternary signaling complex consisting of the dimeric receptor, dimeric CheA, and CheW molecules is stable both in the presence and absence of attractant. Filled circles represent the four adaptation sites on each receptor subunit. Binding of attractant at the periplasmic domain of the receptor and modification of the cytoplasmic adaptation sites serve to modulate the kinase activity of the signaling complex.
The chemotaxis receptors transmit two opposing signals to the bound kinase: (1) attractant binding to the periplasmic domain generates a transmembrane signal that inhibits cytoplasmic kinase activity; and (2) covalent modification of specific adaptation sites on the receptor cytoplasmic domain stimulates kinase activity. These two opposing signals can be termed the attractant and adaptation signals. The adaptation sites of the aspartate receptor are four glutamate side chains located at positions 295, 302, 309, and 491 on the surface of the cytoplasmic 4-helix bundle (Terwilliger and Koshland 1984). The wild-type receptor is expressed with glutamine side chains at the first and third adaptation sites, and, thus, is denoted QEQE. The adaptation enzyme CheB, a hydrolase, deamidates the glutamine residues to glutamate, yielding EEEE (Kehry et al. 1983). The other adaptation enzyme CheR, a methyl transferase, can methyl-esterify any of the four sites, converting it to the methylated state Em (Springer and Koshland 1977). Each methylated adaptation site can then become a substrate of CheB, which hydrolyzes the methyl ester to restore the free E side chain (Hess et al. 1988; Russell et al. 1989). Together, the competing activities of CheB and CheR yield a receptor population containing a statistical mixture of modification states ranging from EEEE to EmEmEmEm, where the average number of methylated sites, Em, increases with attractant concentration (Terwilliger et al. 1986). A given receptor dimer can exhibit up to 162 different modification states since each subunit is modified independently to give 16 different combinations of glutamate and methylated-glutamate residues, and since the symmetry of the dimer is broken by negative cooperativity in attractant binding. The average level of adaptation site modification across the receptor population serves as a memory of previous chemical environments that enables temporal comparisons with the current attractant concentration as sensed by the attractant binding sites. Moreover, the adaptation process allows receptors to adjust their sensitivity so that chemotaxis can occur even in the presence of high background attractant concentrations (Blair 1995; Falke et al. 1997). It is not yet established whether the attractant and adaptation signals regulate receptor output via the same conformational mechanism.
A fundamental question in the study of transmembrane surface receptors is how ligand binding and covalent modification of a receptor shifts the receptor signaling state between active and inactive states. Two extreme models can be proposed. At one extreme, a given receptor could possess just two stable conformations, such that an individual receptor molecule is a simple two-state, on-off toggle-switch (Monod et al. 1965; Leff 1995). In this model, ligand binding or covalent modification shifts the equilibrium between the two conformations, thereby generating intermediate signaling levels with different fractions of the receptor population in the activated and inactivated states. An important feature of this toggle-switch equilibrium is that the observed functional parameters of the receptor population will be weighted averages of the intrinsic parameters of the two conformational states. As a result, the values of different parameters will covary as the ratio of receptors in the two states is changed, providing a useful identifying feature of toggle-switch behavior. By contrast, in the rheostat model, ligand binding events or posttranslational methylation or phosphorylation of a receptor can induce many more than two stable receptor conformational states, each representing a local energy minimum on the potential energy surface of receptor conformational space. An individual aspartate receptor dimer could function as a continuously variable dimmer switch or rheostat, with as many as 162 stable, intermediate structural states or settings arising from covalent modification of the individual receptor subunits. It has been known for some time that proteins can adopt a multitude of distinct structural substates, thus, it is not unreasonable to propose that different ligand occupancies and covalent modifications could trap the receptor in a range of different stable conformations (Frauenfelder et al. 1988). For example, ligand binding to certain gated ion channels has been observed to create new conformational states rather than simply modulating the equilibrium between existing states, as detected by altered single channel conductance levels (Eghbali et al. 1997; Ruiz and Karpen 1997; Rosenmund et al. 1998).
For the bacterial chemotaxis receptors, a two-state equilibrium model was first proposed by Asakura and Honda 1984 and is widely used in current simulations of chemotaxis pathway function (Asakura and Honda 1984; Ames and Parkinson 1988; Barkai and Leibler 1997; Morton-Firth et al. 1999). Studies of a related bacterial phototaxis receptor by Jung and Spudich 1998 have shown that the effects of suppressor mutations are consistent with a two-state equilibrium. However, no experimental tests have been devised yet to ascertain whether covalent modification of multiple taxis receptor adaptation sites simply modulates an equilibrium between two conformational states as predicted by the toggle-switch model, or rather generates new stable conformational states as predicted by the rheostat model. More generally, no previous study has attempted to resolve this issue in a classic sensory receptor that transmits information across membranes via conformational change.
To resolve the toggle-switch and rheostat models of aspartate receptor conformational states, it is necessary to generate homogeneous populations of receptors with well-defined modifications of their adaptation sites. The different methylation states of the receptor can be mimicked by mutating the adaptation Glu residues to Gln, since amidation yields a functional response similar to that of the methyl ester (Park et al. 1990; Dunten and Koshland 1991; Borkovich et al. 1992). The resulting mutant receptors are expressed in a strain lacking the adaptation enzymes CheR and CheB to ensure that the engineered modification state is preserved throughout a homogeneous receptor population. Using this approach, the present study generates all 16 possible homogeneous adaptation states ranging from EEEE to QQQQ. In these 16 mutants, the two subunits are modified identically, so together they comprise a small, but representative subset of the modification states encountered in the native system. Most importantly, the 16 mutants sample the entire range of native modification states. Each mutant is incorporated into the reconstituted receptor-regulated phosphorylation pathway to measure its attractant affinity and its ability to stimulate CheA kinase activity in vitro. The results reveal that modifications of multiple adaptation sites have synergistic effects on relative attractant affinity and kinase activity. As predicted by the toggle-switch model, extremely tight coupling is observed between the ligand binding site and the kinase active site, yielding a high degree of covariance between the effects of adaptation site modification on attractant affinity and maximal kinase activity. Similarly, certain mutations that superactivate the kinase have a strongly correlated effect on attractant affinity. Together, these results provide strong evidence for the toggle-switch model of aspartate receptor activation, wherein the combined attractant and adaptation signals control the equilibrium between just two stable receptor conformations that activate and inactivate the CheA kinase, respectively. The findings also provide a simple procedure to quantitate the ratio of receptors in the two conformational states.
MATERIALS AND METHODS
Materials
The plasmid pSCF6 used to express the S. typhimurium aspartate receptor under control of its native promoter has been described previously (Chervitz et al. 1995), Membranes containing an overexpressed aspartate receptor were isolated from the E. coli strain RP3808 (Δ(cheA-cheZ)DE2209 tsr-1 leuB6 his-4 eda-50 rpsL136 [thi-1 Δ(gal-attl)DE99 ara-14 lacY1 mtl-1 xyl-5 tonA31 tsx-78]/mks/) expressing the aspartate receptor from plasmid pSCF6 or its engineered variants. Tests of receptor function in vivo were performed in the E. coli strain RP8611 (tsr-7028 Δ(tar-tap)DE5201 zbd::Tn5 Δ(trg)DE100 leuB6 his-4 rpsL136 thi-1 ara-14 lacY1 mtl-1 xyl-5 tonA31 tsx-78) containing the plasmid pSCF6 or its variants. Both of these strains of E. coli were provided by Dr. John S. Parkinson (University of Utah, Salt Lake City, UT; Liu and Parkinson 1989). The strains and plasmids expressing the S. typhimurium histidine kinase CheA (HB101/pMO4) and the S. typhimurium coupling protein CheW (HB101/pME5) were provided by Dr. Jeff Stock (Princeton University, Princeton, NJ; Stock et al. 1987, Stock et al. 1988). The strain and plasmid (RBB455/pRBB40) for expression of E. coli CheY was provided by Drs. Bob Bourret and Mel Simon (University of North Carolina, Chapel Hill, NC, and California Institute of Technology, Pasadena, CA, respectively; Bourret et al. 1993). Mutagenic oligos were synthesized by GIBCO BRL. Kunkel mutagenesis reagents (unmodified T7 DNA polymerase and T4 DNA ligase) as well as restriction enzymes were purchased from New England Biolabs. The helper phage M13K07 used to generate single stranded phagemid was obtained from BioRad. The reagents l-aspartate and α-methyl-d,l-aspartate (>99% purity and containing no detectable aspartic acid as assayed by TLC) was purchased from Sigma-Aldrich. The enzyme substrate γ-[32P] ATP (6,000 Ci/mmol) was obtained from NEN Lifesciences.
Protein Engineering
Receptors with modified adaptation sites were generated by oligonucleotide directed site specific mutagenesis of the plasmid pSCF6 according to the method of Kunkel et al. with modifications as previously described (Kunkel et al. 1991; Chervitz et al. 1995). Briefly, plasmid pSCF6 variants encoding the aspartate receptor in the EEEE, QEEE, QEQQ, and QQQQ modification state, as well the wild-type (QEQE) pSCF6 plasmid were used to generate single-stranded phagemid template according to the method of Kunkel et al. 1991. Oligonucleotide primers were then used to simultaneously modify the first three adaptation sites using either the QEQE or QEQQ modification state single-stranded phagemid template to produce DNA encoding all 16 possible combinations of glutamate or glutamine at the four adaptation sites. Plasmid DNA encoding additional mutants containing single cysteine residues in different modification state backgrounds was obtained by Kunkel mutagenesis using single stranded phagemid template encoding the appropriate receptor modification state. Screening of candidate mutagenic plasmids was aided by the inclusion of silent restriction sites incorporated into the mutagenic oligonucleotide primers. All mutations were confirmed by dideoxy plasmid DNA sequencing using Sequitherm DNA polymerase (Epicenter) or by automated fluorescent cycle sequencing using an ABI 377 sequencer (Perkin Elmer).
In Vivo Activity Assays
Chemotaxis swarm assays were performed as described previously (Danielson et al. 1997). Each mutant pSCF6 plasmid was transformed into E. coli RP8611 that is deleted for wild-type aspartate receptor. Vector alone (pBluescript) and vector carrying the wild-type receptor (pSCF6) were used as controls to determine the swarm rates of receptorless cells and cells possessing the native receptor. Swarm rates were assayed on minimal media semisolid agar plates with or without 100 μM l-aspartate. Plates were incubated at 30°C, and colony diameters were measured at 3–4 h intervals ∼24 h after spotting. Swarm rates were determined by least-squares linear best-fit to the slope of diameter as a function of time. The aspartate specific swarm rates were determined by subtracting the (−) aspartate swarm rate from the (+) aspartate swarm rate to correct for pseudotaxis, and any other nonaspartate-specific taxis. The resulting rate was normalized to the wild-type rate for comparison. Cells containing vector alone swarmed at an ∼10-fold lower rate than cells containing wild-type receptor and exhibited no aspartate specific swarming.
Purification of the Engineered Receptors and Cytoplasmic Chemotaxis Components
Plasmids encoding engineered receptors were transformed into the E. coli strain RP3808 that lacks functional major chemotaxis receptors and cytoplasmic chemotaxis components. Subsequent expression and preparation of the engineered receptors in native E. coli membranes was performed as previously described (Bass and Falke 1998). Aspartate receptors constituted ∼10% of total protein in the native membrane preparations. The soluble chemotaxis components CheA, CheW, and CheY were overexpressed and purified in E. coli as described previously (Danielson et al. 1997). The concentration and purity of the engineered receptors in isolated membranes as well as soluble chemotaxis proteins was determined by means of BCA assay and quantitation of protein band intensities on SDS-PAGE gels as described previously (Bornhorst and Falke 2000).
In Vitro Activity Assays
The in vitro receptor–coupled kinase assay was performed essentially as described previously (Borkovich et al. 1989; Ninfa et al. 1991; Bornhorst and Falke 2000). Briefly, a reaction mixture consisting of 2 μm of receptor, 2 μm CheW, 0.5 μm CheA, 10 μm CheY and attractant (if included) was incubated at ambient temperature for 45 min to allow formation of the ternary signaling complex. Incubation of the reaction mixture for 30 min was sufficient to yield maximal measured receptor-stimulated kinase activity for the EEEE, QEQE (WT), and QQQQ receptor adaptation states, revealing that the 45-min incubation time was sufficient for complete formation of the ternary signaling complex for each receptor adaptation state. The receptor-coupled kinase reaction was initiated by the addition of γ-[32P]ATP to a the reaction mixture and quenched after a 10-s incubation period. Receptor-regulated CheA autophosphorylation is the rate determining step in this reaction so that the rate of phosphorylated CheY production is linearly proportional to the receptor-regulated CheA autophosphorylation rate (see Fig. 4 B and discussion; Chervitz et al. 1995; Bornhorst and Falke 2000). The [32P]phospho-CheY was resolved on a 15% Laemmli SDS–polyacrylamide gel and quantitated after gel drying by PhosphorImager analysis (Molecular Dynamics, Inc.) The resulting [32P]phospho-CheY formation rates for the engineered receptors were normalized to that of the wild-type receptor that was isolated and measured in parallel, yielding a receptor-regulated CheA kinase activity relative to that of the wild-type receptor in the absence of attractant. After the in vitro kinase reaction each sample was reanalyzed by SDS-PAGE to ensure that equimolar amounts of different mutant receptors were used in the assay.
Figure 4.
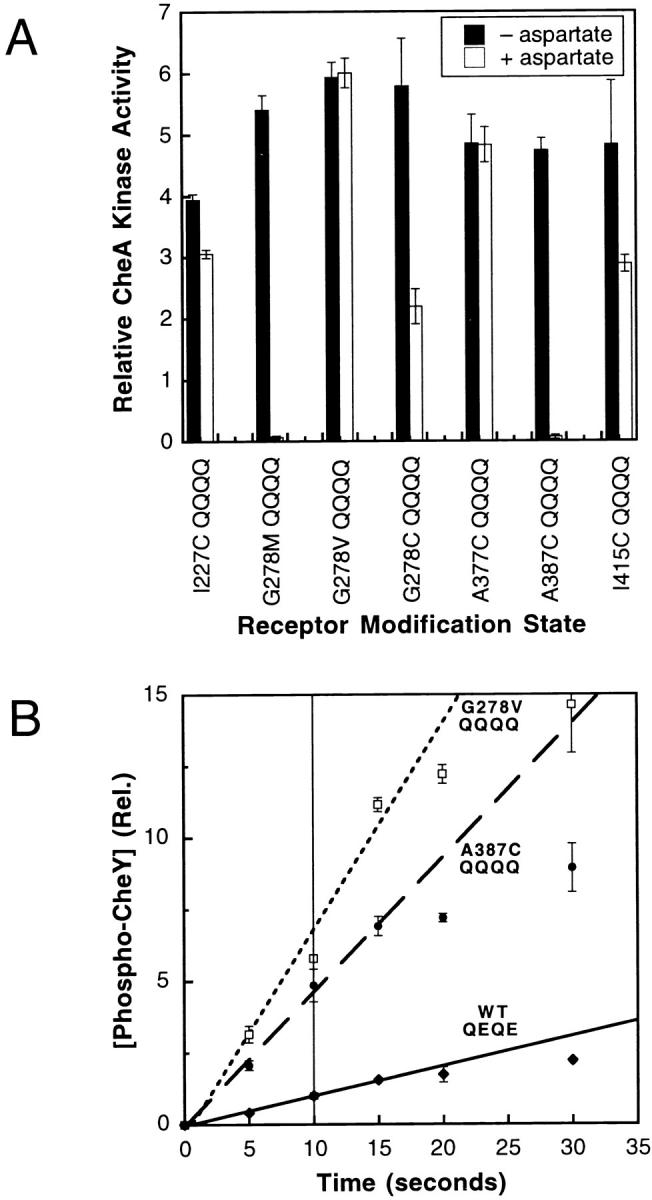
(A) CheA kinase activities stimulated by superactivating receptor mutations in a QQQQ modification background. Seven receptor mutations, selected for their ability to stimulate a high degree of CheA kinase activity in the QEQE modification background, were each incorporated into the QQQQ background. The CheA kinase activity stimulated by each mutant receptor was subsequently measured in the in vitro receptor–coupled kinase assay and normalized to the kinase activity generated by the wild-type receptor (QEQE). Filled bars indicate activity in the absence of attractant and open bars indicate the activity in the presence of 1 mM l-aspartate. The average kinase activity generated by the seven superactivating mutants in the absence of attractant is 5.5 ± 0.7-fold greater than that stimulated by the wild-type receptor (QEQE). (B) Time course of phospho-CheY production stimulated by superactivating and wild-type receptors in the reconstituted receptor–kinase complex. The level of radiolabeled phospho-CheY generated in the standard in vitro receptor–coupled kinase assay was monitored over time for reconstituted complexes containing the either the wild-type receptor (QEQE) or one of the two most superactivating receptors (A387C QQQQ or G278V QQQQ). Shown are the phospho-CheY levels measured at specific time points, as well as the best-fit initial rate lines defined by the 0–15 s time points. Even for the two most superactivating mutants, the initial reactions were linear to at least 15 s. Since the standard in vitro receptor–coupled kinase assay utilizes 10-s time points to define CheA kinase activities, it follows that all kinase activities measured in the present study are valid initial rates measured within the linear range of the kinase reaction.
The Attractant Dependence of Receptor-coupled Kinase Activity
Quantification of the K1/2 and Hill coefficient associated with attractant regulation of receptor-coupled kinase activity was performed using the in vitro receptor–coupled kinase assay (Bornhorst and Falke 2000). For these measurements, the attractant α-methyl-aspartate was used. This attractant was originally used to quantitate the sensitivity of the chemotaxis pathway at low attractant concentrations (Segall et al. 1986). An advantage of α-methyl-aspartate is that its aspartate receptor binding affinity (K d ∼10–100 μM; see below) is significantly less than that of aspartate (K d ∼1 μM; Biemann and Koshland 1994). As a result, it is more straightforward to determine the concentration of free attractant for α-methyl-aspartate than for aspartate when titrations are performed using receptor concentrations in the low micromolar range. Relative receptor-coupled CheA activities were determined over a range of α-methyl-aspartate concentrations for each engineered receptor. A cooperative, multi-site Hill equation () was used to fit the averaged data obtained for each attractant concentration, yielding a best-fit titration curve:
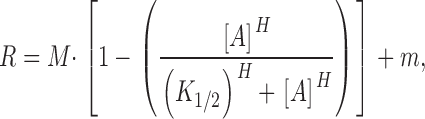 |
1 |
where R is the normalized receptor-regulated CheA kinase rate, M is the maximal rate, m is the background CheA kinase rate at saturating attractant concentration, [A] is the free attractant concentration, K1/2 is the attractant concentration at which the receptor-coupled CheA kinase activity is one-half maximal, and H is the Hill coefficient.
The best fit titration curve to the Hill equation was used to determine both the Hill coefficient and the K1/2 for the attractant response of each engineered receptor–kinase complex. Titration curves were plotted and fit using KaleidaGraph 3.0 software for Macintosh (Synergy Software).
Determination of Correlation Coefficients
All indicated linear correlation coefficients represent the Pearson product moment correlation coefficient, r, as determined using Excel 2001 (Microsoft).
Error Determination
All indicated error bars represent the SD for n ≥ 3.
RESULTS
Generation and Expression of All Possible Modification States
A set of aspartate receptor mutants representing all 16 homogeneous modification states of the receptor was created by generating all possible combinations of glutamate or glutamine at the four receptor adaptation sites. The glutamine residues at the adaptation sites closely mimic the regulatory effects of the native methyl esterification of glutamate as catalyzed by CheR (Park et al. 1990; Dunten and Koshland 1991; Borkovich et al. 1992). Each mutant receptor was expressed in the E. coli strain RP3808 that lacks major chemotaxis receptors and the soluble components of the chemotaxis pathway. The absence of the adaptation enzymes CheR and CheB ensured that no posttranslational modification of the receptor adaptation sites occurred, such that the receptor population was homogeneous, and that the modification states of the two homodimer subunits were identical. In a native cell, independent modification of the two subunits yields many more than 16 receptor modification states (introduction), but the 16 selected mutants provide adequate sampling of the range of native states including the two extremes (EEEE and QQQQ) and representative intermediate states. Preparations of isolated E. coli membranes from cells expressing different mutants yielded roughly equivalent amounts of receptor, indicating that receptor expression and stability are not significantly altered by modification of the adaptation sites.
Effect of Modification State on Receptor Function In Vivo
As a positive control, receptors expressed from plasmids encoding all 16 possible modification states were assayed for the ability to stimulate in vivo swarming activity in response to a self-generated radial aspartate gradient in semisolid agar plates. The E. coli RP8611 strain used in this assay lacks functional major chemotactic receptors, but contains functional soluble components of the chemotactic signaling pathway including the methyl esterase enzyme CheB, which hydrolyzes either glutamine or methyl-esterified glutamate at an adaptation site to yield the free glutamate side chain, and CheR which methyl-esterifies this free glutamate side chain. When plasmids expressing different combinations of glutamines and glutamates at the adaptation sites were expressed in RP8611, all of the modified receptors yielded chemotactic swarm rates similar to the wild-type receptor as shown in Fig. 2 A. It has been shown previously that swarm rates are significantly slowed by mutations preventing the normal methylation and demethylation of any adaptation site (Shapiro et al. 1995). Thus, the observed wild-type swarm rates (Fig. 2 A) suggest that each of the engineered adaptation site glutamines can be efficiently deamidated by CheB, and then reversibly methylated and demethylated by CheR and CheB in vivo, respectively, yielding final receptor populations with similar or identical statistical distributions of modification states regardless of the starting Q and E configuration. Furthermore, these results demonstrate that no unintended changes were introduced into the receptor during mutagenesis.
Figure 2.
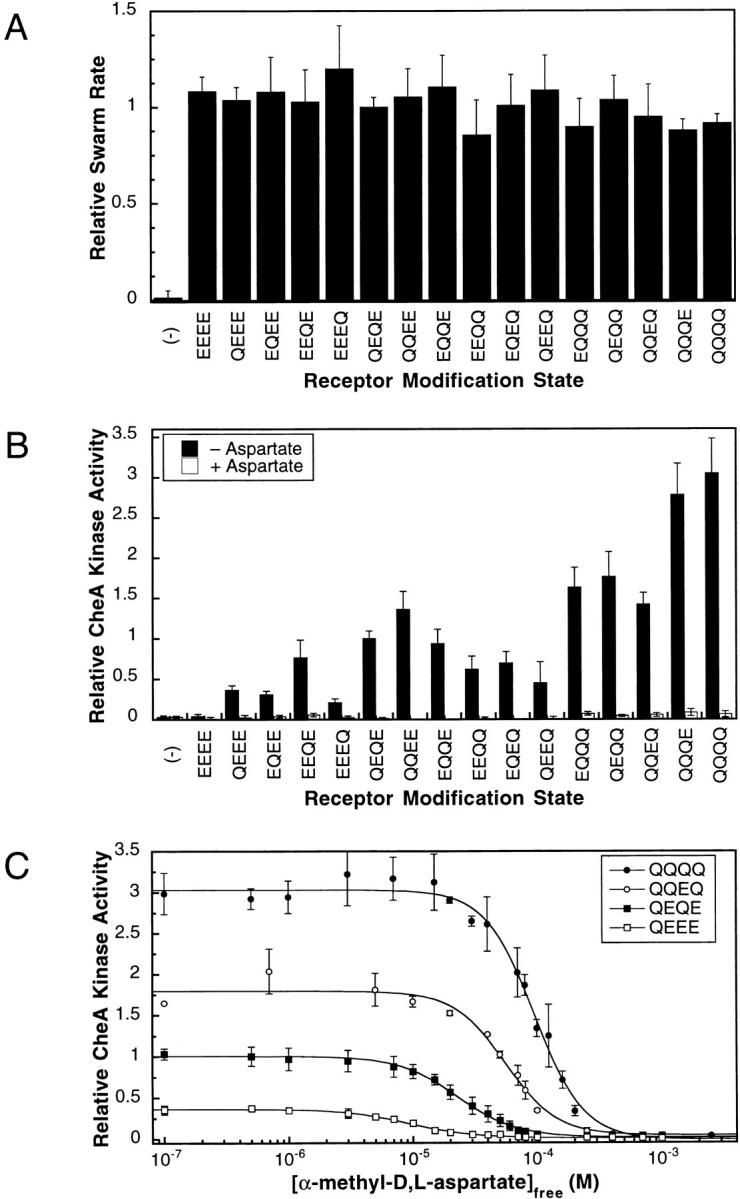
(A) In vivo activity of the engineered receptors. Engineered receptors representing all possible homogeneous modification states of the adaptation sites were expressed in an E. coli strain lacking major chemoreceptors but containing other pathway components, including the adaptation enzymes CheB and CheR. The ability of each engineered receptor to restore cellular taxis up a self-generated aspartate gradient in soft agar was measured by the chemotactic swarm assay (materials and methods). All swarm rates have been normalized to the swarm rate of the wild-type receptor (QEQE). (B) In vitro activity of the engineered receptors. Receptor-coupled CheA kinase activity was measured for each modified receptor in the reconstituted phosphorylation pathway. Native E. coli membranes containing a given receptor were mixed with purified CheA, CheW, and CheY to reconstitute the ternary signaling complex in the presence of excess response regulator CheY. Radiolabeled ATP was added to initiate the phosphotransfer cascade resulting in phosphorylation of CheY. After quenching of the reaction after 10 s, the amount of phospho-CheY was determined and used to quantitate the initial rate of receptor-coupled CheA autophosphorylation. The resulting initial rate was normalized to that of the reconstituted wild-type receptor (QEQE) complex, providing the relative CheA kinase activity. Filled bars indicate activity in the absence of attractant and open bars indicate the activity in the presence of 1 mM l-aspartate. The data shown represents the average activity determined by three independent measurements of two different membrane preparations and is summarized in Table . (C) The effects of increasing attractant concentration on the receptor-coupled kinase activities of the QEEE, QEQE (wild type), QQEQ, and QQQQ receptor modification states. For each engineered receptor the relative CheA kinase activity was measured at different α-methyl-aspartate concentrations. The resulting activities were fit with a multisite Hill model, which provided K1/2 and Hill coefficient values summarized in Table . Each point is the average of three independent measurements.
Effect of Receptor Modification State on Kinase Activation In Vitro
To compare the receptor-coupled CheA kinase activities associated with different receptor modification states, the in vitro receptor–coupled kinase assay was used to measure the kinase activity of the reconstituted signaling complex in the absence of attractant. Since the apo receptor stimulates CheA autophosphorylation whereas the attractant-occupied receptor inhibits CheA, kinase activity measurements performed in the absence of attractant provided quantitation of the maximal kinase activity generated by each receptor modification state. To perform the receptor-coupled kinase assay, engineered receptors in native E. coli membranes were added to purified CheA, CheW, and CheY proteins to reconstitute the active receptor–kinase signaling complex. Upon addition of radiolabeled ATP, the formation of phospho-CheY was followed as a measure of receptor-coupled kinase activity. Excess CheY was used to ensure that the rate-determining step in the overall reaction was the autophosphorylation of CheA, which is the step regulated by the receptor. Table and Fig. 2 B summarize the relative levels of receptor-coupled CheA kinase activity for all 16 possible modification states, measured in the absence of attractant. As has been demonstrated previously by the well-characterized EEEE, QEQE, and QQQQ modification states and their methyl-esterified analogues, increasing modification of the adaptation sites by either amidation or methylation stimulates the autophosphorylation activity of receptor-bound CheA (Ninfa et al. 1991; Borkovich et al. 1992; Bornhorst and Falke 2000). However, this study is the first to examine the effects of all 16 homogeneous receptor modification states on CheA autophosphorylation, which allows for direct quantitation of the relationship between receptor modification and kinase regulation.
Table 1.
Effects of Receptor Adaptation Site Mutations on the Attractant Dependence of Receptor-coupled Kinase Activity
| Adaptation site residues | Measured K1/2 | Measured Hill coefficient | Relative in vitro maximal kinase activity | Estimated fraction of receptor in activated state |
|---|---|---|---|---|
| mM | ||||
| (No receptor) | (–) | (–) | 0.03 ± 0.02 | (–) |
| EEEE | nd | nd | 0.04 ± 0.02 | 0.008 |
| QEEE | 9 ± 1 | 1.7 ± 0.1 | 0.36 ± 0.06 | 0.07 |
| EQEE | 16 ± 4 | 1.6 ± 0.4 | 0.31 ± 0.04 | 0.06 |
| EEQE | 23 ± 2 | 1.8 ± 0.1 | 0.8 ± 0.2 | 0.1 |
| EEEQ | 9 ± 3 | 1.1 ± 0.4 | 0.21 ± 0.05 | 0.04 |
| QEQE | 23 ± 4 | 1.8 ± 0.1 | 1.0 ± 0.1 | 0.2 |
| QQEE | 31 ± 2 | 2.2 ± 0.2 | 1.4 ± 0.2 | 0.3 |
| EQQE | 14 ± 2 | 2.1 ± 0.3 | 0.9 ± 0.2 | 0.2 |
| QEEQ | 19 ± 2 | 2.4 ± 0.5 | 0.5 ± 0.3 | 0.08 |
| EQEQ | 14 ± 1 | 2.9 ± 0.3 | 0.7 ± 0.1 | 0.1 |
| EEQQ | 19 ± 2 | 1.9 ± 0.2 | 0.6 ± 0.2 | 0.1 |
| QQQE | 98 ± 4 | 3.3 ± 0.4 | 2.8 ± 0.4 | 0.5 |
| QQEQ | 54 ± 6 | 2.0 ± 0.5 | 1.4 ± 0.1 | 0.3 |
| QEQQ | 62 ± 3 | 2.2 ± 0.3 | 1.8 ± 0.3 | 0.3 |
| EQQQ | 39 ± 4 | 2.0 ± 0.3 | 1.6 ± 0.2 | 0.3 |
| QQQQ | 97 ± 4 | 2.2 ± 0.2 | 3.1 ± 0.4 | 0.6 |
Hill coefficients and K1/2 values were determined by titration of the attractant α-methyl-aspartate into the receptor-coupled kinase assay as in Fig. 2 C. A negative control (no receptor) used membranes lacking the aspartate receptor when reconstituting the signaling complex. The kinase activity of the signaling complex containing the EEEE receptor was too low to measure K1/2 and Hill coefficient values. The maximal kinase activity for each signaling complex was determined by the in vitro kinase assay in the absence of attractant as in Fig. 2 B. The fraction of receptor in the activated state was calculated from the maximal kinase activity as described in discussion and in Fig. 6 B.
The different adaptation sites are observed to have different propensities to regulate kinase activity (Table and Fig. 2 B). When a single Q is placed in the EEEE background or a single E is placed in the QQQQ background, modification of the fourth site has the smallest effect on kinase activity, whereas modification of the third site has the largest effect on kinase activity. However, the effects of multiple modifications are not always additive. In general, when multiple Q residues are introduced into the EEEE background, the observed kinase activities are greater than the sum of the single Q modifications. For example, if the degree of kinase activation resulting from a single Q modification at each individual adaptation site were additive, the total QQQQ receptor-mediated activity would be 1.6 ± 0.3 times that of QEQE (wild type) receptor-mediated kinase activity. In contrast, the experimentally observed kinase activity for the QQQQ modification state is 3.1 ± 0.4 times that of the QEQE (wild type) receptor. Thus, there are significant synergistic effects between adaptation sites in the regulation of kinase activity. Yet, the data support the general conclusion that in receptors with multiple modifications, the degree of kinase activation depends primarily on the number of modifications at the first three adaptation sites on signaling helix CD1, whereas the effects of modification at the fourth adaptation site on helix CD2 are relatively minor.
Effect of Receptor Modification State on Attractant Affinity In Vitro
To quantitate the relative attractant affinities of modified receptors, the in vitro receptor–coupled kinase assay was used to measure the inhibitory effect of increasing attractant concentrations on receptor-coupled kinase activity as illustrated in Fig. 2 C. The attractant α-methyl aspartate was chosen for titrations because it has been used in cellular behavioral studies and possesses a receptor affinity favorable for quantitation (materials and methods; Segall et al. 1986). As expected, increasing attractant concentration decreased the CheA kinase activity, and the concentration profile of this attractant regulation exhibited limited positive cooperativity as previously observed (Bornhorst and Falke 2000). The attractant dependence of kinase activity was fitted with the Hill model (materials and methods), allowing for accurate quantitation of the attractant concentration yielding half-maximal kinase activity (K1/2) and the Hill coefficient (H) for each receptor modification state. Since the attractant binding equilibrium at the receptor ligand binding site is fast compared with the subsequent catalytic events during CheA autophosphorylation, the K1/2 parameter is a close approximation to the equilibrium dissociation constant (K d) for attractant binding to the active receptor–kinase signaling complex. Experiments have confirmed that the rate-determining step in the in vitro receptor–coupled kinase assay is CheA autophosphorylation, which is slow compared with the aspartate binding equilibrium and to phosphotransfer from CheA to CheY (Danielson et al. 1994; Stewart 1997). Notably, the observed attractant dependence of kinase activity also served to confirm the restoration of functional coupling between the receptor and CheA in the reconstituted signaling complex.
The best-fit α-methyl aspartate K1/2 and Hill coefficient values obtained for the 16 receptor modification states are summarized in Table . No values were determined for the EEEE receptor, which had too little receptor-mediated CheA kinase activity to be accurately quantitated. The observed K1/2 values increased ∼10-fold as the modification level increased from a single glutamine (QEEE, EQEE, EEQE, and EEEQ) to four glutamines (QQQQ). The 10-fold effect of adaptation state on the attractant K1/2 parameter is in agreement with direct in vitro binding measurements of attractant K d values and with in vivo measurements of attractant K1/2 values for stimulation of chemotactic behavior (Dunten and Koshland 1991; Borkovich et al. 1992; Kim et al. 2001). Together, these results further support the close correspondence between the attractant K1/2 value measured in the in vitro receptor–coupled kinase assay and the equilibrium K d for attractant binding to the receptor dimer. The attractant dependence of receptor-coupled kinase activity also provided Hill coefficients ranging from ∼1.5 to ∼3.0 (Table ), which is consistent with limited positive cooperativity between receptor signaling units believed to be individual receptor dimers (Bornhorst and Falke 2000). Overall, there was a slight general increase in measured Hill coefficients as receptor modification level increased from EEEE to QQQQ. Previous studies of selected modification states, although not adequate to reveal the quantitative pattern of modification effects on attractant affinity, provided results qualitatively consistent with the findings summarized in Table (Borkovich et al. 1992; Bornhorst and Falke 2000).
Correlation Between Effects of Receptor Modification on Kinase Activation and Attractant Affinity
When the effects of adaptation site modification on kinase activation and attractant affinity (both measured in the in vitro receptor–coupled kinase assay as described above and summarized in Table ) were compared, a strong correlation emerged. Fig. 3 A summarizes the maximal kinase activities and attractant K1/2 values observed for the 16 modification states excepting EEEE (see above). Fig. 3 B tests the correlation by plotting the K1/2 value for each modified receptor against its maximal kinase activity, revealing a linear relationship with a correlation coefficient of 0.97. Thus, a highly modified receptor such as QQQQ exhibits a high K1/2 value (low attractant affinity) and high kinase activity, whereas, at the other extreme, the QEEE, EQEE, EEQE and EEEQ receptors exhibit low K1/2 values (high attractant affinity) and low kinase activity. This is the behavior predicted for a toggle-switch model in which modification of the adaptation sites shifts an equilibrium between a low attractant affinity, high kinase activity “on” state and a high attractant affinity, low kinase activity “off” state (see discussion).
Figure 3.
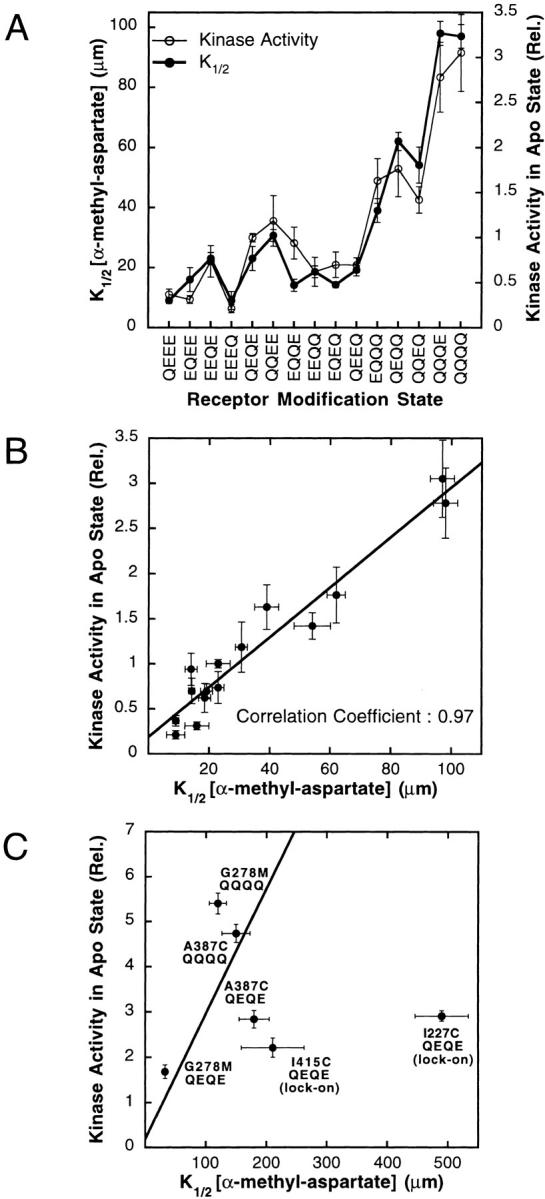
(A) Comparison of the α-methyl-aspartate K1/2 values and receptor-coupled CheA kinase activities measured for different receptor modification states. Shown are the K1/2 value (closed circle) and relative kinase activity (open circle) for each modification state, both measured using the standard in vitro receptor–coupled kinase assay described in the legend to Fig. 2B and Fig. C. (B) Correlation between the receptor-coupled CheA kinase activities and α-methyl-aspartate K1/2 values measured for different receptor modification states. Shown are the data of Fig. 3 A and the best-fit straight line, as well as the Pearson product moment correlation coefficient. (C) Effects of specific mutations on the correlation. Plotted is the kinase activity against the α-methyl-aspartate K1/2 for the indicated mutants, as well as the same correlation line defined by different receptor modification states in Fig. 3 B. The parameters of partial lock-on mutants I415C QEQE and I227C QEQE fall far from the line. Given their measured kinase activities, the attractant affinities of these lock-on mutants are considerably lower than predicted by the correlation of Fig. 3 B.
Some mutations outside of the adaptation sites yield K1/2 values and relative kinase activities that fall near the same correlation line exhibited by the adaptation site modifications (Fig. 3 B), as illustrated in Fig. 3 C for mutations G278M QEQE, G278M QQQQ, A387C QEQE, and A387C QQQQ. By contrast, certain other mutations yield points further from the correlation line as illustrated by I415C QEQE and I227C QEQE. Both behaviors are predicted by the toggle-switch model: some mutations shift the equilibrium between the two fundamental states without altering the characteristics of either state, yielding points falling on the standard correlation line. The remaining mutations significantly alter the characteristics of one or both fundamental states or add additional states, and thus yield points that differ from the standard correlation (see discussion).
Estimating the Upper Limit of Receptor-coupled Kinase Activation
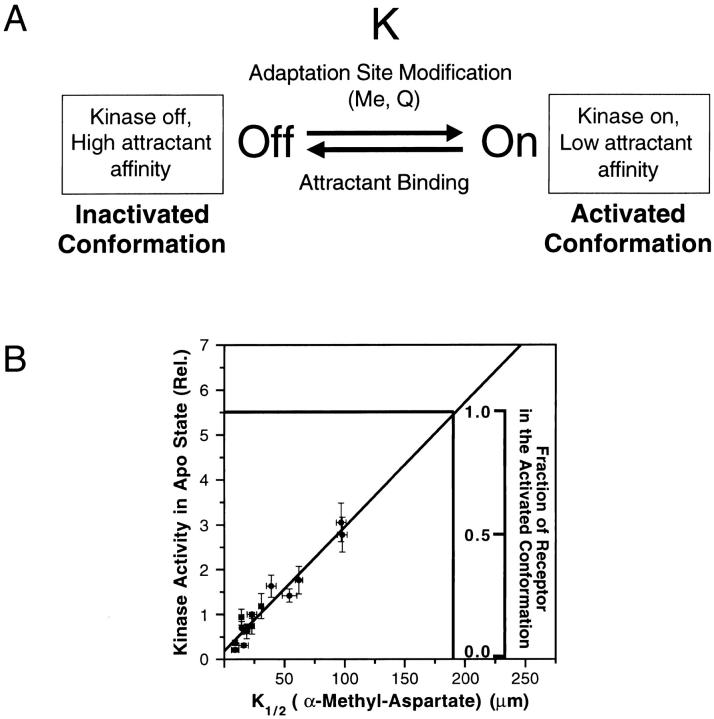
To determine the upper limit of receptor-coupled kinase activation in the absence of attractant, seven mutations previously observed to increase the kinase activity of the QEQE receptor above wild-type levels were moved into the QQQQ receptor background (Bass and Falke 1998; Butler and Falke 1998; Bass et al. 1999; Trammell and Falke 1999). All seven superactivating mutations are located at buried positions in the core of the 4-helix bundle that forms the cytoplasmic domain of the receptor dimer. The effects of these superactivating mutations on kinase activation were quantitatively compared in the in vitro receptor–coupled kinase assay, where they caused variable activity enhancements in the QEQE background. However, in the QQQQ background, all seven yielded super-normal kinase enhancements with similar maximal rates, ∼5.5-fold that stimulated by the WT (QEQE) receptor as shown in Fig. 4 A. Time courses of the kinase reaction revealed that these maximal rates were measured well within the linear, initial-rate range of the assay, illustrated in Fig. 4 B for mutants G278V QQQQ and A387C QQQQ, indicating that the super-normal rate was not being limited by the assay conditions, even when the receptor mutations that had the largest enhancing effects on receptor-mediated CheA kinase activity were tested. Instead, these superactivating mutations appear to fully drive the receptor into the functional state that yields the maximal turnover rate of the bound CheA kinase. In the toggle-switch model, such behavior is observed when the entire receptor population is driven into the kinase-activating conformation. For a toggle-switch system knowledge of this maximal rate is quite significant, since it can be used to calculate the fraction of the receptor population in each of the two fundamental states for any receptor described by the correlation in Fig. 6 B (discussion).
Figure 6.
(A) Schematic toggle-switch model for aspartate receptor signaling, illustrating the two-state equilibrium proposed by the model. Shown are the two conformational states of the receptor with their different attractant binding and kinase activating propensities. The inactivated receptor conformation has high attractant affinity and low receptor-coupled kinase activity, whereas the activated receptor conformation exhibits low attractant affinity and high kinase activity. Modification of the adaptation sites by either methylation or amidation shifts the equilibrium toward the activated conformation, whereas attractant binding shifts the equilibrium toward the inactivated conformation. (B) Method to estimate the fractional populations of the two conformational states. Shown are the receptor-coupled CheA kinase activities of the standard receptor modification states plotted against their α-methyl-aspartate K1/2 values (closed circles), as well as the best-fit straight line defined by these points (diagonal line). These data are the same as those of Fig. 3 B. Added to the figure is a scale used to quantitate the fraction of the receptor population in the activated conformation. This scale, which can be applied to any receptor population described by the standard correlation of Fig. 3 B, is used to interpolate the fraction activated receptor from the relative CheA kinase activity measured for a given receptor population in the in vitro receptor–coupled kinase assay. Note that to carry out this interpolation, the relative CheA kinase activity is the only parameter that need be measured. The method assumes that the inactivated state exhibits an α-methyl-aspartate K1/2 in the micromolar to submicromolar range and little or no activation of CheA autophosphorylation, whereas the activated state exhibits an α-methyl-aspartate K1/2 of 180 μM and a relative CheA kinase activity of 5.5 (see discussion).
Identification of a Mutant Locked in the Kinase-activating State
The present study revealed one mutant receptor, G278V, whose characteristics indicate it is strongly locked in the kinase-activating conformation in a range of modification state backgrounds. The G278 residue is located at a buried regulatory hotspot in the cytoplasmic 4-helix bundle where helix packing is tightly linked to receptor activation (Trammell and Falke 1999). The G278V receptor yields the same maximal kinase activity observed for other superactivating mutants, as noted in Fig. 5 A. However, Fig. 5 A shows that the maximal kinase activity conferred by G278V is insensitive to the modification state of the adaptation sites and is not significantly lowered by very high attractant concentrations with the exception of the EEEE modification state, wherein the mutation allows a partial attractant response. Certain other mutations located at buried cytoplasmic positions (including I227C, S356C, and I415C) were observed previously to partially block normal attractant-triggered inhibition of kinase activity in the QEQE background, but these partial lock-on mutants exhibit considerably more attractant and adaptation regulation of kinase activity than G278V, as illustrated in Fig. 5 B (Bass and Falke 1998; Butler and Falke 1998; Bass et al. 1999). Thus, the G278V mutant is the best example to date of a lock-on mutation that traps the kinase-activating state of the receptor in a manner insensitive to both attractant and adaptation signals. Such results are predicted by the toggle-switch model, wherein it should be possible to generate a lock-on mutant (such as G278V) that strongly drives the equilibrium toward the fundamental “on” or kinase-activating state so that the attractant and adaptation signals lack sufficient free energy to push the equilibrium out of this state (see discussion). The toggle-switch model also explains the greater attractant regulation observed as the modification states of lock-on mutants change from QQQQ to EEEE (Fig. 5A and Fig. B), since the removal of modifications at the adaptation sites drives the toggle-switch equilibrium toward the kinase-inactivating state and thereby reinforces the attractant driving force.
Figure 5.
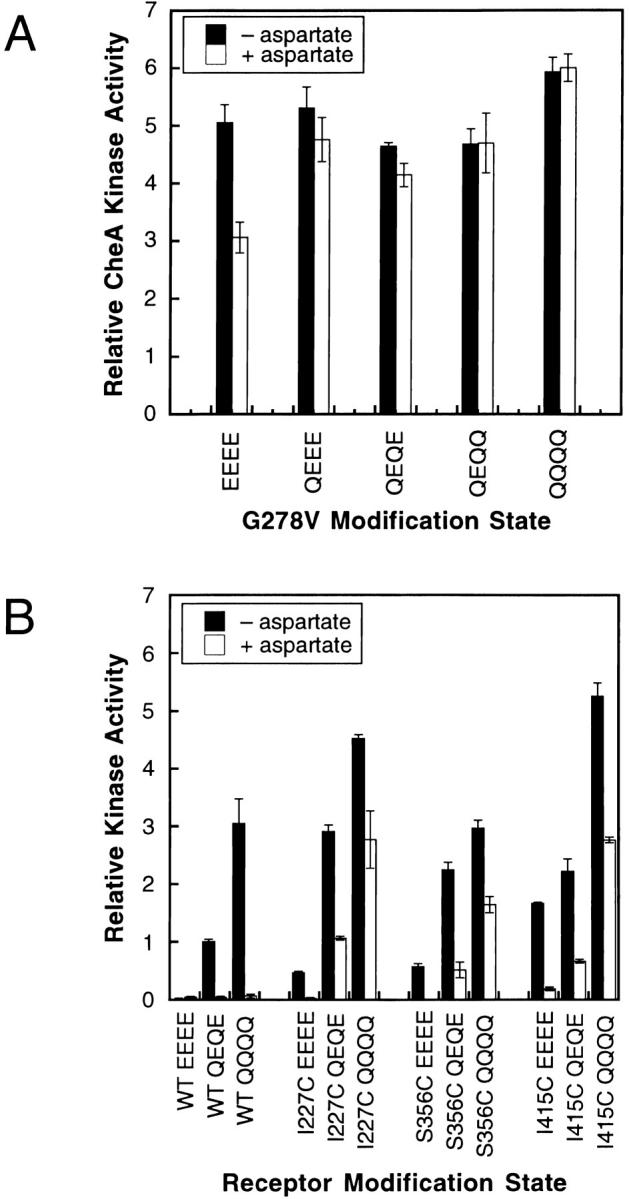
(A) CheA kinase activities stimulated by the lock-on G278V mutation in different modification backgrounds. Shown are receptor-stimulated CheA kinase activities measured in the standard in vitro receptor–coupled kinase assay for the indicated receptor mutants. The G278V mutation is observed to superactivate the kinase and to block the normal attractant and adaptation signals. The normal inhibitory effects of both saturating attractant (1 mM l-aspartate) and glutamate residues at the adaptation sites are largely abolished, except when G287V is moved into the EEEE background where partial attractant sensitivity is restored (see discussion). (B) CheA kinase activities stimulated by partial lock-on receptors in different modification backgrounds. Three representative receptor mutations (I227C, S356C, and I415C) previously found to have partial lock-on character in the QEQE modification background were moved into the EEEE and QQQQ modification states and, using the standard in vitro receptor–coupled kinase assay, tested for their ability to stimulate CheA kinase activity in the presence and absence of 1 mM l-aspartate. All three partial-lock mutations partially block the normal attractant and adaptation signals. However, all three mutations still allow partial inhibition of kinase activity by saturating attractant (1 mM l-aspartate) and by the incorporation of glutamate residues at the adaptation sites. Thus, the lock-on character of these three mutations is weaker than that of the G278V mutation (compare with Fig. 5 A).
DISCUSSION
The present study used an in vitro receptor–coupled kinase assay in which native E. coli membranes containing native or engineered aspartate receptors were reconstituted with purified CheW (coupling protein), CheA (His kinase), and CheY (response regulator). The resulting receptor–kinase signaling complex was used to examine the attractant affinities and kinase stimulating propensities of all possible homogeneous covalent modification states of the receptor adaptation sites. Systematic mutagenesis of the adaptation site glutamates to glutamines, which mimics the native methyl esterification of these regulatory side chains, was found to alter both the attractant K1/2 and receptor-coupled kinase activity. Comparison of the four adaptation sites revealed that modification of the third site usually triggered the largest changes in attractant K1/2 and kinase activity, whereas modification of the fourth site yielded the smallest changes. Notably, previous in vivo studies have shown that the third site is methylated most rapidly and extensively under cellular conditions, whereas the fourth site is methylated most slowly and the least extensively (Terwilliger et al. 1986). Additionally, conversion of the third or fourth adaptation site from glutamate to aspartate, thereby preventing its further modification, yields the largest or smallest effect on in vivo swarming activity, respectively (Shapiro et al. 1995). Taken together, these results present a consistent pattern wherein the third adaptation site is generally most important and the fourth site is least important for receptor-mediated kinase regulation.
Although the adaptation sites exhibited consistent differences between their regulatory potencies and modifications at multiple adaptation sites were largely additive, analysis of all 16 homogeneous modification states revealed subtle deviations from additivity. For example, modification of all four sites from glutamate to glutamine yielded a twofold greater increase in kinase stimulation than predicted by adding the effects of individual site modifications (Fig. 2 B). Thus, synergistic interactions occur when multiple sites are modified. Synergistic interactions are presumably facilitated by the close proximity between the first, second, and third adaptation sites at positions 295, 302, and 309 on the surface of cytoplasmic helix CD1, where they are separated by consecutive heptad repeats of the coiled-coil, whereas the fourth site at position 491 on the surface of cytoplasmic helix CD2 lies near the other three due to the hairpin arrangement of the CD1 and CD2 helices (Bass and Falke 1999; Kim et al. 1999).
An important mechanistic question is whether the different covalent modification states of the receptor adaptation sites yield multiple, stable receptor conformations that each exhibits independent functional parameters. Alternatively, covalent modification could simply modulate an equilibrium between two fundamental conformations. To resolve these possibilities, the present study has asked whether a correlation exists between the effects of modification state changes on two key receptor parameters: attractant affinity and the level of kinase activation, both of which were measured using the in vitro receptor–coupled kinase activity assay. When the modification state was changed from the singly modified state (QEEE, EQEE, EEQE, and EEEQ) to the fully modified state (QQQQ), the measured K1/2 for the attractant α-methyl aspartate increased ∼10-fold (Table ), indicating a significant decrease in attractant affinity as previously noted by more limited studies of receptors in isolated native membranes or chemotaxing cells (Dunten and Koshland 1991; Borkovich et al. 1992; Bornhorst and Falke 2000). Similarly, as the modification state was changed from the singly modified state (QEEE, EQEE, EEQE, and EEEQ) to the fully modified state (QQQQ), the CheA autophosphorylation rate of the reconstituted receptor–kinase complex increased ∼10-fold (Table ). A quantitative analysis of the correlation between modification effects on K1/2 and kinase activity yielded a linear correlation coefficient of 0.97 (Fig. 3A and Fig. B), indicating a strong, highly significant linear relationship between the attractant concentration required for half-maximal kinase inhibition and the maximal kinase activity of the receptor–kinase complex. Extrapolation of the correlation to the unmeasurable EEEE state suggests that both attractant affinity and kinase activity vary over 100-fold ranges between the EEEE and QQQQ states.
The observation of strongly correlated effects of adaptation site modifications on two different functional parameters, attractant K1/2 and kinase activation, is consistent with the predictions of the toggle-switch model for receptor regulation but is difficult to reconcile with the rheostat model. The toggle-switch model proposes that attractant binding and adaptation site modification simply shift an equilibrium between two fundamental receptor conformations: one exhibiting high attractant affinity and low kinase activation, the other exhibiting low attractant affinity and high kinase activation as illustrated in Fig. 6 A. Intermediate states of attractant occupancy or covalent modification yield a mixed receptor population containing both types of fundamental conformations. In such a system, the apparent K1/2 and kinase activation parameters of a given receptor modification state are a weighted average of the individual parameters of the two fundamental conformations, and thus these parameters vary in a correlated way as modification state changes shift the proportions of the population in the two fundamental states. Tight linkage between attractant affinity and receptor-mediated kinase activity is a characteristic feature of an equilibrium between two fundamental states and can be simulated (Morton-Firth et al. 1999). By contrast, it is difficult for the rheostat model to explain the covarying attractant binding and kinase activation parameters since, in this model, each covalent modification state can give rise to a stable, unique receptor conformation with attractant binding and kinase activation parameters independent from all other modification states. To explain the covariance of the attractant K1/2 and kinase activation parameters, the rheostat model would have to propose that the changing modification states ratchet the receptor conformation from one extreme through a series of intermediate conformations to another extreme, and that the resulting stepwise structural changes generate highly correlated changes in the attractant binding site and the kinase active site. This scenario is strongly disfavored by the radically different structures and functions of the attractant binding and kinase active sites.
The dramatic difference between the toggle and rheostat models is emphasized by an analysis of their predictions for the piston-type displacement of the α4/TM2/CD1 signaling helix during transmembrane signaling (Chervitz and Falke 1996). In both models, the “up” state of the piston corresponds to the lowest attractant affinity and highest kinase activation, whereas the “down” state of the piston corresponds to the highest attractant affinity and lowest kinase activity. Structural evidence indicates that the signaling helix is displaced 1–2 Å between the up and down states (Chervitz and Falke 1996). The toggle-switch model proposes that an equilibrium exists between the up and down positions of the piston, and that this equilibrium is shifted from the up state toward the down state as attractant is titrated into the system or as the proportion of adaptation site glutamates increases. The rheostat model proposes that covalent modification of the adaptation sites ratchets the piston through a series of distinct, stable intermediate locations ranging between the up and down positions. To explain the observed covariance of the attractant K1/2 and kinase activation parameters, the rheostat model must propose that the stepwise vertical displacements of the signaling helix cause similar effects in the attractant binding and kinase active sites at the opposite ends of the receptor–kinase complex. It is unlikely that stepwise displacements of the signaling helix with amplitudes of the order of tenths of an angstrom would trigger similar functional effects in these two unrelated sites. By contrast, the shifting equilibrium of the toggle-switch model would cause all the functional parameters of the up and down states to be averaged in the same way, providing a simple explanation for the observed linear covariance. The toggle-switch model further predicts that the attractant and adaptation signals will both drive the same conformational change but in opposite directions, such that attractant binding drives the signaling helix down while modification of the adaptation sites drives the signaling helix up. Thus the model adequately explains the recently observed opposing effects of attractant and adaptation signals on signaling helix position (Beel and Hazelbauer 2001). It should be emphasized that the two states of the toggle-switch are not static; rather, each state exhibits thermal fluctuations away from its stable equilibrium structure, together yielding two different average conformations.
The observed linear relationship between the attractant K1/2 and kinase activation parameters reveals the characteristic parameters for both fundamental conformations of the toggle-switch model, and allows calculation of the fraction of the receptor population in each conformation. This analysis assumes that the attractant K1/2 and kinase activation parameters (Fig. 3 B) vary linearly with the fractional populations of receptors in the two fundamental conformational states. The off-state is found to possess an α-methyl aspartate K1/2 in the submicromolar to micromolar range and a kinase activity approaching zero, as extrapolated from the correlation (Fig. 3 B). The modification state EEEE drives the receptor population almost completely into this off-state. The on-state is found to have an α-methyl aspartate K1/2 of 180 μM and a kinase activity 5.5-fold that of the wild-type (QEQE) receptor, where this upper limit is defined by the maximal kinase activity observed for multiple superactivating receptor mutants (Fig. 4 A). As Fig. 6 B illustrates, the line connecting these points for the pure on- and off-states allows calculation of the fractional occupancies of the fundamental on- and off-states for a native receptor population exposed to arbitrary attractant and modification conditions. To carry out such a calculation, the receptor-coupled kinase activity of the receptor population is measured via the standard in vitro assay, then the scale on Fig. 6 B is used to interpolate the fractional occupancy of the on-state. Any mutant receptor adequately described by the standard correlation can also be analyzed in this manner (although it should be noted that not all mutant receptors satisfy this requirement; see below).
The fractional populations in the on- and off-conformations depend on both the receptor modification level and the attractant concentration. Notably, in the absence of attractant the EEEE modification state shifts the equilibrium nearly completely toward the off-position of the toggle-switch but the QQQQ state shifts the equilibrium only 60% toward the on-position. This observation leaves open the possibility of further kinase activation (e.g., by repellent binding) in the fully modified state (Fig. 6 B). Alternatively, the native modification of the adaptation sites by methyl esterification may shift the equilibrium more strongly toward the on-position than amidation, such that the EmEmEmEm state might be fully activated. Overall, the conversion of the EEEE state to the fully activated state increases kinase activity at least 140-fold, indicating a corresponding increase in the population of the on-state. It follows that the free energy difference between the EEEE and fully activated states is ∼3 kcal mol−1 at physiological temperature. Binding of the native attractant l-aspartate provides a free energy driving force exceeding 8 kcal mol−1 toward the off-state (given a binding constant of 106 M−1 for the QQQQ receptor; unpublished data), thereby explaining the observation that saturating aspartate is able to reduce the kinase activities of even the most highly activated receptors to unmeasurable levels.
Mutations at sites other than the adaptation sites can also shift the equilibrium between the on- and off-conformations, or can alter the intrinsic parameters of one or both fundamental states. Mutations of the former type yield attractant K1/2 and kinase activation parameters that fall on the standard correlation, whereas mutations of the latter type yield parameters distant from the standard correlation (Fig. 3 C). Findings to date indicate that lock-on mutations typically shift parameters away from the standard correlation (examples are illustrated in Fig. 3 C). However, even these mutations may still be described as toggle-switches biased toward the on-state, since the EEEE modification state typically restores at least partial attractant sensitivity as expected for a modification that biases the toggle-switch back toward the off-state (Fig. 5A and Fig. B). The most dramatic lock-on mutation discovered thus far is G278V, which drives the toggle-switch toward the on-state so strongly that both the attractant and adaptation signals are unable to inactivate the kinase except in the EEEE background where saturating attractant provides partial inactivation (Fig. 5 A). Similarly, the sensitivity of weaker lock-on mutants to attractant increases as the modification level is decreased from QQQQ to EEEE (Fig. 5 B). The greater sensitivity of the EEEE background to attractant is expected since this background exhibits the highest attractant affinity, such that attractant binding provides the maximal driving force toward the off-state. Overall, the results indicate that lock-on mutations stabilize the on-state by as much as 8 kcal mol−1, but also modify the native parameters of the on- or off-state so the standard correlation between attractant K1/2 and kinase activation no longer applies.
The design of the toggle-switch equilibrium has important implications for the biology of chemotaxis. As a cell chemotaxes toward increasing attractant concentrations, the average modification state of the receptor population gradually increases (corresponding to a shift toward the activated conformation in Fig. 6 A). Concomitantly, this modification change causes the apparent K1/2 for attractant binding to increase, thereby allowing the sensitivity of the chemotaxis receptor to be appropriately adjusted for the ambient attractant concentration. Such tuning of the attractant affinity allows the cell to chemotax in a much larger range of attractant concentrations than one would expect for a receptor population with a fixed K1/2 value. Moreover, computational studies have suggested that a two-state equilibrium-type receptor can successfully reproduce various experimentally observed characteristics of the chemotaxis signaling system (Morton-Firth et al. 1999; Yi et al. 2000). For example, a receptor well-described by the toggle-switch model is able to adapt to changing attractant concentrations in a robust manner that is insensitive to variations in parameters such as the chemotaxis protein concentrations used in the simulation (Barkai and Leibler 1997; Alon et al. 1999). These computational studies further underscore the biological significance of a toggle-switch receptor of the type first systematically characterized in the present study. Future simulations of the chemotaxis pathway will be facilitated by the attractant K1/2 and kinase activation parameters measured in the present study.
The impressive sensitivity of the chemotaxis pathway at low attractant concentrations (Segall et al. 1986), together with the observed clustering of chemoreceptors (Maddock and Shapiro 1993; Kim et al. 1999), has led to the development of models proposing extensive cooperative interactions between receptors that are maximal at low attractant concentrations (Bray et al. 1998). However, such models are not supported by the present findings. As previously noted (Bornhorst and Falke 2000), the Hill coefficients obtained for attractant titrations ranged between ∼1.5 and 3, and generally increased as the aspartate receptor modification level increased from EEEE to QQQQ (Table ). A parallel study of the serine receptor also observed that the Hill coefficient of the attractant regulation increased with the receptor modification level (Li and Weis 2000). By contrast, models proposing increased cooperativity at low attractant concentrations incorrectly predict that the Hill coefficient will be largest for the unmodified receptor, which predominates in low attractant conditions. The Hill coefficients measured for the aspartate receptor are consistent either with strong cooperative interactions between receptor dimers within a structural unit composed of three dimers (Kim et al. 1999) or with weak cooperative interactions between dimers in larger arrays (Maddock and Shapiro 1993). The simplest version of the toggle-switch model proposes that each receptor dimer functions as a signaling unit that exhibits an equilibrium between well-defined on- and off-states, and that this equilibrium is modulated by attractant binding, modification of the adaptation sites, and cooperative interactions with nearby dimers. The internal toggle-switch responsible for two-state behavior appears to dominate the dimer output signal, whereas interactions between dimers appear less important since attractant binding is able to fully switch off the receptor in a wide range of modification states that exhibit varying degrees of dimer–dimer cooperativity.
More broadly, two-state behavior is observed in other components of the chemotaxis pathway and is widespread in signaling biology. NMR studies have revealed that periplasmic binding proteins of the chemotaxis system exhibit on- and off-conformations modulated by ligand binding, whereas the response regulator CheY displays an equilibrium between on- and off-states modulated by phosphorylation and protein–protein interactions (Luck and Falke 1991a,Luck and Falke 1991b; Schuster et al. 2001). Similarly, two-state behavior has been detected in bacterial phototaxis transducers (Jung and Spudich 1998), and, in the prokaryotic nitrogen-sensing pathway, phosphorylation of the response regulator NtrC has been demonstrated to shift a preexisting equilibrium between the on- and off-conformations (Volkman et al. 2001). In eukaryotic systems, G protein–coupled receptors have been proposed to exhibit two-state behavior (Leff 1995). Although rare examples are known of ligand binding events that yield new conformational states (introduction), in most systems, ligand binding simply modulates an equilibrium between two existing conformational states (Monod et al. 1965; Leff 1995). Similarly, covalent modification of regulatory sites on ion channels has been observed previously to modulate an equilibrium between existing states, as illustrated by single-channel recordings of calcium-gated potassium channels in which channel phosphorylation simply shifts the equilibrium between open and closed conformations (Chung et al. 1991). The present findings for a representative cell-surface sensory receptor indicate that ligand binding and covalent modification of the adaptation sites both modulate the equilibrium between two existing conformational states rather than creating new states. Thus, current evidence suggests covalent modification of signaling proteins generally regulates a preexisting equilibrium between the smallest possible number of stable signaling conformations.
Acknowledgments
We thank Sandy Parkinson and his laboratory for bacterial strains, helpful conversations, and comments on the manuscript. We also thank Chris Miller and Gary Yellen for helpful discussions. Finally, we are grateful to past and current members of the Falke lab, including Randal Bass, Eric Nalefski, Susy Kohout, Ryan Mehan, Matt Trammell, Gina Westhoff, and Noah White, for helpful advice and assistance.
Support for this study was provided by National Institutes of Health Grant GM R01-40731 (to J.J. Falke) and by NIH Training Grant T32-GM08759 (to J.A. Bornhorst).
References
- Alon U., Surette M.G., Barkai N., Leibler S. Robustness in bacterial chemotaxis. Nature. 1999;397:168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- Ames P., Parkinson J.S. Transmembrane signaling by bacterial chemoreceptorsE. coli transducers with locked signal output. Cell. 1988;55:817–826. doi: 10.1016/0092-8674(88)90137-7. [DOI] [PubMed] [Google Scholar]
- Armitage J.P. Bacterial tactic responses. Adv. Microb. Physiol. 1999;41:229–289. doi: 10.1016/s0065-2911(08)60168-x. [DOI] [PubMed] [Google Scholar]
- Asakura S., Honda H. Two-state model for bacterial chemoreceptor proteins. The role of multiple methylation. J. Mol. Biol. 1984;176:349–367. doi: 10.1016/0022-2836(84)90494-7. [DOI] [PubMed] [Google Scholar]
- Barkai N., Leibler S. Robustness in simple biochemical networks. Nature. 1997;387:913–917. doi: 10.1038/43199. [DOI] [PubMed] [Google Scholar]
- Bass R.B., Falke J.J. Detection of a conserved alpha-helix in the kinase-docking region of the aspartate receptor by cysteine and disulfide scanning. J. Biol. Chem. 1998;273:25006–25014. doi: 10.1074/jbc.273.39.25006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass R.B., Falke J.J. The aspartate receptor cytoplasmic domainin situ chemical analysis of structure, mechanism and dynamics. Struct. Fold Des. 1999;7:829–840. doi: 10.1016/s0969-2126(99)80106-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass R.B., Coleman M.D., Falke J.J. Signaling domain of the aspartate receptor is a helical hairpin with a localized kinase docking surfacecysteine and disulfide scanning studies. Biochemistry. 1999;38:9317–9327. doi: 10.1021/bi9908179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner J.W., Kim C., Brissette R.E., Inouye M., Park C., Hazelbauer G.L. Transmembrane signalling by a hybrid proteincommunication from the domain of chemoreceptor Trg that recognizes sugar-binding proteins to the kinase/phosphatase domain of osmosensor EnvZ. J. Bacteriol. 1994;176:1157–1163. doi: 10.1128/jb.176.4.1157-1163.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beel B.D., Hazelbauer G.L. Signalling substitutions in the periplasmic domain of chemoreceptor Trg induce or reduce helical sliding in the transmembrane domain. Mol. Microbiol. 2001;40:824–834. doi: 10.1046/j.1365-2958.2001.02446.x. [DOI] [PubMed] [Google Scholar]
- Bibikov S.I., Biran R., Rudd K.E., Parkinson J.S. A signal transducer for aerotaxis in Escherichia coli . J. Bacteriol. 1997;179:4075–4079. doi: 10.1128/jb.179.12.4075-4079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biemann H.P., Koshland D.E., Jr. Aspartate receptors of Escherichia coli and Salmonella typhimurium bind ligand with negative and half-of-the-sites cooperativity. Biochemistry. 1994;33:629–634. doi: 10.1021/bi00169a002. [DOI] [PubMed] [Google Scholar]
- Blair D.F. How bacteria sense and swim. Annu. Rev. Microbiol. 1995;49:489–522. doi: 10.1146/annurev.mi.49.100195.002421. [DOI] [PubMed] [Google Scholar]
- Borkovich K.A., Kaplan N., Hess J.F., Simon M.I. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc. Natl. Acad. Sci. USA. 1989;86:1208–1212. doi: 10.1073/pnas.86.4.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich K.A., Alex L.A., Simon M.I. Attenuation of sensory receptor signaling by covalent modification. Proc. Natl. Acad. Sci. USA. 1992;89:6756–6760. doi: 10.1073/pnas.89.15.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornhorst J.A., Falke J.J. Attractant regulation of the aspartate receptor-kinase complexlimited cooperative interactions between receptors and effects of the receptor modification state. Biochemistry. 2000;39:9486–9493. doi: 10.1021/bi0002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret R.B., Drake S.K., Chervitz S.A., Simon M.I., Falke J.J. Activation of the phosphosignaling protein CheY. II. Analysis of activated mutants by 19F NMR and protein engineering. J. Biol. Chem. 1993;268:13089–13096. [PMC free article] [PubMed] [Google Scholar]
- Bray D., Levin M.D., Morton-Firth C.J. Receptor clustering as a cellular mechanism to control sensitivity. Nature. 1998;393:85–88. doi: 10.1038/30018. [DOI] [PubMed] [Google Scholar]
- Bren A., Eisenbach M. How signals are heard during bacterial chemotaxisprotein-protein interactions in sensory signal propagation. J. Bacteriol. 2000;182:6865–6873. doi: 10.1128/jb.182.24.6865-6873.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler S.L., Falke J.J. Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. Biochemistry. 1998;37:10746–10756. doi: 10.1021/bi980607g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervitz S.A., Falke J.J. Lock on/off disulfides identify the transmembrane signaling helix of the aspartate receptor. J. Biol. Chem. 1995;270:24043–24053. doi: 10.1074/jbc.270.41.24043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervitz S.A., Falke J.J. Molecular mechanism of transmembrane signaling by the aspartate receptora model. Proc. Natl. Acad. Sci. USA. 1996;93:2545–2550. doi: 10.1073/pnas.93.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervitz S.A., Lin C.M., Falke J.J. Transmembrane signaling by the aspartate receptorengineered disulfides reveal static regions of the subunit interface. Biochemistry. 1995;34:9722–9733. doi: 10.1021/bi00030a010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S.K., Reinhart P.H., Martin B.L., Brautigan D., Levitan I.B. Protein kinase activity closely associated with a reconstituted calcium-activated potassium channel. Science. 1991;253:560–562. doi: 10.1126/science.1857986. [DOI] [PubMed] [Google Scholar]
- Danielson M.A., Biemann H.P., Koshland D.E., Jr., Falke J.J. Attractant- and disulfide-induced conformational changes in the ligand binding domain of the chemotaxis aspartate receptora 19F NMR study. Biochemistry. 1994;33:6100–6109. doi: 10.1021/bi00186a009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson M.A., Bass R.B., Falke J.J. Cysteine and disulfide scanning reveals a regulatory alpha-helix in the cytoplasmic domain of the aspartate receptor. J. Biol. Chem. 1997;272:32878–32888. doi: 10.1074/jbc.272.52.32878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic S., Stock A.M. Structural analysis of bacterial chemotaxis proteinscomponents of a dynamic signaling system. J. Struct. Biol. 1998;124:189–200. doi: 10.1006/jsbi.1998.4034. [DOI] [PubMed] [Google Scholar]
- Dunten P., Koshland D.E., Jr. Tuning the responsiveness of a sensory receptor via covalent modification. J. Biol. Chem. 1991;266:1491–1496. [PubMed] [Google Scholar]
- Eghbali M., Curmi J.P., Birnir B., Gage P.W. Hippocampal GABA(A) channel conductance increased by diazepam. Nature. 1997;388:71–75. doi: 10.1038/40404. [DOI] [PubMed] [Google Scholar]
- Falke J.J., Kim S.H. Structure of a conserved receptor domain that regulates kinase activitythe cytoplasmic domain of bacterial taxis receptors. Curr. Opin. Struct. Biol. 2000;10:462–469. doi: 10.1016/s0959-440x(00)00115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falke J.J., Hazelbauer G.L. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 2001;26:257–265. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falke J.J., Bass R.B., Butler S.L., Chervitz S.A., Danielson M.A. The two-component signaling pathway of bacterial chemotaxisa molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu. Rev. Cell Dev. Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauenfelder H., Parak F., Young R.D. Conformational substates in proteins. Annu. Rev. Biophys. Chem. 1988;17:451–479. doi: 10.1146/annurev.bb.17.060188.002315. [DOI] [PubMed] [Google Scholar]
- Gegner J.A., Graham D.R., Roth A.F., Dahlquist F.W. Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell. 1992;70:975–982. doi: 10.1016/0092-8674(92)90247-a. [DOI] [PubMed] [Google Scholar]
- Grebe T.W., Stock J. Bacterial chemotaxisthe five sensors of a bacterium. Curr. Biol. 1998;8:R154–R157. doi: 10.1016/s0960-9822(98)00098-0. [DOI] [PubMed] [Google Scholar]
- Hess J.F., Oosawa K., Kaplan N., Simon M.I. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell. 1988;53:79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- Jung K.H., Spudich J.L. Suppressor mutation analysis of the sensory rhodopsin I-transducer complexinsights into the color-sensing mechanism. J. Bacteriol. 1998;180:2033–2042. doi: 10.1128/jb.180.8.2033-2042.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehry M.R., Bond M.W., Hunkapiller M.W., Dahlquist F.W. Enzymatic deamidation of methyl-accepting chemotaxis proteins in Escherichia coli catalyzed by the cheB gene product. Proc. Natl. Acad. Sci. USA. 1983;80:3599–3603. doi: 10.1073/pnas.80.12.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C., Jackson M., Lux R., Khan S. Determinants of chemotactic signal amplification in Escherichia coli . J. Mol. Biol. 2001;307:119–135. doi: 10.1006/jmbi.2000.4389. [DOI] [PubMed] [Google Scholar]
- Kim K.K., Yokota H., Kim S.H. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- Kolodziej A.F., Tan T., Koshland D.E., Jr. Producing positive, negative, and no cooperativity by mutations at a single residue located at the subunit interface in the aspartate receptor of Salmonella typhimurium . Biochemistry. 1996;35:14782–14792. doi: 10.1021/bi961481v. [DOI] [PubMed] [Google Scholar]
- Krikos A., Conley M.P., Boyd A., Berg H.C., Simon M.I. Chimeric chemosensory transducers of Escherichia coli . Proc. Natl. Acad. Sci. USA. 1985;82:1326–1330. doi: 10.1073/pnas.82.5.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T.A., Bebenek K., McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- Le Moual H., Kirkland D.E., Jr. Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J. Mol. Biol. 1996;261:568–585. doi: 10.1006/jmbi.1996.0483. [DOI] [PubMed] [Google Scholar]
- Le Moual H., Quang T., Koshland D.E., Jr. Methylation of the Escherichia coli chemotaxis receptorsintra- and interdimer mechanisms. Biochemistry. 1997;36:13441–13448. doi: 10.1021/bi9713207. [DOI] [PubMed] [Google Scholar]
- Leff P. The two-state model of receptor activation. Trends Pharmacol. Sci. 1995;16:89–97. doi: 10.1016/s0165-6147(00)88989-0. [DOI] [PubMed] [Google Scholar]
- Li G., Weis R.M. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli . Cell. 2000;100:357–365. doi: 10.1016/s0092-8674(00)80671-6. [DOI] [PubMed] [Google Scholar]
- Li J., Li G., Weis R.M. The serine chemoreceptor from Escherichia coli is methylated through an inter-dimer process. Biochemistry. 1997;36:11851–11857. doi: 10.1021/bi971510h. [DOI] [PubMed] [Google Scholar]
- Liu J.D., Parkinson J.S. Role of CheW protein in coupling membrane receptors to the intracellular signaling system of bacterial chemotaxis. Proc. Natl. Acad. Sci. USA. 1989;86:8703–8707. doi: 10.1073/pnas.86.22.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck L.A., Falke J.J. 19F NMR studies of the d-galactose chemosensory receptor. 1. Sugar binding yields a global structural change Biochemistry. 30 1991. 4248 4256a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck L.A., Falke J.J. Open conformation of a substrate-binding cleft19F NMR studies of cleft angle in the d-galactose chemosensory receptor Biochemistry. 30 1991. 6484 6490b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lybarger S.R., Maddock J.R. Polarity in actionasymmetric protein localization in bacteria. J. Bacteriol. 2001;183:3261–3267. doi: 10.1128/JB.183.11.3261-3267.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddock J.R., Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- Milburn M.V., Prive G.G., Milligan D.L., Scott W.G., Yeh J., Jancarik J., Koshland D.E., Jr., Kim S.H. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 1991;254:1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- Milligan D.L., Koshland D.E., Jr. Site-directed cross-linking. Establishing the dimeric structure of the aspartate receptor of bacterial chemotaxis. J. Biol. Chem. 1988;263:6268–6275. [PubMed] [Google Scholar]
- Moe G.R., Bollag G.E., Koshland D.E., Jr. Transmembrane signaling by a chimera of the Escherichia coli aspartate receptor and the human insulin receptor. Proc. Natl. Acad. Sci. USA. 1989;86:5683–5687. doi: 10.1073/pnas.86.15.5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod J., Wyman J., Changeux J. On the nature of allosteric transitionsa plausible model. J. Mol. Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- Morton-Firth C.J., Shimizu T.S., Bray D. A free-energy-based stochastic simulation of the Tar receptor complex. J. Mol. Biol. 1999;286:1059–1074. doi: 10.1006/jmbi.1999.2535. [DOI] [PubMed] [Google Scholar]
- Ninfa E.G., Stock A., Mowbray S., Stock J. Reconstitution of the bacterial chemotaxis signal transduction system from purified components. J. Biol. Chem. 1991;266:9764–9770. [PubMed] [Google Scholar]
- Nishiyama S., Maruyama I.N., Homma M., Kawagishi I. Inversion of thermosensing property of the bacterial receptor Tar by mutations in the second transmembrane region. J. Mol. Biol. 1999;286:1275–1284. doi: 10.1006/jmbi.1999.2555. [DOI] [PubMed] [Google Scholar]
- Pakula A.A., Simon M.I. Determination of transmembrane protein structure by disulfide cross-linkingthe Escherichia coli Tar receptor. Proc. Natl. Acad. Sci. USA. 1992;89:4144–4148. doi: 10.1073/pnas.89.9.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C., Dutton D.P., Hazelbauer G.L. Effects of glutamines and glutamates at sites of covalent modification of a methyl-accepting transducer. J. Bacteriol. 1990;172:7179–7187. doi: 10.1128/jb.172.12.7179-7187.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J.S. Signal transduction schemes of bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- Perazzona B., Spudich J.L. Identification of methylation sites and effects of phototaxis stimuli on transducer methylation in Halobacterium salinarum . J. Bacteriol. 1999;181:5676–5683. doi: 10.1128/jb.181.18.5676-5683.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenmund C., Stern-Bach Y., Stevens C.F. The tetrameric structure of a glutamate receptor channel. Science. 1998;280:1596–1599. doi: 10.1126/science.280.5369.1596. [DOI] [PubMed] [Google Scholar]
- Ruiz M.L., Karpen J.W. Single cyclic nucleotide-gated channels locked in different ligand-bound states. Nature. 1997;389:389–392. doi: 10.1038/38744. [DOI] [PubMed] [Google Scholar]
- Russell C.B., Stewart R.C., Dahlquist F.W. Control of transducer methylation levels in Escherichia coliinvestigation of components essential for modulation of methylation and demethylation reactions. J. Bacteriol. 1989;171:3609–3618. doi: 10.1128/jb.171.7.3609-3618.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M., Silversmith R.E., Bourret R.B. Conformational coupling in the chemotaxis response regulator CheY. Proc. Natl. Acad. Sci. USA. 2001;98:6003–6008. doi: 10.1073/pnas.101571298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster S.C., Swanson R.V., Alex L.A., Bourret R.B., Simon M.I. Assembly and function of a quaternary signal transduction complex monitored by surface plasmon resonance. Nature. 1993;365:343–347. doi: 10.1038/365343a0. [DOI] [PubMed] [Google Scholar]
- Segall J.E., Block S.M., Berg H.C. Temporal comparisons in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA. 1986;83:8987–8991. doi: 10.1073/pnas.83.23.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro M.J., Chakrabarti I., Koshland D.E., Jr. Contributions made by individual methylation sites of the Escherichia coli aspartate receptor to chemotactic behavior. Proc. Natl. Acad. Sci. USA. 1995;92:1053–1056. doi: 10.1073/pnas.92.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slocum M.K., Halden N.F., Parkinson J.S. Hybrid Escherichia coli sensory transducers with altered stimulus detection and signaling properties. J. Bacteriol. 1987;169:2938–2944. doi: 10.1128/jb.169.7.2938-2944.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer W.R., Koshland D.E., Jr. Identification of a protein methyltransferase as the cheR gene product in the bacterial sensing system. Proc. Natl. Acad. Sci. USA. 1977;74:533–537. doi: 10.1073/pnas.74.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart R.C. Kinetic characterization of phosphotransfer between CheA and CheY in the bacterial chemotaxis signal transduction pathway. Biochemistry. 1997;36:2030–2040. doi: 10.1021/bi962261k. [DOI] [PubMed] [Google Scholar]
- Stock A., Mottonen J., Chen T., Stock J. Identification of a possible nucleotide binding site in CheW, a protein required for sensory transduction in bacterial chemotaxis. J. Biol. Chem. 1987;262:535–537. [PubMed] [Google Scholar]
- Stock A., Chen T., Welsh D., Stock J. CheA protein, a central regulator of bacterial chemotaxis, belongs to a family of proteins that control gene expression in response to changing environmental conditions. Proc. Natl. Acad. Sci. USA. 1988;85:1403–1407. doi: 10.1073/pnas.85.5.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R.V., Simon M.I. Signal transduction. Bringing the eukaryotes up to speed. Curr. Biol. 1994;4:234–237. doi: 10.1016/s0960-9822(00)00052-x. [DOI] [PubMed] [Google Scholar]
- Tatsuno I., Lee L., Kawagishi I., Homma M., Imae Y. Transmembrane signalling by the chimeric chemosensory receptors of Escherichia coli Tsr and Tar with heterologous membrane-spanning regions. Mol. Microbiol. 1994;14:755–762. doi: 10.1111/j.1365-2958.1994.tb01312.x. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C., Koshland D.E., Jr. Sites of methyl esterification and deamination on the aspartate receptor involved in chemotaxis. J. Biol. Chem. 1984;259:7719–7725. [PubMed] [Google Scholar]
- Terwilliger T.C., Wang J.Y., Koshland D.E., Jr. Kinetics of receptor modification. The multiply methylated aspartate receptors involved in bacterial chemotaxis. J. Biol. Chem. 1986;261:10814–10820. [PubMed] [Google Scholar]
- Trammell M.A., Falke J.J. Identification of a site critical for kinase regulation on the central processing unit (CPU) helix of the aspartate receptor. Biochemistry. 1999;38:329–336. doi: 10.1021/bi981964u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsumi R., Brissette R.E., Rampersaud A., Forst S.A., Oosawa K., Inouye M. Activation of bacterial porin gene expression by a chimeric signal transducer in response to aspartate. Science. 1989;245:1246–1249. doi: 10.1126/science.2476847. [DOI] [PubMed] [Google Scholar]
- Volkman B.F., Lipson D., Wemmer D.E., Kern D. Two-state allosteric behavior in a single-domain signaling protein. Science. 2001;291:2429–2433. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- Weerasuriya S., Schneider B.M., Manson M.D. Chimeric chemoreceptors in Escherichia colisignaling properties of Tar-Tap and Tap-Tar hybrids. J. Bacteriol. 1998;180:914–920. doi: 10.1128/jb.180.4.914-920.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurgler-Murphy S.M., Saito H. Two-component signal transducers and MAPK cascades. Trends Biochem. Sci. 1997;22:172–176. doi: 10.1016/s0968-0004(97)01036-0. [DOI] [PubMed] [Google Scholar]
- Yi T.M., Huang Y., Simon M.I., Doyle J. Robust perfect adaptation in bacterial chemotaxis through integral feedback control. Proc. Natl. Acad. Sci. USA. 2000;97:4649–4653. doi: 10.1073/pnas.97.9.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]