Abstract
The bacterial toxin colicin Ia forms voltage-gated channels in planar lipid bilayers. The toxin consists of three domains, with the carboxy-terminal domain (C-domain) responsible for channel formation. The C-domain contributes four membrane-spanning segments and a 68-residue translocated segment to the open channel, whereas the upstream domains and the amino-terminal end of the C-domain stay on the cis side of the membrane. The isolated C-domain, lacking the two upstream domains, also forms channels; however, the amino terminus and one of the normally membrane-spanning segments can move across the membrane. (This can be observed as a drop in single-channel conductance.) In longer carboxy-terminal fragments of colicin Ia that include ≤169 residues upstream from the C-domain, the entire upstream region is translocated. Presumably, a portion of the C-domain creates a pathway for the polar upstream region to move through the membrane. To determine the size of this translocation pathway, we have attached “molecular stoppers,” small disulfide-bonded polypeptides, to the amino terminus of the C-domain, and determined whether they could be translocated. We have found that the translocation rate is strongly voltage dependent, and that at voltages ≥90 mV, even a 26-Å stopper is translocated. Upon reduction of their disulfide bonds, all of the stoppers are easily translocated, indicating that it is the folded structure, rather than some aspect of the primary sequence, that slows translocation of the stoppers. Thus, the pathway for translocation is ≥26 Å in diameter, or can stretch to this value. This is large enough for an α-helical hairpin to fit through.
Keywords: α-conotoxin, apamin, charybdotoxin, disulfides, molecular stoppers
INTRODUCTION
Colicin Ia is a 626-residue bactericidal protein that forms voltage-gated channels in the inner membrane of sensitive E. coli bacteria and in planar lipid bilayers. The protein consists of three domains (Wiener et al., 1997), with the 10–α-helix colicin Ia carboxy-terminal domain (C-domain)* (residues 451–626) forming the lethal channel. (For a general review of channel-forming colicins and colicin Ia in particular, see Jakes et al., 1999). Channel formation by whole colicin Ia can be conceptually divided into two steps: (1) voltage-independent insertion of the hydrophobic hairpin formed by helices 8 and 9 (Kienker et al., 1997) and (2) voltage-dependent insertion of portions of helix 1 and helices 6–7 (Qiu et al., 1996). Associated with the voltage-dependent step is the translocation across the membrane of ∼68 residues corresponding to helices 2–5 (Qiu et al., 1996). The open channel formed by whole colicin Ia thus consists of four membrane-spanning segments,1 with the upstream T and R domains remaining on the cis side (the side to which the colicin was added) (Fig. 1 C).
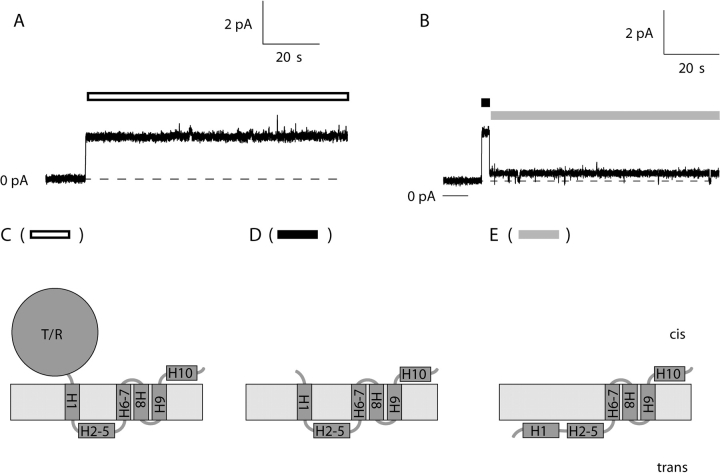
Figure 1.
Comparison of single channels formed by whole colicin Ia and its carboxy-terminal domain (Kienker et al., 2000). (A) A whole colicin Ia channel opened at +50 mV with its characteristic, relatively high conductance (39 pS) and, as is typical at this voltage, stayed at this level. (B) A C-domain channel (residues 438–626) opened at +50 mV with a comparable conductance (44 pS), but then dropped to a state of smaller conductance (7 pS). (C) Schematic model of the whole colicin Ia channel in its open state, with four membrane-spanning segments. The 10 α-helices of the C-domain are designated H1–H10; T/R represents the upstream T and R domains. The part of H1 spanning the membrane is roughly from residues 451–467. (D) Model of the C-domain channel in its transient, higher conductance state, also with four membrane-spanning segments. (E) Model of the C-domain channel in its small-conductance open state, with only three membrane-spanning segments, and helix H1 translocated across the membrane. The white bar in A indicates the open channel state that is diagrammed in C. In B, the black bar indicates when the channel is in the transient open state of D, and the gray bar indicates when it is in the small-conductance open state of E. The zero-current level in A and B is labeled 0 pA. There are two small-conductance channels already open at the beginning of the record in B. The solution on both sides of the membrane for A and B was 1 M KCl, 5 mM CaCl2, 1 mM EDTA, 20 mM MES, pH 6.2.
Channel formation by the isolated C-domain, or by carboxy-terminal fragments containing as many as 345 residues (residues 282–626), is similar to channel formation by whole colicin, except that the portion of helix 1 (H1) that was in the membrane, and everything upstream from it, is also translocated across the membrane, resulting in a small-conductance channel having only three transmembrane segments (Fig. 1 E) (Kienker et al., 2000). On the way to this small-conductance state, the channel initially opens to a state of relatively normal conductance (Kienker et al., 2000) (Fig. 1 B). We have good reason to believe (see results) that the initial normal-conductance channel corresponds to the channel with four transmembrane segments, like that formed by the whole colicin (Fig. 1, compare C with D). The transition to the small-conductance channel marks the passage of the fourth segment (H1) (and everything upstream of it) across the membrane, and thus the conversion to a channel with only three transmembrane segments (Fig. 1 E).
What is the nature of the translocation passageway that the colicin creates for the fourth segment to cross the membrane? We know that this passageway allows the movement through it of the very polar and highly charged sequences that make up the upstream parts of colicin Ia carboxy-terminal fragments (Kienker et al., 2000), suggesting that it provides a high-dielectric–constant milieu, but how wide is it? We previously showed that if the amino terminus of the C-domain is biotinylated and then preincubated with streptavidin, the resulting channels open to the normal-conductance state and remain there, failing to drop to the small-conductance state typical of C-domain channels (Kienker et al., 2000). Presumably, streptavidin holds the amino terminus on the cis side and prevents its translocation across the membrane. Thus, we know from these earlier experiments that the translocation pathway for the amino terminus is not wide enough to accommodate the 61-Å-diameter streptavidin tetramer. Does the peptide chain have to unfold to fit through it, or can its secondary structure, and even its tertiary structure, be preserved during its passage? To address these questions, we attached a series of molecular “stoppers” to the amino terminus of the C-domain and determined how large a stopper could still cross the membrane. The stoppers consisted of peptides locked into a more or less fixed conformation by one or more disulfide bonds (see materials and methods). We reasoned that if the channel passed from the normal-conductance to the small-conductance state (as in Fig. 1 B), then the stopper had passed through, whereas if it remained in the normal-conductance state (as in Fig. 1 A), the stopper was too large to pass through. Using this approach we determined that the passageway can be at least 26 Å wide at its narrowest point.
MATERIALS AND METHODS
Construction of the Stoppers
Four separate molecular stopper constructs were prepared, all with the stopper attached near the amino-terminal end of the colicin Ia carboxy-terminal fragment 438–626 (called CT-M in Kienker et al., 2000). pKSJ155 has residues 438–626 of colicin Ia inserted downstream of the 6 histidine tag (His6-tag) sequence in the Novagen vector pET-15b. The three smaller stoppers were inserted genetically, by site-directed mutagenesis, so that their sequences were inserted immediately upstream of the beginning of the colicin sequence, before either residue 438 or 439. All of the insertions were made using Stratagene's QuikChange site-directed mutagenesis kit. The largest stopper, charybdotoxin (CTX), was attached chemically to the side chain of a cysteine introduced at residue 439.
Peptide U,
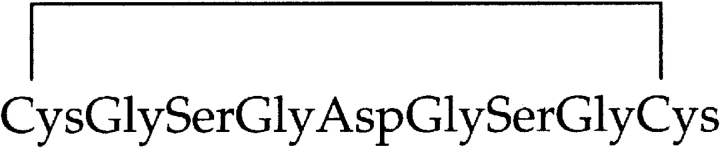 ,
,
which has a single disulfide bond, was inserted before residue 439 of colicin Ia, using the mutagenic oligonucleotide 5′-GCGGCAGCCATATGCTCGACTGTGGAAGTGGTGACGGAAGCG-GCTGC GAGAAAAGAAAACAGGATGAACTGAAGGC-3′ and its reverse complement. The inserted sequence is in boldfaced type, the upstream sequence from the pET-15b vector is in standard type, and colicin sequence beginning with residue 439 is in italics.
The twelve amino acids of α-conotoxin ImI, which has two disulfide bonds,
(Maslennikov et al., 1999), were inserted in a single round of mutagenesis before residue 438 of the colicin Ia sequence in pKSJ155. The mutagenic oligonucleotides used for the insertion were 5′-GCAGCCATATGCTCGGCTGTTGTTCCGACCCGCGCTGTGCTTGGCGTTGT GAGGAGAAAAGAAAACAGGATG-3′ and its reverse complement. Vector sequence is in standard type, the inserted sequence is in boldfaced type, and colicin Ia sequence is in italics.
The eighteen residues of apamin, which has two disulfide bonds,
(Pease and Wemmer, 1988), were inserted before residue 438 of pKSJ155 in two steps of mutagenesis, first inserting the last nine residues, and then inserting the first nine residues immediately before the first insertion. The mutagenic oligonucleotides for the first round of insertions were 5′-GCGCGGCAGCCATATGCTCCTGTGCGCCCGTCGCTGCCAGCAGCAT GAGGAGAAAAGAAAACAGGATGAACTGAAGG-3′ and its reverse complement. The mutagenic oligonucleotides used for the subsequent round were 5′-GCGCGGCAGCCATATGCTCTGCAACTGCAAAGCGCCGGAAACCGCG CTGTGCGCCCGTCGCTGC-3′ and its reverse complement. pET-15b sequences are shown in standard type, apamin inserted sequences are in boldfaced type and in underlined type, and colicin sequence is in italics. The products of all mutagenesis reactions were sequenced to confirm the correct insertion sequences.
The proteins were expressed and purified from the plasmids bearing the insertions in BL21 (DE3) cells induced with 1 mM isopropyl-β-D-thiogalactoside (Labscientific, Inc.). Cultures of 400–500 ml were grown and the proteins were purified on His-Bind (Novagen) nickel affinity columns, as described previously (Kienker et al., 2000). The yields were ∼4 mg for the peptide U insertion protein, 16 mg for the α-conotoxin insertion protein, and 11 mg for the protein with apamin inserted.
The His6-tag was removed from each of the three proteins containing genetically inserted stoppers by incubating the protein with thrombin (Novagen) for 5 h at room temperature in 20 mM Tris-Cl, pH 8.5, 150 mM NaCl. The thrombin/protein ratio was 1:1,000 by weight. The reaction was stopped with 1 mM PMSF. In one set of experiments with peptide U (see RESULTS), the His6-tag was not removed.
CTX, with 37 amino acids, has the sequence H-PyrPheThrAsnValSerCys7ThrThrSerLysGluCys13TrpSerValCys17GlnArgLeu-HisAsn-ThrSerArgGlyLysCys28MetAsnLysLysCys33ArgCys35Tyr-Ser, with three disulfide bonds, between cysteine residues 7–28, 13–33, and 17–35 (Bontems et al., 1992). CTX was genetically modified and chemically attached to colicin Ia residue 439, which had been mutated from glutamic acid to cysteine in pKSJ155, to yield pKSJ156 (Kienker et al., 2000), as follows. Three of the four lysines of CTX were mutated (K11Q/K31R/K32R) to generate a toxin bearing a single amino group (K27). Mutation, expression, amino-terminal pyroglutamate (Pyr) blocking, and purification of this toxin were as previously reported (Stampe et al., 1994). N-succinimidyl 3-maleimidopropionate (SMP) was synthesized as described (Nielsen and Buchardt, 1991) and was used to introduce maleimide functionality to the toxin via chemical modification of K27. Specifically, 2 ml of a solution containing 1.7 mM SMP in acetonitrile were added to a 10-ml solution containing 50 μM CTX and 50 mM sodium phosphate buffer, pH 7.2. The resultant solution was stirred at room temperature for 30 min and then centrifuged under reduced pressure for ∼3 min to decrease the acetonitrile concentration to 5–10%. The maleimido-CTX was purified using a C18 column (YMC, Inc.) with a linear, water–acetonitrile gradient (Waters HPLC system, Waters Corp.), and assayed on a Mariner electrospray/TOF mass spectrometer (PE Biosystems/Perkin Elmer). Eluates from several preparative HPLC runs were pooled, evaporated under reduced pressure to a final volume of ∼5 ml, and adjusted to pH 7.2 via the addition of sodium phosphate to a concentration of 25 mM. The sample was frozen and lyophilized to a volume of ∼0.5 ml, and the volume was then increased to 1.8 ml by the addition of sodium phosphate buffer. The composition of this final solution was 150 μM maleimido-CTX in 150 mM sodium phosphate buffer. Purified material was >95% pure as judged by reinjection on an analytic C18 column while monitoring absorbance at 280 nm.
The His6-tag was removed from E439C colicin Ia C-domain protein by digestion with thrombin, using the same conditions as described above, except that the thrombin/protein ratio was 1:2,000 by weight. After the reaction was stopped with PMSF, the protein was reduced by adding 1 M DTT to a final concentration of 20 mM and incubating 1 h at room temperature. DTT was removed by dialysis, first against degassed 10 mM MES, pH 6.0, 2 mM EDTA, 0.1 mM tris (2-carboxyethyl)phosphine hydrochloride (TCEP) (Pierce Chemical Co.), and then against the same degassed buffer without EDTA. The reduced, dialyzed protein was transferred to argon-filled tubes and stored under argon at −80°C. 7.5 μg of this protein in 3 μl (3 × 10−10 moles) was added to 37 μl of maleimido-CTX (5.5 × 10−9 moles) in 150 mM sodium phosphate, pH 7.2, an ∼18-fold excess of the CTX over colicin peptide. The reaction was incubated for 30 min at 30°C and then stopped by addition of excess β-mercaptoethanol. Approximately 80–90% of the colicin peptide was judged to have reacted with the maleimido-CTX by analysis on a 15% polyacrylamide-SDS gel. Wild-type colicin Ia C-domain was treated with thrombin, reduced, dialyzed, and reacted with maleimido-CTX in exactly the same way as the E439C mutant, as a control for nonspecific reaction of the maleimido-CTX with residues other than cysteine. There was no apparent reaction of the wild-type C-domain peptide with the reagent. The CTX moiety attached to colicin (CTX–C-domain) was unfolded by adding 1/25 volume of 1.5 M Tris-Cl, pH 8.8, to raise the pH to ∼8.0, and making the protein solution 10 mM in DTT. The protein was then heated at 70°C for 10 min and quenched on ice (Naini et al., 1996).
Assessment of Stopper Folding
The proteins containing the three smaller stoppers had a tendency to dimerize or multimerize via their multiple cysteine sulfhydryl groups. Keeping the proteins at concentrations below ∼1 mg/ml throughout the purification procedure reduced the ratio of multimers to monomers. We were also concerned that there might be improperly folded stoppers that had a free cysteine. To assay for free cysteines, the proteins were reacted with N-[6′-(biotinamido) hexyl]-3′-(2′-pyridyldithio) propionamide (biotin-HPDP) (Pierce Chemical Co.) without prior reduction (Qiu et al., 1996). Any free cysteines were thus biotinylated, and could be detected by a gel-shift assay with streptavidin (Qiu et al., 1994). Both the peptide U and α-conotoxin stoppers were judged to have measurable free cysteine by this method, ∼10% of peptide U and nearly 50% of α-conotoxin (unpublished data). The biotinylated protein was then removed from the preparation by chromatography on monomeric avidin, as described previously (Qiu et al., 1996). This procedure appeared to remove any protein that had a free cysteine that had reacted with biotin-HPDP, as measured by gel-shift by streptavidin (Qiu et al., 1994). The preparations still contained a small amount of dimers and higher multimers, as assessed by SDS-polyacrylamide gel.
Purification of Streptavidin- and Avidin-bound C-domain
We prepared and purified streptavidin-bound C-domain as follows. The mutant E439C of C-domain (residues 438–626) was biotinylated and had its His6-tag removed as previously described (Kienker et al., 2000); 15 μg of this protein was incubated with 800 μg of streptavidin (an 18-fold molar excess of streptavidin tetramer) at room temperature for 15 min in a total volume of 120 μl. The mixture was centrifuged to remove any particulates, applied to a BioSil 250 sizing column (Bio-Rad Laboratories), and eluted in 100 mM NaCl, 50 mM HEPES, pH 7.4. The absorbance at 280 nm was monitored. Three peaks were seen: first a small peak containing streptavidin-bound C-domain, then a large streptavidin peak, and finally a small peak containing unbound C-domain. The first two peaks were also observed with streptavidin alone; apparently the commercial streptavidin contains some high molecular weight material that happens to run together with the streptavidin-bound C-domain.
The purification of avidin-bound C-domain was similar, except that avidin was incubated with biotinylated C-domain for 60 min. Avidin alone had only one broad peak. No peak was visible for avidin-bound C-domain, so we used a fraction from the leading edge of the avidin peak.
Estimation of Stopper Diameters
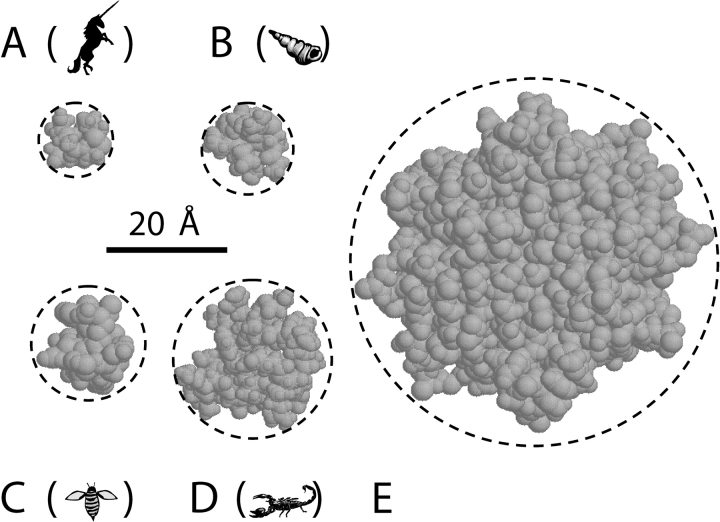
Three-dimensional structures of peptide U (with its two cysteine residues forming a disulfide bond) were calculated by T. Newman using an AMBER force field (Cornell et al., 1995), with water treated as a continuum solvent. The 16 lowest-energy structures were displayed as space-filling models in RasMol (v2.6-ucb), and rotated by eye to minimize the diameter of the two-dimensional projection on the computer screen. This minimal cross-sectional diameter is defined by the smallest circle that can be drawn around the image of the molecule (Fig. 2 A). The peptide U model structure with the 4th-lowest energy had the smallest diameter, 12.3 Å.
Figure 2.
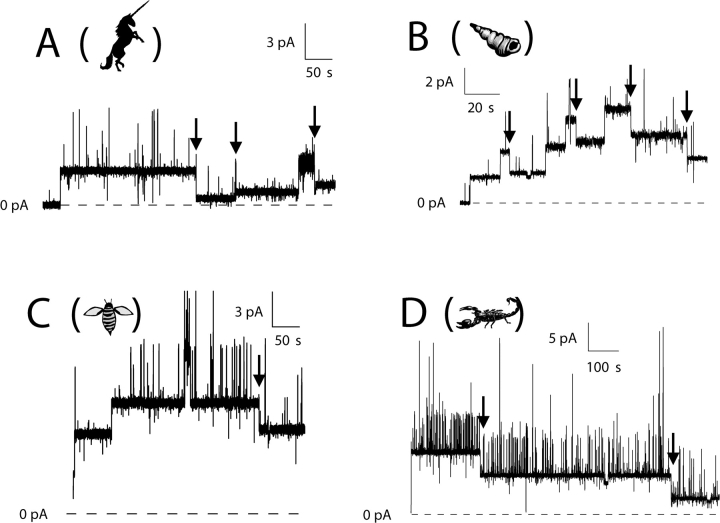
Structures of the stopper peptides. Each stopper is viewed along its longest axis, so that the smallest cross-sections are shown. The cross-sectional diameter, defined by the circumscribed circle (dashed line) is given for each stopper. (A) Peptide U, 12 Å. (B) α-Conotoxin ImI, 15 Å. (C) Apamin, 19 Å. (D) CTX, 26 Å. (E) Streptavidin tetramer bound to biotin, 61 Å. Details about the stopper structures and the estimation of their diameters are given in materials and methods. The pictures of animals in this and subsequent figures are provided to aid in matching each record with the corresponding stopper. In A–D, respectively, the animal represents the natural source of the stopper: unicorn–peptide U; cone snail–α-conotoxin; honeybee–apamin; scorpion–CTX.
Structures of α-conotoxin ImI were obtained from the Protein Data Bank (Berman et al., 2000) files 1IMI (Maslennikov et al., 1999), 1IM1 (Rogers et al., 1999), 1CNL (Gehrmann et al., 1999), and 1G2G (Lamthanh et al., 1999), and inspected as above. The smallest diameter, 15.2 Å, was found in 1CNL, model 7 (Fig. 2 B).
Six sets of coordinates for the structure of apamin were kindly provided by D.E. Wemmer (Pease and Wemmer, 1988). The structure in model 6 had the smallest diameter, 17.7 Å. Because this model did not include hydrogen atoms, we added 1 Å to get 18.7 Å (Fig. 2 C).
The CTX structures came from Protein Data Bank file 2CRD (Bontems et al., 1992); the smallest diameter was 26.4 Å in model 9 (Fig. 2 D). The structures of the streptavidin tetramer bound to two or four biotin molecules were from files 1SWD and 1SWE, respectively (Freitag et al., 1997). Both structures had a minimal diameter of 59.7 Å; adding 1 Å to account for the missing hydrogen atoms gives 60.7 Å (Fig. 2 E).
Of course, the stopper peptides are somewhat flexible, so they may be able to assume conformations with a smaller diameter than those we have estimated above. For instance, allowing the projecting Arg and Lys side chains to bend out of the way might reduce the CTX diameter by a few angstroms. However, the reason we chose to use stoppers with a disulfide-bonded core was that we expected their dimensions would be reasonably well defined. This expectation was supported by inspection of the available structures. The standard deviation of the minimal diameter was less than or equal to 1.0 Å for the calculated peptide U structures, the apamin and CTX NMR structures, and the two streptavidin crystal structures. This was also true for the structures in each of the α-conotoxin files; however, the mean diameter varied by up to 3.2 Å among these files. Note that our CTX stopper contains three mutations, whereas the structure in Fig. 2 D is wild-type CTX.
Visual inspection of the stopper structures using RasMol showed them to be quite polar molecules, with no significant hydrophobic patches on the surface. Thus, these stoppers are well suited for probing a passageway that we know can translocate parts of the very polar colicin Ia protein.
Planar Bilayer Experiments
Folded planar lipid (asolectin) bilayer membranes (100–150 μm in diameter) were formed at room temperature as previously described (Kienker et al., 2000). The membrane separated two 1-ml compartments, each containing a solution of 100 mM KCl, 5 mM CaCl2, 1 mM EDTA, 20 mM MES, pH 6.2. For single-channel experiments, 1 M KCl was used. The voltage-clamp recording setup was as described previously (Kienker et al., 2000). Voltages are given as the potential of the cis compartment, defined as the side to which colicin Ia C-domain was added, with respect to that of the opposite trans compartment. Streptavidin and avidin were obtained from Calbiochem. The antibody to the His6-tag was mouse Penta-His mAb (QIAGEN), mouse anti–β-galactosidase mAb was from Promega, rabbit anti-CTX polyclonal antibody was from Alomone Labs, and rabbit anti-HA.11 polyclonal antibody was from Covance.
Analysis of Normal-State Dwell Times
C-domain channels initially open to a state of “normal” conductance (like that of whole colicin Ia); they may subsequently make an abrupt transition to a small-conductance open state (Fig. 1 B). This transition is closely associated with the translocation of the amino terminus of the C-domain across the membrane to the trans side (Fig. 1 E). Thus, the dwell time in the normal state contains information about how readily the amino-terminal end is translocated. As we shall see, similar conductance transitions can be observed in channels formed by C-domain with an amino-terminal stopper peptide. In this case, the dwell time in the normal state reflects the rate of stopper translocation.
We show the distribution of normal-state dwell times in a survival plot, in which the ordinate is the percentage of events observed with a dwell time greater than the time t plotted on the abscissa. For example, if four channels made transitions in times t 1, t 2, t 3, and t 4, ordered from shortest to longest, then we would plot symbols at (0, 100%), (t 1, 75%), (t 2, 50%), (t 3, 25%), and (t 4, 0%). As another example, if two channels made transitions in times t 1 and t 2, and the other two channels did not make a transition within the maximum time T allowed, then we would plot symbols at (0, 100%), (t 1, 75%), (t 2, 50%), and (T, 50%). The small-conductance channel can be returned to the original closed state by negative voltages (Kienker et al., 2000); thus, we could make several measurements of the normal-state dwell time from an individual channel. In general, however, each survival plot contains lifetimes of several different channels, each measured multiple times.
If all the channels are identical and the normal state is a single Markov state, then the normal-state lifetimes should be distributed as a single decaying exponential. In reality, a single exponential function is generally inadequate to fit the survival plots for C-domain with or without a stopper (see below). Thus, we usually fit the survival plots using a sum of two exponential components: A exp(−t/τ1) + (100% − A) exp(−t/τ2).
RESULTS
Validity of the Translocation Assay
The conclusions regarding the translocation pathway that are derived from the results to follow rest on the evidence that the transition of C-domain channels from a normal-conductance state (similar to that of channels formed by whole colicin Ia) to a small-conductance state (Fig. 1 B) represents the translocation of the amino terminus from the cis to the trans side of the membrane, and thus the conversion from a channel with four transmembrane segments to one with only three (Fig. 1, D and E). It is therefore appropriate, before proceeding, to review this evidence. We have shown previously that in channels formed by whole colicin Ia, the amino-terminal region is on the cis side of the membrane, and that there are four membrane-spanning segments, with the most upstream segment formed by part of helix 1 (Qiu et al., 1996). In channels formed by the isolated C-domain, on the other hand, the amino terminus ends up on the trans side, leaving only three membrane-spanning segments (Fig. 1 E) (Kienker et al., 2000). Initially the C-domain channel opens to a conductance similar to that of the whole colicin Ia channel (Fig. 1 B), consistent with its having four membrane-spanning segments. It drops to a conductance that is the same as that generated in whole colicin Ia channels when trypsin added on the trans side of the membrane cleaves the colicin in the loop between helices 2 and 3, thereby leaving only three membrane-spanning segments (Kienker et al., 2000).
All of this is consistent with the transition from the normal to the small conductance level being a consequence of the movement of the membrane-spanning portion of helix 1, along with its amino terminus, across the membrane to the trans side. Confirming this interpretation is the fact that when the amino terminus of the C-domain is held on the cis side of the membrane by streptavidin, the transition to the small conductance level does not occur; i.e., when the upstream membrane-spanning segment formed by part of helix 1 cannot move across the membrane, the conductance stays at the higher level (Kienker et al., 2000). These observations convinced us that, for a C-domain protein with a stopper appended to its amino terminus, the presence or absence of transitions from a normal to small channel conductance state would provide a way of knowing whether the stopper had traversed the membrane.
Direct Demonstration of Stopper Translocation
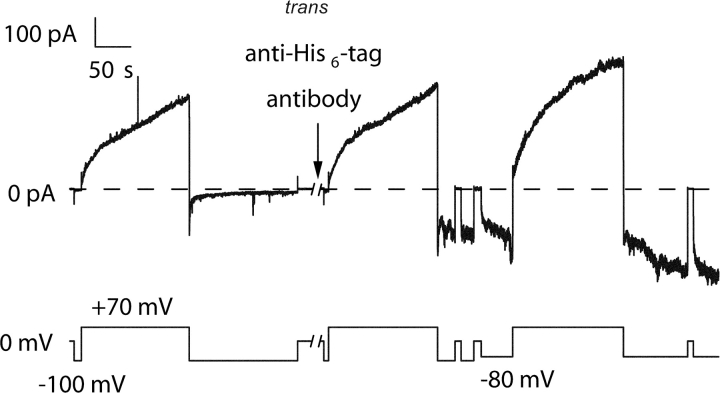
With two of the stoppers, peptide U and CTX, we directly demonstrated, without having to rely on our interpretation of the conductance transitions, that they could be translocated across the membrane. In experiments with the peptide U stopper, we found that the amino terminal His6-tag (which had not been removed for these experiments) was accessible to anti–His6-tag antibody in the trans compartment. Thus, although channels formed by this construct showed the normal gating pattern of turning on at positive voltages and off at negative voltages, a new conductance pattern developed when mouse anti–His6-tag mAb was added to the trans compartment, in which there was a rise in conductance at negative voltages (Fig. 3) . (This was observed in n = 3 separate experiments.) The reason why channels formed by the isolated C-domain appear to reverse orientation when bound by trans anti–His6-tag antibody or trans streptavidin (if they are biotinylated near the amino terminus) is not clear, but it is a phenomenon we have reported previously for several C-domain constructs (Kienker et al., 2000). Whatever the mechanism, the important point in this instance is that the amino terminal His6-tag, along with the peptide U stopper, has been translocated across the membrane. In control experiments, mouse anti–β-galactosidase mAb added to the trans compartment did not affect channel gating.
Figure 3.
The effect of trans anti–His6-tag antibody on channels formed by C-domain with a peptide U stopper and an amino-terminal His6-tag. Before the start of the record, 56 ng of C-domain with a peptide U stopper and an amino-terminal His6-tag were added to the cis compartment. Normal gating was observed, with the conductance turning on at +70 mV and off at voltages from −50 to −100 mV. During the 2-min break in the record, 2 μg of anti–His6-tag antibody were added to the trans compartment. A new conductance rapidly developed that turned on at negative voltages (−100 or −80 mV) and off at 0 mV or positive voltages—the reverse of the normal voltage dependence. This demonstrates that the amino-terminal His6-tag was accessible to trans antibody, and indicates that the peptide U stopper is translocated to the trans side. The solution on both sides of the membrane was 100 mM KCl, 5 mM CaCl2, 1 mM EDTA, 20 mM MES, pH 6.2. The record was filtered at 30 Hz.
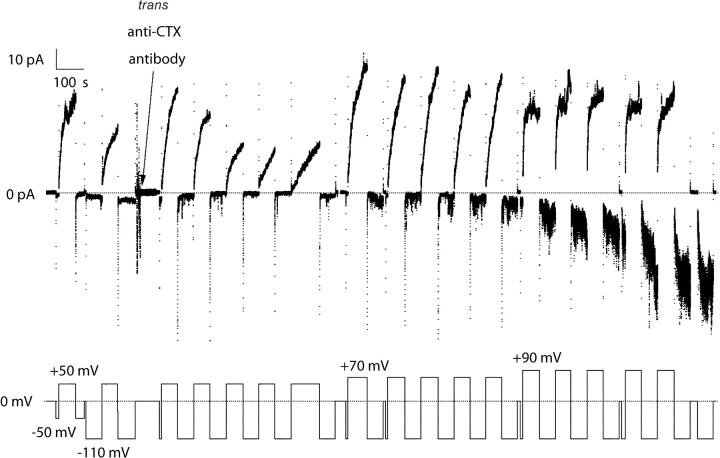
In experiments with the CTX stopper, rabbit polyclonal anti-CTX added to the trans compartment caused a “reverse turn-on” conductance at negative voltage similar to that described in the previous paragraph (Fig. 4) (n = 7). Interestingly, this effect did not develop if the channels were turned on by small positive voltages (+50 or +70 mV), but only if they were turned on at larger positive voltages (+90 to +110 mV) (Fig. 4). It appears that large voltages are required for the translocation of the CTX stopper. This observation is confirmed in the single channel conductance transition experiments described in a later section. The addition of a control antibody (rabbit polyclonal anti-HA.11) to the trans compartment had no effect on channel gating.
Figure 4.
The effect of trans antibody to CTX on C-domain with a CTX stopper, and the dependence of the effect on the turn-on voltage. Before the start of the record, 9 ng of C-domain with a CTX stopper and without a His6-tag were added to the cis compartment. Normal gating was seen, with the conductance turning on at +50 mV and off at −50 or −110 mV. At the arrow, 3 μg of anti-CTX antibody were added to the trans compartment. Conductance that was subsequently turned on at +50 mV continued to turn off normally at −110 mV. With a turn-on voltage of +70 mV, a small amount of reverse turn-on developed at −110 mV. With a turn-on voltage of +90 mV, a substantial reverse turn-on developed at −110 mV. Thus, the amino-terminal CTX stopper was accessible to trans antibody when a large turn-on voltage was used. The solution on both sides of the membrane and the filtering were the same as in Fig. 3. The membrane was reformed after the colicin addition and before the start of the record; this apparently introduced a small amount of colicin to the trans side, as evidenced by the small conductance at negative voltages at the beginning of the record. These channels washed away shortly after the antibody was stirred in. (Similar effects of trans anti-CTX were observed in experiments without a reformed membrane.)
Although these macroscopic experiments with antibody to His6 and CTX do not yield the kind of quantitative data presented in the following sections, they clearly establish that the translocation pathway is (or can be made) large enough to accommodate peptide U, our smallest stopper, and CTX, our largest stopper.
Single-Channel Analysis of Folded Stopper C-Domain Proteins
The three-dimensional structures of the four stoppers that were attached to the end of colicin Ia C-domain are depicted in Fig. 2, A–D. Their diameters range from 12–26 Å. The colicin Ia C-domain proteins, with their amino-terminal stoppers, were added to planar bilayers, as described in materials and methods. Fig. 5 shows representative single-channel records from these experiments. Channels formed by C-domains with the 12- and 15-Å-diameter amino-terminal stoppers (peptide U and α-conotoxin, respectively) opened in the normal-conductance state and rapidly dropped to the small-conductance state (Fig. 5, A and B), indicating that these stoppers are readily translocated at +50 to +70 mV. Channels formed by C-domain with the 19-Å-diameter amino-terminal stopper (apamin) occasionally, and after considerable time, dropped from the normal to the small conductance state at +70 mV (Fig. 5 C). Channels formed at +70 mV by C-domain with the 26-Å-diameter amino-terminal stopper (CTX) remained locked in the large conductance state (unpublished data). Fig. 6 shows more systematically the effect of stopper size on the transition rate at +40 mV. Each curve is a survival plot from a single experiment, showing the percentage of channels whose dwell time in the normal-conductance state was greater than the time plotted on the abscissa (see materials and methods). In most cases, the distribution of dwell times for a given condition is fit by a sum of two exponential components. Plots are shown for C-domain without a stopper and with the peptide U, α-conotoxin, and apamin stoppers. These plots confirm our initial judgment that, at a given voltage, larger stoppers make slower transitions than do smaller stoppers.
Figure 5.
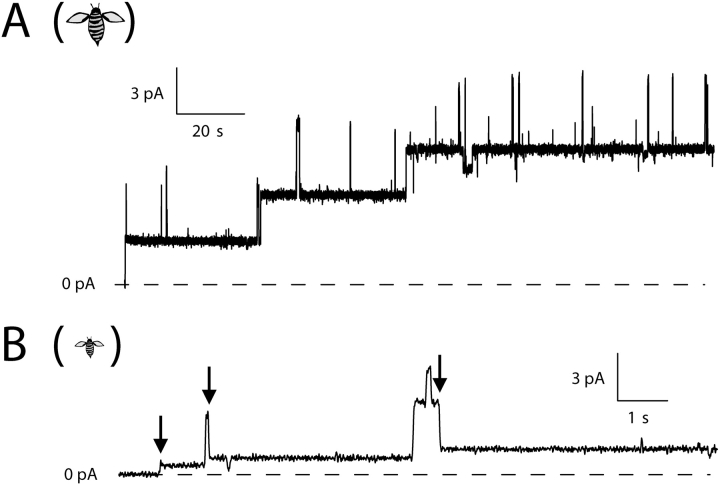
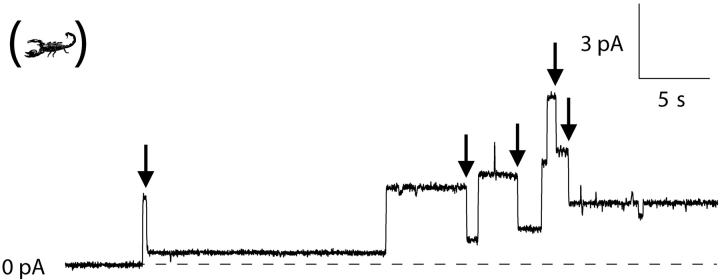
Representative single-channel records for C-domain with different amino-terminal stoppers. (A) With a 12 Å stopper (peptide U), channels readily made transitions to the small-conductance state. Three channels in succession opened at +70 mV with approximately normal conductance (33–42 pS) and dropped to the small-conductance state (each 8 pS), as noted by the arrows. The second channel made its transition quickly, so that on the slow time scale of the figure the normal-conductance state appears as a brief spike. (B) With a 15-Å stopper (α-conotoxin), channels also made transitions readily. Of the five channels that opened at +50 mV with normal conductance (39–49 pS), four made transitions (at the arrows) to the small-conductance state (6–9 pS). (C) With a 19-Å stopper (apamin), channels made transitions to the small-conductance state very slowly or not at all at +70 mV. Shown are three channels, which opened at +70 mV with normal conductance (38–51 pS) (the second channel opened ∼2 s after the first). After more than 4 min at this level, one of the channels dropped in conductance by 32 pS, as noted by the arrow, to a small-conductance state. The other two channels did not make transitions during the record. (D) With a 26-Å stopper (CTX), it was necessary to go to +90 mV to see conductance transitions at an appreciable rate. The record shows two channels that opened quickly at +90 mV with normal conductance (60–63 pS). After long delays (4–14 min), each channel made a transition (noted by the arrows) to a small-conductance state (14–16 pS). Solutions were the same as described in Fig. 1. The amount of stopper–C-domain protein added to the cis solution for each experiment was as follows: A, 3.4 ng; B, 44 ng; C, 90 pg; D, 2 ng. In each of the records, channels also made brief transients to a high-conductance state (60–70 pS above the normal-conductance level); these events are not considered in this paper. Animals are as described in Fig. 2.
Figure 6.
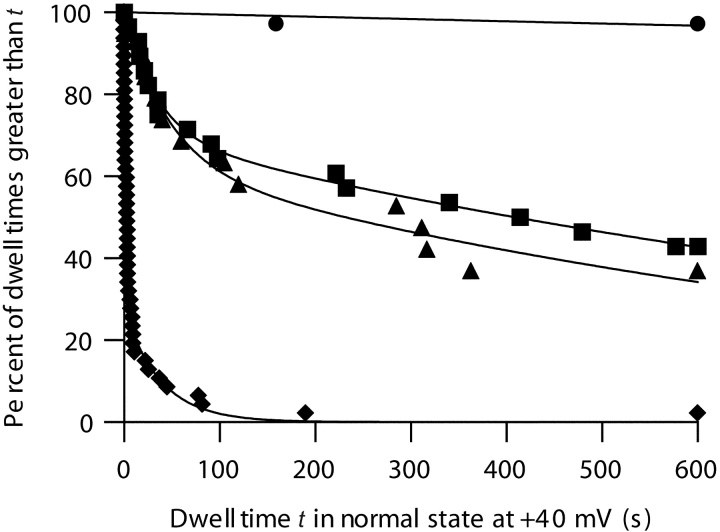
The effect of stopper size on the transition rate of C-domain channels at +40 mV. Survival plots, each from a single membrane, show the distribution of dwell times in the normal-conductance state before a transition to the small-conductance state. Each curve is fit by a sum of two decaying exponential components, with time constants τ1 and τ2 and amplitudes A and (100% − A), respectively. The exception is the apamin curve, which is fit by a single exponential. n is the number of channels measured. No stopper, ♦, n = 47, τ1 = 2.5 s, τ2 = 38 s, A = 73%. Peptide U stopper, ▴, n = 19, τ1 = 46 s, τ2 = 980 s, A = 37%. α-Conotoxin stopper, ▪, n = 28, τ1 = 34 s, τ2 = 1200 s, A = 30%. Apamin stopper, •, n = 36, τ = 18000 s. Transitions are fastest with no stopper, and become progressively slower as larger stoppers are used. Solutions were the same as described in Fig. 1.
From these experiments at voltages up to +70 mV, it would appear that the translocation pathway for the amino terminus is ∼19 Å wide at its narrowest part and that the 26-Å-diameter CTX, like the much-larger streptavidin discussed in the introduction, cannot pass through the transmembrane passageway. This conclusion, however, is erroneous. The translocation rate is strongly voltage dependent, so that although at +70 mV it appears that the apamin stopper reluctantly crosses the membrane and the CTX stopper does not cross at all, at larger voltages, both of these stoppers readily cross the membrane (Figs. 5 D and 7). Fig. 7 presents survival plots for the four stoppers at several different voltages. These plots show unambiguously that all of the stoppers make faster transitions at larger positive voltages (n = 2–3 for each stopper). Perhaps the most dramatic illustration of the voltage dependence is for the apamin stopper (Fig. 7 C): at +110 mV, all 25 channels made transitions in <25 s, whereas at +40 mV, only 1 channel out of 36 made a transition in <600 s.
Figure 7.
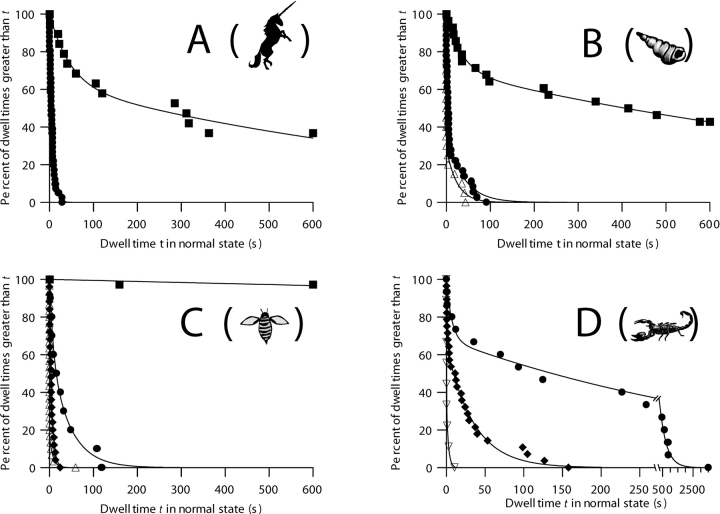
Survival plots illustrating the effects of voltage and disulfide bond reduction on the transition rate of C-domain with each of the peptide stoppers. Each curve represents a series of dwell times measured at the same voltage. The measurements at different voltages for a given unreduced stopper are all from a single membrane; the measurements for each reduced stopper were obtained on a different single membrane. Unreduced stoppers: ▪, +40 mV; •, +90 mV; ♦, +110 mV. Reduced stoppers: ▵, +70 mV; ▿, +80 mV. Parameters have the same meanings as described in Fig. 6. The data at +40 mV in A, B, and C are the same as described in Fig. 6, so the fitting parameters are not repeated here. (A) Peptide U. +90 mV, n = 41, τ = 6.33 s. (See Fig. 8 for the effect of disulfide bond reduction.) (B) α-Conotoxin. +90 mV, n = 36, τ1 = 2.6 s, τ2 = 38 s, A = 63%. +70 mV, reduced, n = 20, τ1 = 0.07 s, τ2 = 24 s, A = 71%. (C) Apamin. +90 mV, n = 10, τ1 = 8 s, τ2 = 44 s, A = 37%. +110 mV, n = 25, τ1 = 0.1 s, τ2 = 5.6 s, A = 12%. +70 mV, reduced, n = 30, τ1 = 0.045 s, τ2 = 8 s, A = 84%. The +90 mV apamin data may be biased toward shorter dwell times, because several transitions that occurred with multiple open channels in the normal state were excluded from the analysis. (D) CTX. Note the change of scale at the break in the time axis. +90 mV, n = 15, τ1 = 7 s, τ2 = 440 s, A = 32%. +110 mV, n = 28, τ1 = 1.3 s, τ2 = 35 s, A = 35%. +80 mV, reduced, n = 9, τ1 = 0.19 s, τ2 = 2 s, A = 64%. For all of the stoppers, the rate of transition to the small-conductance state is conspicuously voltage dependent, with faster transitions at larger positive voltages. Disulfide bond reduction, which allows the stoppers to unfold, also greatly increased the transition rates. For the folded stoppers, the solutions were the same as described in Fig. 1; for the unfolded stoppers, both the cis and trans solutions also contained 1–20 mM TCEP.
Just as we had observed for C-domain without a stopper (Kienker et al., 2000), it is also true with a stopper that the small-conductance state is stable, in the sense that small channels do not revert to the normal-conductance state while the positive voltage is maintained. At a sufficiently large negative voltage, the small channels can be driven back to the original closed state (i.e., with the amino terminus back on the cis side), often passing through some higher-conductance states on the way. Although we have not systematically studied the closing rate, it is clearly voltage dependent, being quite slow (several minutes) at −50 mV and fast (seconds) at −200 mV.
We have shown previously that binding streptavidin near the amino terminus of C-domain prevents the transition to the small-conductance state at +50 mV (Kienker et al., 2000); that is, the streptavidin “stopper” is not translocated. In light of the strong effect of voltage on the transition rate of the other stoppers, we repeated the streptavidin experiments at larger voltages. C-domain with a biotin near the amino terminus and without a His6-tag was incubated with streptavidin, and unbound C-domain was then removed using a sizing column (see materials and methods). This material was added to the cis compartment in a bilayer experiment. Seven channels were each held at +110 mV for over an hour without making any transitions to the small-conductance state (unpublished data) (n = 5). (At +120 mV, the channels began to slowly inactivate—that is, to enter a completely closed state—so we were unable to use larger voltages.) Results with avidin-bound C-domain were similar, but were more difficult to analyze due to a slow “blocking” and “unblocking” of the channels (n = 5). So, at least at voltages up to +110 mV, the 61-Å-diameter streptavidin and avidin stoppers cannot be translocated.
Effect of Unfolding the Stoppers
The validity of the above results, and hence the conclusion about pathway width drawn from them, is predicated on two assumptions: (1) that these stoppers, which we claim traverse the membrane, are internally disulfide bonded and therefore have the conformations depicted in Fig. 2; and (2) that the slower rates (at a given voltage) at which the large diameter stoppers (CTX and apamin) traverse the amino-terminal translocation pathway are a consequence of steric constraints, rather than being due to some property intrinsic to their sequences, such as their numerous positive charges. The following results address these issues.
We have removed any detectable C-domain molecules having free –SH groups in their stoppers (materials and methods). Presumably, therefore, the molecules that we tested on the bilayers all had the internal disulfide bonds formed in their stoppers, with the exception of minor contamination from dimers and higher multimers. As confirmation of both this and the validity of our entire approach, we found that when these disulfide bonds were reduced, thereby unfolding the stoppers, it became easier for them to traverse the pathway, as evidenced by the shortening of dwell times in the normal-conductance state. Fig. 8 shows an example of this. Several minutes after the addition of the disulfide-reducing agent TCEP, the transition rate from the normal-conductance to the small-conductance state of C-domain channels with the amino-terminal peptide U stopper significantly increased (n = 4). The effect of reduction was particularly pronounced on C-domain channels having the amino-terminal apamin stopper (Fig. 9) , which has two disulfide bonds (n = 6). Prior to TCEP addition, the dwell times in the open state at +70 mV were of the order of minutes, whereas following reduction they were merely several seconds or shorter.
Figure 8.
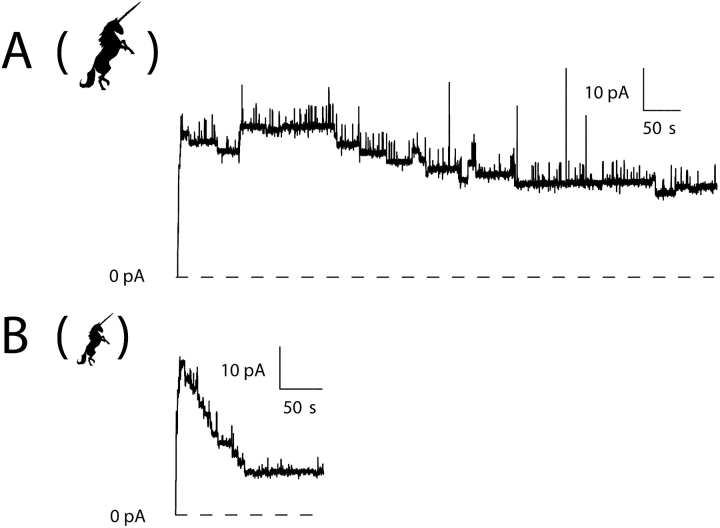
Comparison of translocation rates of peptide U stopper attached to C-domain before and after disulfide bond reduction. Each record shows many channels (∼20), which opened at +70 mV with normal conductance and then dropped to the small-conductance state. Records with most of the channels opening at the beginning of the voltage pulse were selected, so that the rate of transition to the small-conductance state is reflected in the decay of the current. (A) Channels formed by unreduced peptide U–C-domain dwelled in the normal-conductance state for a relatively long time (τ ∼200 s) before making the transition to the small-conductance state. (B) Later in the same experiment, after TCEP was added to reduce the disulfide bond in peptide U, channels made their transitions much more quickly (τ ∼30 s). This demonstrates that the cysteines in the stoppers of the channels making transitions in A were in fact disulfide bonded. The concentration of TCEP was 9 mM on the cis side and 5 mM on the trans side. The transition rate increased markedly within a few minutes of TCEP addition; trace B happens to begin 25 min after TCEP was added. Channels in the small-conductance state could be closed by a brief pulse to −200 mV (not shown), after which they were ready to reopen through the normal-conductance state in response to a subsequent positive voltage pulse. Thus, the population of channels in trace B is more or less the same as that in trace A. The unicorn icon in B is reduced in size to indicate that a reducing agent was added. Solutions were the same as described in Fig. 1, except that TCEP was added in B. The amount of peptide U–C-domain added to the cis solution at the beginning of the experiment was 1.4 ng.
Figure 9.
Comparison of apamin stopper translocation by C-domain before and after disulfide bond reduction. (A) Three channels formed by unreduced apamin–C-domain opened at +70 mV with normal conductance (42–43 pS) and made transitions to small conductance very slowly or, as in this record, not at all. (B) Later in the same experiment, after TCEP was added to reduce the disulfide bonds in apamin, channels made rapid transitions (τ < 1 s), indicated by the arrows. The first channel made its transition so quickly that the transient, normal-conductance state was not resolved. The concentration of TCEP was 5 mM on both the cis and trans sides. The transition rate increased markedly within a few minutes of TCEP addition; trace B happens to begin 45 min later. Solutions were the same as described in Fig. 1, except that TCEP was added in B. The amount of apamin–C-domain added to the cis solution at the beginning of the experiment was 3.6 ng.
In the case of CTX, it is not possible to reduce its three disulfide bonds by simple TCEP treatment at room temperature, as was done for the other stoppers. However, it is known that incubating the toxin for 10 min at 70°C in 10 mM DTT reduces these bonds (Naini et al., 1996). C-domain with its amino-terminal CTX survived this treatment and produced channels that now rapidly underwent the transition from the normal to the small conductance state at +70 mV (Fig. 10) (n = 3), a voltage at which no transitions were observed with unreduced CTX. Thus, unfolded CTX is fully capable of traversing the translocation pathway for the amino terminus.
Figure 10.
Fast translocation of reduced CTX stopper by C-domain. (A corresponding experiment with unreduced CTX is shown in Fig. 5 D.) CTX–C-domain was reduced as described in materials and methods before 100 ng of it were added to the cis solution. The record shows five channels, which opened at +70 mV with normal conductance (37–39 pS), and quickly dropped to the small-conductance state (6–8 pS), as highlighted by the arrows. Both the cis and trans solutions contained 1.0 mM TCEP to keep the sulfhydryl groups reduced. In this experiment, 10 μg of octylglucoside were added to the cis solution to promote channel activity, but in general, channels and transitions could be observed without octylglucoside. Solutions were the same as described in Fig. 1, except for the addition of TCEP.
Survival plots for the reduced α-conotoxin, apamin, and CTX stoppers are shown in Fig. 7 B–D (▵ and ▿). In each case, the transition rate with the reduced stopper is faster than with the unreduced stopper, even though more moderate voltages were used with the reduced stoppers. This confirms that the unfolded, reduced stoppers can be translocated more easily than the folded, unreduced stoppers.
DISCUSSION
We describe here a method of characterizing the size of a translocation pathway for protein across a lipid bilayer membrane. We attached a series of small peptides, held in a fixed conformation by one or more disulfide bonds, very near the amino-terminus of the colicin Ia channel-forming (C) domain, and found that all of these stoppers, whose minimal diameters ranged from 12 to 26 Å (see materials and methods), crossed the membrane. There was a strong voltage dependence to their translocation, such that at +40 to +70 mV, the larger stoppers crossed slowly, if at all; at +90 to +110 mV they crossed readily (Figs. 5–7). That the size of the stoppers was the relevant parameter governing their relative rates of translocation in our experiments (rather than some other factors, such as charge or specific amino acid composition) was illustrated by the much greater translocation rates after their unfolding by disulfide bond reduction (Figs. 7–10). The translocation pathway is thus large enough to allow the passage of an α-helix, or even a pair of α-helices packed together side by side. In our earlier experiments with carboxy-terminal fragments of colicin Ia (Kienker et al., 2000), it is therefore likely that all of helix 1 (residues 359–467) was translocated as an α-helix. The R domain (residues 282–385) has a larger cross-section than CTX, however (Wiener et al., 1997), so it must have been at least partially unfolded in those experiments in which it moved across the membrane at voltages up to +70 mV.
A possible concern about the sizes we have assigned to the stoppers is that they may not be folded in the configurations shown in Fig. 2. Although we are reasonably confident that we have removed stoppers with free sulfhydryls (see materials and methods), it is possible that the pairing of the cysteines is different from that in the native structures. The stoppers that were inserted genetically also have their ends constrained by the rest of the molecule into which they were inserted, and therefore may not fold as the free stopper peptides do. This is obviously not a concern for peptide U, which has only two cysteines. Nor is it a concern for the recombinant, bacterially expressed CTX. Although it has six cysteines, it has been shown to fold into its native conformation and block potassium channels in the same way as the native scorpion toxin (Park et al., 1991). It is conceivable that the α-conotoxin and apamin stoppers, each of which has four cysteines, could have formed nonnative disulfide bonds, in which case the stopper diameters would be somewhat different from those given. However, at least for apamin, it appears that rather drastic alterations in the peptide sequence are required to change the disulfide pairing (Volkman and Wemmer, 1997).
In our description of the C-domain with its stoppers, we have implied that the stoppers are located exactly at the amino terminus. This is not precisely correct. After removal of the amino-terminal histidine tags from the expressed constructs, there remain five (in the case of peptide U) or six (in the case of α-conotoxin and apamin) residues amino-terminal to the stoppers. Depending on the details of the stopper structures (i.e., on the proximity of the amino and carboxy termini), this short extension may lie next to the stopper during translocation, in which case the effective diameter of the stopper would be somewhat larger. The situation is even more complicated with the CTX stopper, which was not inserted genetically into the C-domain construct, but rather was chemically linked to residue 439 of the colicin Ia C-domain (see materials and methods). Thus, CTX is attached via a linker near its own middle to a residue six positions from the amino terminus of the C-domain. Unfolding CTX by reducing its disulfide bonds therefore yields a structure with considerable branching near its amino terminus. Nevertheless, the translocation pathway is able to accommodate its passage.
The distribution of dwell times of the channel at the normal-conductance level (in the state with four membrane-spanning segments) before undergoing a transition to the small-conductance level (the state with three membrane-spanning segments) is fit by two (conceivably more) exponentials (Figs. 6 and 7), implying that the channel has at least two normal-conductance open states. This is seen for each of the stoppers and also for the carboxy-terminal fragment 438–626, which lacks a stopper (Figs. 6 and 7). It is our impression that a given channel has only one normal-conductance open state—it either always has a short dwell time distribution or always has a long dwell time distribution—so that the two exponentials reflect two different channel populations. Since we are dealing with one particular protein (for a given stopper), this is somewhat surprising. One possibility is that a given molecule associated with the bilayer can be locked into two or more particular configurations that are not readily interconvertible. For instance, different colicin molecules might have the proline at residue 566 (the only proline residue in the C-domain) in either the cis or the trans isomer, with the resulting channels having different conformations. Further work is required to clarify this issue.
Voltage Dependence of Translocation Rate
A striking characteristic of the stopper translocation kinetics we measured is their strong voltage dependence—the larger the positive voltage, the more rapid the translocation (Fig. 7). The mechanism of this voltage dependence is not clear. Given the positive charge of the conotoxin, apamin, and CTX stoppers, an obvious explanation is a simple electrophoretic effect. This cannot explain, however, the same sort of voltage dependence seen with the peptide U stopper, which carries a negative charge. A possible mechanism is that the applied electric field is acting directly on the translocation pathway and affecting its size. Thus, the diameter of this pathway may not be fixed, but rather may be a function of the applied transmembrane potential. Such an effect of potential is seen on the size of lipid pores (Abidor et al., 1979; Chernomordik et al., 1983). A feature of this type of mechanism is that large voltages of either sign should increase the pore diameter and, in our case, increase the translocation rate. (Because the normal-conductance channels rapidly turn off at negative voltages, this aspect of the mechanism is difficult to test. We are presently attempting to design experiments that get around this problem.) There is also evidence for protein translocation pores with a variable diameter in the endoplasmic reticulum translocon (Hamman et al., 1997, 1998) and the mitochondrial inner membrane (Schwartz and Matouschek, 1999).
Comparison with Other Protein Translocation Pathways
The sizes of various conducting channels have been estimated in countless studies, dating to antiquity (Hille, 1971); some of these channels are permeable to large molecules, such as polyethylene glycols and single-stranded nucleic acids (Bezrukov, 2000). In what follows, however, we shall restrict our discussion to pores or other pathways whose function is to allow proteins to cross a membrane.
Self-inserting translocators
Several bacterial protein toxins consist of a toxic enzymatic domain that is attached (usually covalently) to a domain that is responsible for translocating the enzyme across a lipid membrane into the cytosol of the target cell, where it exerts its toxic effect. In many cases, the translocation domain has been shown to form a conducting channel, raising the possibility that the enzyme might be translocated through the channel.2 For both diphtheria toxin (Oh et al., 1999) and botulinum neurotoxin (Koriazova and Montal, 2003), no additional proteins are required for translocation across planar lipid bilayers. Interestingly, the membrane topology of the diphtheria toxin channel is similar to that of the colicin Ia C-domain small channel (Kienker et al., 2000; Senzel et al., 2000). Anthrax toxin also makes a channel and translocates two separate enzymes; there is indirect evidence from planar bilayer experiments that they may be translocated through the conducting channel (Finkelstein, 1994). Most likely, the enzymatic domains of these toxins are translocated in an unfolded conformation (Wesche et al., 1998), so that the sizes of their translocation pathways are not clear.
Other translocation domains may transport their passenger domains in a folded conformation. The bacterial autotransporters constitute one class of such proteins, in which one domain is thought to export another domain across the bacterial outer membrane by forming a β-barrel pore through which the passenger domain passes (Henderson et al., 1998). One example is the IgA protease of Neisseria gonorrhoeae, whose pore domain is capable of exporting an attached passenger protein containing two disulfide bonds. As we have shown here for colicin Ia C-domain, the passenger protein was translocated more readily in its reduced state, but there was, nevertheless, significant transport of the unreduced, folded protein (Veiga et al., 1999). Another autotransporter, Antigen 43 of Escherichia coli, can export a separate passenger protein containing a disulfide bond (Kjærgaard et al., 2002). Streptolysin O belongs to another class of bacterial proteins that form conducting channels. It seems that its main function is to translocate the enzyme NAD-glycohydrolase across eukaryotic cell membranes (Madden et al., 2001). If the large (300-Å) pores visualized by electron microscopy (Palmer et al., 1998) represent the functional channel, then the 52-kD enzyme could easily be translocated as a folded protein. Finally, the bacteriophage λ holin protein forms a pore in the inner membrane of the target bacterium that allows the passage of an endolysin protein. The purified holin is sufficient to cause calcein release from liposomes (Smith et al., 1998). In vivo, endolysin fused to β-galactosidase can still pass through the holin pore; it was argued that the β-galactosidase portion was translocated as a folded tetramer of 480 kD (Wang et al., 2003).
For most, if not all, of these proteins, the detailed mechanisms of protein translocation have not yet been elucidated; thus, it remains to be seen whether any of them (with the possible exception of diphtheria toxin) operates by a mechanism similar to that of colicin Ia.
Endogenous translocation pathways
There have been many studies of protein translocation across eukaryotic intracellular and bacterial membranes (for review see Schnell and Hebert, 2003). Probably the most extensive studies on the size of a translocation pore have been on the protein import channel of the mitochondrial outer membrane. It has been shown that branched polypeptides (Vestweber and Schatz, 1988b) and a protein loop (Wiedemann et al., 2001) can be imported into mitochondria. A gold cluster 20 Å in diameter can also be imported, whereas a 26-Å cluster cannot (Schwartz and Matouschek, 1999). Bovine pancreatic trypsin inhibitor, which contains three disulfide bonds, is not imported (Vestweber and Schatz, 1988a); its minimal diameter is ∼30 Å. Furthermore, the import of dihydrofolate reductase (∼40–50 Å) is blocked when its folded state is stabilized by the binding of methotrexate (Eilers and Schatz, 1986). Thus, proteins can pass through the outer membrane as an extended strand or as a hairpin, but folded proteins are not normally imported (Schwartz et al., 1999).
Another well-characterized pore is the translocon of the endoplasmic reticulum. Its diameter has been estimated to be 40–60 Å when the ribosome is bound (Hamman et al., 1997) and 9–15 Å in the ribosome-free state (Hamman et al., 1998). Despite the large diameter of the ribosome-bound pore, it appears that nascent secretory proteins traverse the membrane in an extended, α-helical form (Plath et al., 1998).
There are also pores in cell membranes that translocate folded proteins such as those we have used in this study (and even larger proteins). Proteins in the endoplasmic reticulum can be transported back to the cytosol, a process termed dislocation. It has been shown that folded dihydrofolate reductase can be dislocated, indicating that the translocation pathway has a diameter of at least 40 Å (Tirosh et al., 2003). The twin-arginine translocation (Tat) system of bacteria apparently functions principally to move proteins that bind cofactors, and thus must fold in the cytoplasm, across the inner bacterial membrane. The Tat apparatus can translocate proteins as large as the formate dehydrogenase-N GH subcomplex, indicating a diameter of at least 60–70 Å. (Berks et al., 2000; Palmer and Berks, 2003). However, unlike colicin Ia, diphtheria toxin, and botulinum neurotoxin, which are believed to be monomeric, these translocation pores are large, multimeric complexes. Thus, their structures are unlikely to explain how the colicin translocates protein.
On the Structure of the Translocation Pathway
The translocation pathway followed by the amino-terminal stoppers is probably not identical to the ion-conducting channel. The diameter of the conducting channel has been estimated in two ways: by the size of ions that can pass through the narrowest part of the channel and by the size of nonelectrolytes that can fit into some part of the channel. No upper limit has been determined from the ion permeation experiments, but a lower limit of 8–9 Å has been established for colicins E1, A, and Ib (Raymond et al., 1985; Bullock et al., 1992; Bullock and Kolen, 1995), and 7–8 Å for colicin Ia, based on its permeability to bis-tris propane and tetraethyl ammonium (Jakes et al., 1999; Slatin and Kienker, 2003). From the experiments using nonelectrolytes, it was concluded that the cis and trans entrances of the colicin Ia channel are ∼18 and 10 Å in diameter, respectively, with a 7-Å constriction in between (Krasilnikov et al., 1998). It is hard to see how any of our stopper peptides (12–26 Å) could fit through such a pore. Alternatively, one might expect that a stopper would follow C-domain helix 1, to which it is attached, through the membrane; that is, the space normally occupied by helix 1 would serve as the translocation pathway. Conceivably, the smaller stoppers (12–15 Å) could fit through such a pathway, although they would presumably be exposed to lipid. By combining the helix 1 space with the conducting channel, we might get a large enough pathway for apamin (19 Å) to pass through the membrane. But even this combined pathway is apparently much too small to allow the CTX stopper (26 Å) to be translocated, unless the diameter of the conducting channel has been substantially underestimated.
We have not addressed the question of what kind of structure of three transmembrane segments, formed from ∼85 amino acid residues, could mediate the translocation of a passenger as large as the 26-Å CTX. This question is made more problematic by the fact that the colicin channel appears to be a monomer, based on the linear dependence of channel formation on colicin concentration, as well as on the colicin's monomeric state in aqueous solution above pH 4. (See Cavard et al., 1988; Levinthal et al., 1991; Mel and Stroud, 1993; and references therein.) A similar problem exists in the case of the diphtheria toxin T-domain, whose channels appear to be monomeric based on the number of site-specific chemical reactions (one) observed on single channels and on the absence of hybrid channels formed from mixtures of mutant T-domains with distinct gating properties, as discussed by Gordon and Finkelstein (2001). Thus, the only real evidence for a multimeric channel is common sense: how could such a small amount of protein possibly make such a large channel? Although we have no model for the channel structure, it is not unlikely that lipid is involved in the translocation pathway (and in the conducting channel, for that matter).
There is experimental evidence of interaction of both secretory proteins and integral membrane proteins with lipids adjacent to the proteinaceous translocon pore (Martoglio et al., 1995; Mothes et al., 1997; Plath et al., 1998; Heinrich et al., 2000). Since these studies also found interaction of the signal sequences of these proteins with the Sec61 protein, the pathway for protein export in eukaryotes appears to involve both protein and lipid.
It has been reported that a number of “protein transduction domains” are internalized by cells, without the involvement of receptors, transporters, or endocytosis, and that attached proteins and other objects are also carried across the membrane. These domains include relatively short, very basic sequences from the Drosophila Antennapedia transcription factor, the HIV-1 TAT protein (not to be confused with the bacterial Tat pathway discussed above) and herpes simplex virus-1 DNA-binding protein VP22 (Schwarze et al., 2000). It was hypothesized that the Antennapedia domain may translocate itself and an attached passenger by inducing the formation of an inverted micelle in the membrane (Derossi et al., 1996), and likewise for the HIV TAT domain (Vivès et al., 1997). It is tempting to consider this as a model for translocation of stoppers by the colicin C-domain. However, evidence in favor of the translocation of fluorescently-labeled Antennapedia domain in pure lipid bilayers (Thorén et al., 2000) has been balanced by contrary evidence (Drin et al., 2001a,b). It has also been reported that labeled HIV TAT domain does not translocate into liposomes (Krämer and Wunderli-Allenspach, 2003). Furthermore, there has recently been some discussion as to whether the HIV TAT and VP22 domains are truly able to efficiently carry cargo across cell membranes (Falnes et al., 2001; Lundberg and Johansson, 2002; Leifert et al., 2002; Richard et al., 2003; Krämer and Wunderli-Allenspach, 2003).
Other models for a transmembrane pore lined by protein and lipid have been proposed (for review see Gilbert, 2002). Streptolysin O, whose role as a translocation pore was discussed above, has been reported to form large circular pores lined by oligomerized protein in erythrocyte membranes, as visualized by electron microscopy. Mixing of native streptolysin O with a modified, oligomerization-deficient mutant resulted in hybrid oligomers that formed pore structures partially lined by an arc of protein, with the remainder apparently lined by a straight boundary of lipid (Palmer et al., 1998). Presumably, the lipid head groups face into the pore. As evidence for the functional relevance of these structures, the hybrid pores appeared to have a smaller size cutoff for permeant solutes than the native pores. A somewhat different model comes from experimental and theoretical studies of antimicrobial peptides such as magainin, tachyplesin, and melittin (Matsuzaki, 1999; Lin and Baumgaertner, 2000; Huang, 2000; Yang et al., 2001; Zemel et al., 2003). In this picture, the insertion of the peptides into the membrane destabilizes the packing of the bilayer lipids, inducing them to form a toroidal structure, with the peptide monomers separated from one another by lipid. Similar principles may be at work in the induction by polypeptides of lipidic fusion pores (Basañez, 2002). Perhaps the passageway that colicin Ia C-domain creates for its first transmembrane segment (helix 1) and the amino-terminal stoppers we have attached will turn out to have a similar architecture, with the difference that the membrane-spanning segments of the colicin are tethered together by the extramembrane segments.
Acknowledgments
The authors thank Tom Newman for modeling the peptide U structure, David Wemmer for providing the apamin coordinates, and Jessica Muoio for the BioSil chromatography.
This work was supported by National Institutes of Health grants GM29210 (A. Finkelstein) and GM31768 (C. Miller).
Olaf S. Anderson served as editor.
R.O. Blaustein's present address is Molecular Cardiology Research Institute, Tufts–New England Medical Center, Boston, MA 02111.
Footnotes
Abbreviations used in this paper: C-domain, colicin Ia carboxy-terminal domain; CTX, charybdotoxin; H1, helix 1 of the C-domain; His6-tag, 6 histidine tag; Tat, twin-arginine translocation; TCEP, tris (2-carboxyethyl)phosphine hydrochloride.
More precisely, four membrane-spanning segments are contributed by each colicin Ia molecule in the channel. Surprisingly, it appears that there is only one colicin molecule per channel. (See Jakes et al., 1999, for references.)
This may be contrasted with the channel-forming polyene-like antibiotic monazomycin, in which the whole molecule both participates in an oligomeric channel and is subsequently translocated (Heyer et al., 1976).
References
- Abidor, I.G., V.B. Arakelyan, L.V. Chernomordik, Yu.A. Chizmadzhev, V.F. Pastushenko, and M.R. Tarasevich. 1979. Electric breakdown of bilayer lipid membranes. I. The main experimental facts and their qualitative discussion. Bioelectrochem. Bioenerg. 6:37–52. [Google Scholar]
- Basañez, G. 2002. Membrane fusion: the process and its energy suppliers. Cell. Mol. Life Sci. 59:1478–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berks, B.C., F. Sargent, and T. Palmer. 2000. The Tat protein export pathway. Mol. Microbiol. 35:260–274. [DOI] [PubMed] [Google Scholar]
- Berman, H.M., J. Westbrook, Z. Feng, G. Gilliland, T.N. Bhat, H. Weissig, I.N. Shindyalov, and P.E. Bourne. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezrukov, S.M. 2000. Ion channels as molecular Coulter counters to probe metabolite transport. J. Membr. Biol. 174:1–13. [DOI] [PubMed] [Google Scholar]
- Bontems, F., B. Gilquin, C. Roumestand, A. Menez, and F. Toma. 1992. Analysis of side-chain organization on a refined model of charybdotoxin: structural and functional implications. Biochemistry. 31:7756–7764. [DOI] [PubMed] [Google Scholar]
- Bullock, J.O., and E.R. Kolen. 1995. Ion selectivity of colicin E1: III. Anion permeability. J. Membr. Biol. 144:131–145. [DOI] [PubMed] [Google Scholar]
- Bullock, J.O., E.R. Kolen, and J.L. Shear. 1992. Ion selectivity of colicin E1: II. Permeability to organic cations. J. Membr. Biol. 128:1–16. [DOI] [PubMed] [Google Scholar]
- Cavard, D., P. Sauve, F. Heitz, F. Pattus, C. Martinez, R. Dijkman, and C. Lazdunski. 1988. Hydrodynamic properties of colicin A. Existence of a high-affinity lipid-binding site and oligomerization at acid pH. Eur. J. Biochem. 172:507–512. [DOI] [PubMed] [Google Scholar]
- Chernomordik, L.V., S.I. Sukharev, I.G. Abidor, and Yu.A. Chizmadzhev. 1983. Breakdown of lipid bilayer membranes in an electric field. Biochim. Biophys. Acta. 736:203–213. [Google Scholar]
- Cornell, W.D., P. Cieplak, C.I. Bayly, I.R. Gould, K.M. Merz, Jr., D.M. Ferguson, D.C. Spellmeyer, T. Fox, J.W. Caldwell, and P.A. Kollman. 1995. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117:5179–5197. [Google Scholar]
- Derossi, D., S. Calvet, A. Trembleau, A. Brunissen, G. Chassaing, and A. Prochiantz. 1996. Cell internalization of the third helix of the Antennapedia homeodomain is receptor-independent. J. Biol. Chem. 271:18188–18193. [DOI] [PubMed] [Google Scholar]
- Drin, G., H. Déméné, J. Temsamani, and R. Brasseur. 2001. a. Translocation of the pAntp peptide and its amphipathic analogue AP-2AL. Biochemistry. 40:1824–1834. [DOI] [PubMed] [Google Scholar]
- Drin, G., M. Mazel, P. Clair, D. Mathieu, M. Kaczorek, and J. Temsamani. 2001. b. Physico-chemical requirements for cellular uptake of pAntp peptide: role of lipid-binding affinity. Eur. J. Biochem. 268:1304–1314. [DOI] [PubMed] [Google Scholar]
- Eilers, M., and G. Schatz. 1986. Binding of a specific ligand inhibits import of a purified precursor protein into mitochondria. Nature. 322:228–232. [DOI] [PubMed] [Google Scholar]
- Falnes, P.Ø., J. Wesche, and S. Olsnes. 2001. Ability of the Tat basic domain and VP22 to mediate cell binding, but not membrane translocation of the diphtheria toxin A-fragment. Biochemistry. 40:4349–4358. [DOI] [PubMed] [Google Scholar]
- Finkelstein, A. 1994. The channel formed in planar lipid bilayers by the protective antigen component of anthrax toxin. Toxicology. 87:29–41. [DOI] [PubMed] [Google Scholar]
- Freitag, S., I. Le Trong, L. Klumb, P.S. Stayton, and R.E. Stenkamp. 1997. Structural studies of the streptavidin binding loop. Protein Sci. 6:1157–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrmann, J., N.L. Daly, P.F. Alewood, and D.J. Craik. 1999. Solution structure of α-conotoxin ImI by 1H nuclear magnetic resonance. J. Med. Chem. 42:2364–2372. [DOI] [PubMed] [Google Scholar]
- Gilbert, R.J.C. 2002. Pore-forming toxins. Cell. Mol. Life Sci. 59:832–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, M., and A. Finkelstein. 2001. The number of subunits comprising the channel formed by the T domain of diphtheria toxin. J. Gen. Physiol. 118:471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamman, B.D., J.-C. Chen, E.E. Johnson, and A.E. Johnson. 1997. The aqueous pore through the translocon has a diameter of 40-60 Å during cotranslational protein translocation at the ER membrane. Cell. 89:535–544. [DOI] [PubMed] [Google Scholar]
- Hamman, B.D., L.M. Hendershot, and A.E. Johnson. 1998. BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell. 92:747–758. [DOI] [PubMed] [Google Scholar]
- Heinrich, S.U., W. Mothes, J. Brunner, and T.A. Rapoport. 2000. The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell. 102:233–244. [DOI] [PubMed] [Google Scholar]
- Henderson, I.R., F. Navarro-Garcia, and J.P. Nataro. 1998. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 6:370–378. [DOI] [PubMed] [Google Scholar]
- Heyer, E.J., R.U. Muller, and A. Finkelstein. 1976. Inactivation of monazomycin-induced voltage-dependent conductance in thin lipid membranes. II. Inactivation produced by monazomycin transport through the membrane. J. Gen. Physiol. 67:731–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille, B. 1971. The permeability of the sodium channel to organic cations in myelinated nerve. J. Gen. Physiol. 58:599–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H.W. 2000. Action of antimicrobial peptides: two-state model. Biochemistry. 39:8347–8352. [DOI] [PubMed] [Google Scholar]
- Jakes, K.S., P.K. Kienker, and A. Finkelstein. 1999. Channel-forming colicins: translocation (and other deviant behaviour) associated with colicin Ia channel gating. Q. Rev. Biophys. 32:189–205. [DOI] [PubMed] [Google Scholar]
- Kienker, P.K., K.S. Jakes, and A. Finkelstein. 2000. Protein translocation across planar bilayers by the colicin Ia channel-forming domain. Where will it end? J. Gen. Physiol. 116:587–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienker, P.K., X.-Q. Qiu, S.L. Slatin, A. Finkelstein, and K.S. Jakes. 1997. Transmembrane insertion of the colicin Ia hydrophobic hairpin. J. Membr. Biol. 157:27–37. [DOI] [PubMed] [Google Scholar]
- Kjærgaard, K., H. Hasman, M.A. Schembri, and P. Klemm. 2002. Antigen 43-mediated autotransporter display, a versatile bacterial cell surface presentation system. J. Bacteriol. 184:4197–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koriazova, L.K., and M. Montal. 2003. Translocation of botulinum neurotoxin light chain protease through the heavy chain channel. Nat. Struct. Biol. 10:13–18. [DOI] [PubMed] [Google Scholar]
- Krämer, S.D., and H. Wunderli-Allenspach. 2003. No entry for TAT(44-57) into liposomes and intact MDCK cells: novel approach to study membrane permeation of cell-penetrating peptides. Biochim. Biophys. Acta. 1609:161–169. [DOI] [PubMed] [Google Scholar]
- Krasilnikov, O.V., J.B. Da Cruz, L.N. Yuldasheva, W.A. Varanda, and R.A. Nogueira. 1998. A novel approach to study the geometry of the water lumen of ion channels: colicin Ia channels in planar lipid bilayers. J. Membr. Biol. 161:83–92. [DOI] [PubMed] [Google Scholar]
- Lamthanh, H., C. Jegou-Matheron, D. Servent, A. Ménez, and J.-M. Lancelin. 1999. Minimal conformation of the α-conotoxin ImI for the α7 neuronal nicotinic acetylcholine receptor recognition: correlated CD, NMR and binding studies. FEBS Lett. 454:293–298. [DOI] [PubMed] [Google Scholar]
- Leifert, J.A., S. Harkins, and J.L. Whitton. 2002. Full-length proteins attached to the HIV tat protein transduction domain are neither transduced between cells, nor exhibit enhanced immunogenicity. Gene Ther. 9:1422–1428. [DOI] [PubMed] [Google Scholar]
- Levinthal, F., A.P. Todd, W.L. Hubbell, and C. Levinthal. 1991. A single tryptic fragment of colicin E1 can form an ion channel: Stoichiometry confirms kinetics. Proteins. 11:254–262. [DOI] [PubMed] [Google Scholar]
- Lin, J.-H., and A. Baumgaertner. 2000. Stability of a melittin pore in a lipid bilayer: A molecular dynamics study. Biophys. J. 78:1714–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg, M., and M. Johansson. 2002. Positively charged DNA-binding proteins cause apparent cell membrane translocation. Biochem. Biophys. Res. Commun. 291:367–371. [DOI] [PubMed] [Google Scholar]
- Madden, J.C., N. Ruiz, and M. Caparon. 2001. Cytolysin-mediated translocation (CMT): a functional equivalent of type III secretion in Gram-positive bacteria. Cell. 104:143–152. [DOI] [PubMed] [Google Scholar]
- Martoglio, B., M.W. Hofmann, J. Brunner, and B. Dobberstein. 1995. The protein-conducting channel in the membrane of the endoplasmic reticulum is open laterally toward the lipid bilayer. Cell. 81:207–214. [DOI] [PubMed] [Google Scholar]
- Maslennikov, I.V., Z.O. Shenkarev, M.N. Zhmak, V.T. Ivanov, C. Methfessel, V.I. Tsetlin, and A.S. Arseniev. 1999. NMR spatial structure of α-conotoxin ImI reveals a common scaffold in snail and snake toxins recognizing neuronal nicotinic acetylcholine receptors. FEBS Lett. 444:275–280. [DOI] [PubMed] [Google Scholar]
- Matsuzaki, K. 1999. Why and how are peptide-lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim. Biophys. Acta. 1462:1–10. [DOI] [PubMed] [Google Scholar]
- Mel, S.F., and R.M. Stroud. 1993. Colicin Ia inserts into negatively charged membranes at low pH with a tertiary but little secondary structural change. Biochemistry. 32:2082–2089. [DOI] [PubMed] [Google Scholar]
- Mothes, W., S.U. Heinrich, R. Graf, I.M. Nilsson, G. von Heijne, J. Brunner, and T.A. Rapoport. 1997. Molecular mechanism of membrane protein integration into the endoplasmic reticulum. Cell. 89:523–533. [DOI] [PubMed] [Google Scholar]
- Naini, A.A., E. Shimony, E. Kozlowski, T. Shaikh, W. Dang, and C. Miller. 1996. Interaction of Ca2+-activated K+ channels with refolded charybdotoxins mutated at a central interaction residue. Neuropharmacology. 35:915–921. [DOI] [PubMed] [Google Scholar]
- Nielsen, O., and O. Buchardt. 1991. Facile synthesis of reagents containing a terminal maleimido ligand linked to an active ester. Synthesis. 10:819–821. [Google Scholar]
- Oh, K.J., L. Senzel, R.J. Collier, and A. Finkelstein. 1999. Translocation of the catalytic domain of diphtheria toxin across planar phospholipid bilayers by its own T domain. Proc. Natl. Acad. Sci. USA. 96:8467–8470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, M., R. Harris, C. Freytag, M. Kehoe, J. Tranum-Jensen, and S. Bhakdi. 1998. Assembly mechanism of the oligomeric streptolysin O pore: the early membrane lesion is lined by a free edge of the lipid membrane and is extended gradually during oligomerization. EMBO J. 17:1598–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, T., and B.C. Berks. 2003. Moving folded proteins across the bacterial cell membrane. Microbiology. 149:547–556. [DOI] [PubMed] [Google Scholar]
- Park, C.-S., S.F. Hausdorff, and C. Miller. 1991. Design, synthesis, and functional expression of a gene for charybdotoxin, a peptide blocker of K+ channels. Proc. Natl. Acad. Sci. USA. 88:2046–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease, J.H., and D.E. Wemmer. 1988. Solution structure of apamin determined by nuclear magnetic resonance and distance geometry. Biochemistry. 27:8491–8498. [DOI] [PubMed] [Google Scholar]
- Plath, K., W. Mothes, B.M. Wilkinson, C.J. Stirling, and T.A. Rapoport. 1998. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 94:795–807. [DOI] [PubMed] [Google Scholar]
- Qiu, X.-Q., K.S. Jakes, A. Finkelstein, and S.L. Slatin. 1994. Site-specific biotinylation of colicin Ia: a probe for protein conformation in the membrane. J. Biol. Chem. 269:7483–7488. [PubMed] [Google Scholar]
- Qiu, X.-Q., K.S. Jakes, P.K. Kienker, A. Finkelstein, and S.L. Slatin. 1996. Major transmembrane movement associated with colicin Ia channel gating. J. Gen. Physiol. 107:313–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond, L., S.L. Slatin, and A. Finkelstein. 1985. Channels formed by colicin E1 in planar lipid bilayers are large and exhibit pH-dependent ion selectivity. J. Membr. Biol. 84:173–181. [DOI] [PubMed] [Google Scholar]
- Richard, J.P., K. Melikov, E. Vives, C. Ramos, B. Verbeure, M.J. Gait, L.V. Chernomordik, and B. Lebleu. 2003. Cell-penetrating peptides: a reevaluation of the mechanism of cellular uptake. J. Biol. Chem. 278:585–590. [DOI] [PubMed] [Google Scholar]
- Rogers, J.P., P. Luginbühl, G.S. Shen, R.T. McCabe, R.C. Stevens, and D.E. Wemmer. 1999. NMR solution structure of α-conotoxin ImI and comparison to other conotoxins specific for neuronal nicotinic acetylcholine receptors. Biochemistry. 38:3874–3882. [DOI] [PubMed] [Google Scholar]
- Schnell, D.J., and D.N. Hebert. 2003. Protein translocons: multifunctional mediators of protein translocation across membranes. Cell. 112:491–505. [DOI] [PubMed] [Google Scholar]
- Schwartz, M.P., S. Huang, and A. Matouschek. 1999. The structure of precursor proteins during import into mitochondria. J. Biol. Chem. 274:12759–12764. [DOI] [PubMed] [Google Scholar]
- Schwartz, M.P., and A. Matouschek. 1999. The dimensions of the protein import channels in the outer and inner mitochondrial membranes. Proc. Natl. Acad. Sci. USA. 96:13086–13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarze, S.R., K.A. Hruska, and S.F. Dowdy. 2000. Protein transduction: unrestricted delivery into all cells? Trends Cell Biol. 10:290–295. [DOI] [PubMed] [Google Scholar]
- Senzel, L., M. Gordon, R.O. Blaustein, K.J. Oh, R.J. Collier, and A. Finkelstein. 2000. Topography of diphtheria toxin's T domain in the open channel state. J. Gen. Physiol. 115:421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatin, S.L., and P.K. Kienker. 2003. Colicin channels and protein translocation. Parallels with diphtheria toxin. Pore-forming Peptides and Protein Toxins. G. Menestrina, M. Dalla Serra, and P. Lazarovici, editors. Taylor & Francis, London. 102–131.
- Smith, D.L., D.K. Struck, J.M. Scholtz, and R. Young. 1998. Purification and biochemical characterization of the lambda holin. J. Bacteriol. 180:2531–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stampe, P., L. Kolmakova-Partensky, and C. Miller. 1994. Intimations of K+ channel structure from a complete functional map of the molecular surface of charybdotoxin. Biochemistry. 33:443–450. [DOI] [PubMed] [Google Scholar]
- Thorén, P.E.G., D. Persson, M. Karlsson, and B. Nordén. 2000. The Antennapedia peptide penetratin translocates across lipid bilayers—the first direct observation. FEBS Lett. 482:265–268. [DOI] [PubMed] [Google Scholar]
- Tirosh, B., M.H. Furman, D. Tortorella, and H.L. Ploegh. 2003. Protein unfolding is not a prerequisite for endoplasmic reticulum-to-cytosol dislocation. J. Biol. Chem. 278:6664–6672. [DOI] [PubMed] [Google Scholar]
- Veiga, E., V. de Lorenzo, and L.A. Fernández. 1999. Probing secretion and translocation of a β-autotransporter using a reporter single-chain Fv as a cognate passenger domain. Mol. Microbiol. 33:1232–1243. [DOI] [PubMed] [Google Scholar]
- Vestweber, D., and G. Schatz. 1988. a. A chimeric mitrochondrial precursor protein with internal disulfide bridges blocks import of authentic precursors into mitochondria and allows quantitation of import sites. J. Cell Biol. 107:2037–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestweber, D., and G. Schatz. 1988. b. Mitochondria can import artificial precursor proteins containing a branched polypeptide chain or a carboxy-terminal stilbene disulfonate. J. Cell Biol. 107:2045–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivès, E., P. Brodin, and B. Lebleu. 1997. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J. Biol. Chem. 272:16010–16017. [DOI] [PubMed] [Google Scholar]
- Volkman, B.F., and D.E. Wemmer. 1997. Deletion of a single amino acid changes the folding of an apamin hybrid sequence peptide to that of endothelin. Biopolymers. 41:451–460. [DOI] [PubMed] [Google Scholar]
- Wang, I.-N., J. Deaton, and R. Young. 2003. Sizing the holin lesion with an endolysin-β-galactosidase fusion. J. Bacteriol. 185:779–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesche, J., J.L. Elliott, P.Ø. Falnes, S. Olsnes, and R.J. Collier. 1998. Characterization of membrane translocation by anthrax protective antigen. Biochemistry. 37:15737–15746. [DOI] [PubMed] [Google Scholar]
- Wiedemann, N., N. Pfanner, and M.T. Ryan. 2001. The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J. 20:951–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener, M., D. Freymann, P. Ghosh, and R.M. Stroud. 1997. Crystal structure of colicin Ia. Nature. 385:461–464. [DOI] [PubMed] [Google Scholar]
- Yang, L., T.A. Harroun, T.M. Weiss, L. Ding, and H.W. Huang. 2001. Barrel-stave model or toroidal model? A case study on melittin pores. Biophys. J. 81:1475–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemel, A., D.R. Fattal, and A. Ben-Shaul. 2003. Energetics and self-assembly of amphipathic peptide pores in lipid membranes. Biophys. J. 84:2242–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]