Abstract
Salmonella typhimurium employs the specialized type III secretion system encoded in pathogenicity island 1 (SPI1) to translocate effector proteins into host cells and to modulate host cell signal transduction. The SPI1 type III system and the effector proteins are conserved among all salmonellae and are thought to be acquired by horizontal gene transfer. The genetic mechanisms mediating this horizontal transfer are unknown. Here, we describe that SopE, a SPI1-dependent translocated effector protein, is present in relatively few S. typhimurium isolates. We have isolated a temperate phage that encodes SopE. Phage morphology and DNA hybridization, as well as partial sequence information, suggest that this phage (SopEΦ) is a new member of the P2 family of bacteriophages. By lysogenic conversion this phage can horizontally transfer genes between different S. typhimurium strains. Strikingly, most of the isolates harboring SopEΦ belong to the small group of epidemic strains of S. typhimurium that have been responsible for a large percentage of human and animal salmonellosis and have persisted for a long period of time. Our data suggest that horizontal transfer of type III dependent effector proteins by lysogenic infection with bacteriophages (lysogenic conversion) may provide an efficient mechanism for fine-tuning the interaction of Salmonella spp. with their hosts.
Infection with nontyphoid Salmonella spp. is one of the leading causes of diarrhea in developed countries. Epidemiological studies have revealed that the bulk of nontyphoidal Salmonella infections at any one time is caused by only one or relatively few Salmonella strains (1–5). The alleles that may determine the epidemiologic success of certain strains have remained largely unknown. So far, genetic elements conferring resistance to antibiotics commonly used in animal production represent the only exception to this rule.
There is abundant evidence for horizontal gene transfer contributing to the virulence of Salmonella spp. (6, 7). Often, these virulence functions are encoded in chromosomal “pathogenicity islands” or on mobile genetic elements such as transposons or plasmids. In addition, Salmonella spp. are known to harbor a multitude of phages. Most of these phages belong to the P22 family and are able to facilitate horizontal transfer of bacterial genes by transduction (8). However, little is known about the contribution of phage-encoded genes to Salmonella virulence.
Salmonella spp. employ a large array of mechanisms to colonize, replicate, and survive within hosts (7). The specialized type III secretion system encoded in SPI1 is important during the gut-associated stages of the infection (9–11). Disruption of the SPI1 type III system leads to less pronounced induction of fluid secretion in the bovine ileal loop model (11) and to a 50-fold-increased LD50 in oral BALB/c mouse infection experiments (10). Evidence from tissue culture experiments indicates that the SPI1 type III system of Salmonella spp. exerts its function by translocating a set of at least eight different proteins into the cytosol of host cells (12–19). The type III secretion apparatus as well as most effector proteins are highly conserved among all Salmonella spp. and have been acquired by horizontal gene transfer (20). The genetic mechanisms mediating this horizontal transfer are unknown.
SopE is translocated into host cells by the SPI1 type III secretion system and activates the RhoGTPases CDC42 and Rac1 (21). Thereby, SopE can promote efficient entry of the bacterium into tissue culture cells (17, 22). The role of SopE in Salmonella pathogenesis, however, has not been determined yet. Interestingly, sopE is flanked by sequences resembling tail and tail-fiber genes of P2-like phages and was identified in one S. typhimurium strain and absent from another (22).
In the present study, we have analyzed sopE-loci from S. typhimurium isolates in more detail. We have isolated a temperate P2-like sopE-encoding phage (SopEΦ). Experiments using a derivative of SopEΦ implicate lysogenic conversion as an important mechanism facilitating horizontal transfer of type III-dependent effector proteins. We discuss the implications of our results for the evolution of Salmonella as a pathogen and the emergence of new epidemic strains.
MATERIALS AND METHODS
Recombinant DNA Techniques and Southern Hybridization.
Cloning of DNA fragments was performed according to standard protocols (23). For Southern hybridization, we used the enhanced chemiluminescence random-primed labeling and detection system, as recommended by the manufacturer (Amersham Pharmacia). Hybridization was performed at 65°C (probes derived from S. typhimurium sequences) or 55°C (probes derived from Salmonella typhi sequences) in a buffer containing 0.75 M NaCl, 75 mM sodium-citrate, pH 7, 0.1% SDS, 5% dextran sulfate, and 100 μg/ml salmon sperm DNA. Probes were prepared by PCR using S. typhi X3744 (a sopE+ strain) DNA as template and the following primers: probe I, 5′-TGATGTACAAAACCGACCAG and 5′-TTTAGCACCACCTTTAGCC (33×: 94°C for 30 sec, 50°C for 30 sec, 72°C for 8 min); probe II, 5′-TTCTCTCCCATTTTCAACG and 5′-GGTCCAGTTTTGCGTAGG (33×: 94°C for 30 sec, 50°C for 30 sec, 72°C for 8 min); probe III, 5′-CTGGCAAACCGTAAGCA and 5′-CAGCCAGTCATCAACCTTCT (32×: 94°C for 30 sec, 50°C for 30 sec, 72°C for 10 min); probe IV, 5′-TGGCGCTGGCACTTG and 5′-CAATAGACGATGCCACAAAT (30×: 94°C for 30 sec, 53°C for 30 sec, 72°C for 5 min). We also used fragments of cloned DNA from S. typhimurium SL1344RDNC;E+: probe V, a 720-nt fragment of pSB1136 (22) corresponding to the ORF of sopE; probe VI, a 700-nt HindIII/PvuI fragment of pSB1119 (W.-D.H. and J. E. Galán, unpublished data) corresponding to the C-terminal part of orfJ; probe VII, a 2.6-kb SacII/NdeI fragment comprising the C terminus of orfG and downstream sequences (W.-D.H. and J. E. Galán, unpublished data); and probe VIII, 1.6-kb EcoRV/NdeI fragment of pM36 (Fig. 2). pM36 carries a 4.5-kb NcoI/EcoRV fragment of M106SL1344;aphT-Φ comprising the right end of sopE∷aphT-Φ and flanking chromosomal sequence. It was isolated in a filter-lift assay by using probe VII from a M106SL1344;aphT-Φ DNA library.
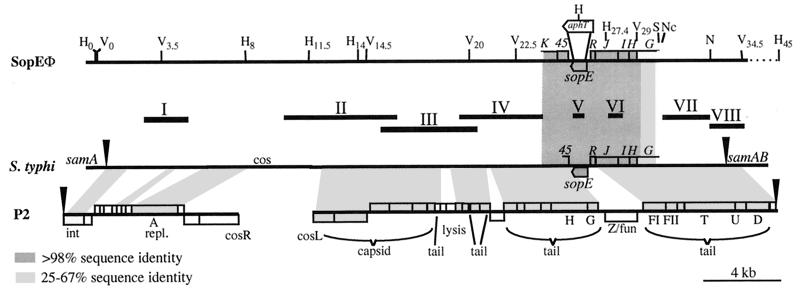
Figure 2.
Restriction map of SopEΦ. Alignment of the SopEΦ-pro-phage of S. typhimurium 3351/78DT204;E+ with the sopE-region from S. typhi (boxed region: our own sequencing results = accession no. AF153829; the rest is part of a contig produced by the Pathogen Sequencing Unit at the Sanger Centre; http://www.sanger.ac.uk/Projects/S_typhi; arrowheads, predicted integration site in samA) and phage P2 (accession no. AF063097; split at the attachment site = arrowheads); solid bars, probes for Southern analysis of the SopEΦ-pro-phage (see Table 2). The restriction map of the SopEΦ-pro-phage includes all recognition sites for H (HindIII) and V (EcoRV) and some recognition sites for S (SacII), Nc (NcoI), and N (NdeI). Subscript numbers indicate their relative positions (in kb). aphT, position of the resistance cassette in sopE∷aphT-Φ. The sequence similarity (tblastx server at the Sanger Center) between ORFs of P2 or the sequenced part of the SopEΦ-pro-phage (boxed region) to predicted polypeptides encoded by the S. typhi sequence is indicated: white (no significant similarity), light gray (25–67% identity), or dark gray (>98% identity). The shaded trapezoids indicate the location and the degree of similarity of similar regions.
Pulsed-field gel electrophoresis (PFGE) analysis was performed according to standard protocols by using a Chef DRIII apparatus (Bio-Rad; switch time, 5–35 sec; run time, 22 h; angle, 120°; voltage gradient, 6 V/cm in 0.5× TBE at 8°C).
The sopE region from S. typhi X3744 (sopE+) was cloned by integrating the suicide vector pSB377 (K. Kaniga, unpublished data) carrying nucleotides 165–615 of S. typhimurium sopE into the sopE gene of S. typhi X3744. The integrated vector and flanking chromosomal sequences were retrieved with Acc65I and XbaI (or SmaI), and a 4,938-nt fragment comprising sopE was sequenced.
The 2.7-kb sopE region from S. typhimurium 3351/78DT204;E+ was amplified by PCR (primers +1722, 5′-CCGTGGAACGATTGACTG, and −1017, 5′-AGCCATTAGCAGCAAGGT; 30×: 94°C for 30 sec, 53°C for 30 sec, 72°C for 3 min; 2.7-kb product) and sequenced.
Bacterial Strains.
Bacterial strains were obtained from the Robert Koch Institut, the Max von Pettenkofer-Institut, or from the Salmonella reference collection A (SARA) (24) and analyzed by phage typing (see refs. 25–27). S. typhimurium isolates are referred to as “strain numberphage type;sopE status”. Artificial derivatives are referred to as “strain numberparent strain;presence of sopE∷aphT-Φ” (e.g., M4A36;aphT-Φ). Strains S. typhimurium SL1344RDNC;E+ and S. typhi X3744 (sopE+) were provided by J. E. Galán (Yale University, New Haven, CT). Escherichia coli strain SM10λpir has been described (28).
S. typhimurium A36DT36;E− has been described (25). A36DT36;E− derivatives WR1173A36 (fhuA) and WR1174A36 (tonB) were constructed by phage transduction of markers from AR895 or AIR36 (29, 30). S. typhimurium 3351/78DT204;E+ was isolated in 1978 from a calf with severe gastroenteritis on a farm in Saxony Anhalt, Germany. M1063351/78;aphT-Φ was constructed by allelic exchange as described for SB856 (22) by using the suicide vector pSB1134 (sopE∷aphT; sacAB; ref. 22). Thereby, sopE was replaced with a 1.2-kb aphT cassette including a terminator.
Serological Procedures and Western Blot Analysis.
A polyclonal rabbit antiserum (IM1) was raised against a recombinant 78- to 240-aa fragment of SopE from S. typhimurium SL1344RDNC;E+. Detection of secreted SopE in 50 μl of Salmonella culture supernatant by Western blot analysis using IM1 (dilution, 1:30,000) was performed as described (22).
Induction, Detection, and Propagation of SopEΦ.
To produce SopEΦ or sopE∷aphT-Φ lysates, 150 μl of a fresh, overnight culture of the lysogen was diluted in 1.5 ml of LB supplemented with 2 μg/ml mitomycin C (Sigma) and grown for 6 h at 37°C. Lysates were processed by centrifugation (10,000 × g, 5 min, 4°C) and 0.45-μm membrane filtration.
For detection of SopEΦ, 5 μl of each lysate was streaked on S. typhimurium A36DT36;E− embedded in EBU-top agarose (0.7% agarose; EBU: 1% tryptone/0.5% yeast extract/0.5% NaCl/0.25% glucose/0.25% K2HPO4/12.5 mg/liter Evans blue/25 mg/liter sodium fluorescein). Plaque DNA was transferred onto nitrocellulose membranes and probed by hybridization with probe V (see above). Phages were recovered from the top-agarose by elution in λ buffer (100 mM NaCl/10 mM MgSO4/35 mM Tris⋅Cl, pH 7.5) at 4°C. Purification, determination of titers, and preparation of plate lysates on A36DT36;E− were performed according to standard protocols for phage λ (23). Plate lysates typically had titers of 107 pfu/ml. CaCl2 and EGTA did not affect the titers.
For preparation of lysogens, 106 plaque-forming units (pfu) of sopE∷aphT-Φ were incubated for 15 min at 21°C with 4 × 107 bacteria in a total of 100 μl λ buffer. The mixture was plated on LB (50 μg/ml kanamycin), and colonies resistant to kanamycin were counted after a 12-h incubation at 37°C. The lysogens were subjected to three rounds of purification.
The average number of pfu released from an infected bacterium was determined in single-burst experiments (31). Briefly, 1 ml of a WR1174A36 (tonB) overnight culture was inoculated for 10 min with 3 × 105 pfu and diluted 100-fold. Forty aliquots of 50 μl each were incubated at 37°C for an additional 30 min and titered by plating in soft agar. Statistical analysis showed that 8 ± 2 pfu were released per infected cell.
Electron Microscopy.
Phage lysates (5 × 106 pfu/ml) were cleared by centrifugation, 0.45-μm ultrafiltration, and dialysis against λ buffer (50-kDa molecular mass cutoff). Phages were adsorbed onto thin, carbon support films, washed with TE buffer (10 mM Tris/1 mM EDTA, pH 8; ref. 23), stained with 4% uranyl acetate, pH 4.5, and examined in a Zeiss transmission electron microscope (TEM910).
RESULTS
Distribution of sopE Among S. typhimurium Isolates.
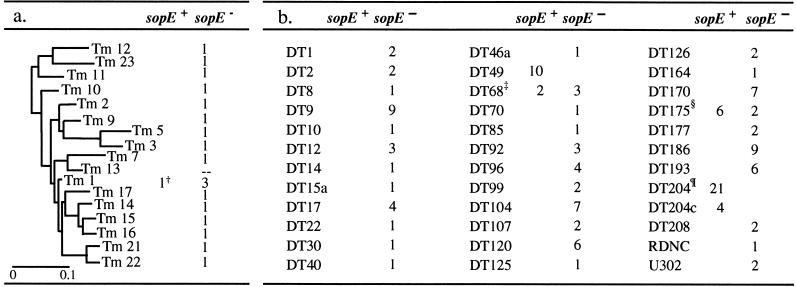
Of the two S. typhimurium clones analyzed previously, only SL1344RDNC;E+ was found to harbor the sopE gene (22). To analyze the distribution of sopE among S. typhimurium strains in more detail, we screened the SARA collection (24). DNA-hybridization experiments revealed that a large majority of the S. typhimurium isolates were lacking sopE (Table 1, a). Interestingly, one phylogenetic group (Tm 1) contained sopE− and sopE+ isolates (Table 1, a).
Table 1.
Distribution of sopE among S. typhimurium isolates*
Presence of sopE was analyzed by DNA hybridization (probe V; Materials and Methods). Isolates were part of the SARA collection (a) or clinical isolates from Germany (b). The unrooted phylogenetic tree in a was adapted from ref. 18.
SARA4DT20;E+.
624/96DT68;E−, DT68 reference strainDT68;E−, 11075/97DT68;E−, 11635/98DT69;E+, 1061/97DT68;E+.
305/70DT175;E−, 1/82DT175;E+, 198/82DT175;E+, 1646/85DT175;E+, 1662/82DT175;E+, 533/82DT175;E+, 1357/80DT175;E+, DT175 reference strainDT175;E−. --, not determined.
In addition, we screened a representative set of S. typhimurium isolates from Germany. For epidemiologic purposes, these isolates had been characterized by phage typing (Materials and Methods). Again, sopE was absent from the majority of isolates tested (Table 1, b). We found sopE− and sopE+ isolates of phage types DT68 and DT175 (Table 1, b). Furthermore, each isolate of the phage types DT49, DT204, and DT204c (10, 21, or 4 isolates tested) carried the sopE gene. S. typhimurium strains of the phage types DT49 and especially of DT204 and DT204c belong to the small group of epidemic strains that have accounted for a large percentage of human and animal infections and have persisted for long periods of time (3, 4, 32, 33). The incidence of S. typhimurium isolates of phage type DT175, although lower, has been significant over the past 30 years, whereas strains of phage type DT68 only recently have been isolated in Germany (W.R., unpublished observation).
Lytic Induction of a sopE-Encoding Phage from Epidemic S. typhimurium Strains.
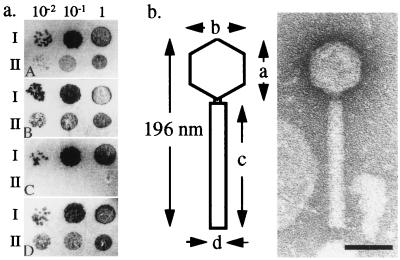
The P2 phage-like flanking sequences (22) and the distribution of sopE within the serovar Typhimurium suggested that sopE might be transferred horizontally by lysogenic conversion. To screen for sopE-encoding phages, we incubated 12 sopE+ S. typhimurium isolates (SL1344RDNC;E+, 3351/78DT204;E+, 1153/72DT204;E+, 646/96DT204c;E+, 660/96DT204c;E+, 1/82DT175;E+, 198/82DT175;E+, 11635/98DT68;E+, 1061/97DT68;E+, 838/88DT49;E+, 409/88DT49;E+, SARA4DT20;E+) with mitomycin C (see Materials and Methods). In line with earlier observations (refs. 8 and 34; H. Schmieger, personal communication), 11 isolates produced supernatants forming plaques of various morphology on S. typhimurium A36DT36;E−. Lysates from strains 3351/78DT204;E+, 1/82DT175;E+, 11635/98DT68;E+, 838/88DT49;E+, and 409/88DT49;E+ yielded sopE+ plaques as determined by DNA hybridization in a filter-lift assay (see Materials and Methods). Compared with other plaques from natural Salmonella isolates or with the well characterized phage P22HTint (35), the number and size of the sopE+ plaques were very small (Fig. 1a). This was probably a result of the low yield of 8 ± 2 pfu per infected bacterial cell, which we had determined in later single-burst experiments (see Materials and Methods). Therefore, the strong lytic growth of other phages may have interfered with detection of a similar sopE+ phage in the remaining lysates. sopE+ plaques from the epidemic strain 3351/78DT204;E+ were chosen for further analysis. The phage particles were eluted and purified to homogeneity in two further rounds of plating on S. typhimurium A36DT36;E− (Materials and Methods). Chloroform or high concentrations of DNase 1 or ribonuclease A had no effect on the titer (Fig. 1a) or on the sopE+ hybridization signal associated with each plaque (data not shown). In contrast, heat treatment (70°C) completely abolished plaque formation. These data demonstrate that sopE is an integral part of a phage particle. We have named this phage SopEΦ.
Figure 1.
Properties of SopEΦ from S. typhimurium 3351/78DT204;E+. (a) Heat sensitivity and DNase/RNase resistance of SopEΦ. Phage P22HTint (I) or SopEΦ (II) was incubated for 30 min in λ buffer (5 × 105 pfu/ml) on ice (A), at 37°C in the presence of RNase A (250 μg/ml) (B), at 70°C (C), or at 37°C in the presence of DNase1 (250 μg/ml) (D). After pretreatment, 5 μl of the appropriate phage dilution (10−2, 10−1, 1) was applied to a lawn of A36DT36;E−. Plaque formation was evaluated after 7 h at 37°C. (b) Electron microscopy analysis of the SopEΦ morphology. The particle dimensions are averages determined from at least 20 phages: a, head height = 58 ± 2 nm; b, head width = 54 ± 2 nm; c, tail length = 133 ± 5 nm; d, tail width = 19 ± 1 nm. (Bar = 50 nm.)
S. typhimurium strains 2138/96DT120;E−, 3805/96DT186;E−, 1509/96DT193;E− were also found to be sensitive to SopEΦ (data not shown), demonstrating that SopEΦ is able to infect a range of S. typhimurium strains. No lytic growth of SopEΦ was observed on S. typhimurium 3351/78DT204;E+, 838/88DT49;E+, 11635/98DT68;E+, 1/82DT175;E+, and 646/96DT204c;E+.
SopEΦ Is a New Member of the P2 Family.
Most phages found in S. typhimurium belong to the P22 family and have characteristic short tails. However, the 5 kb of DNA flanking sopE in S. typhimurium SL1344RDNC;E+ that was sequenced previously shows sequence similarity to tail and tail-fiber genes from P2-like phages (22). Because phage genomes are organized in a modular fashion and hybrids between otherwise unrelated phages frequently are observed (36–38), additional data were needed to assign SopEΦ to one of the known bacteriophage families. Electron microscopy analysis revealed that SopEΦ particles had a morphology quite different from that of the P22-like phages commonly found in Salmonella spp. They have icosahedral heads of 58 ± 2-nm diameter and straight tails of 133 ± 5-nm length (Fig. 1b), which are similar to those of phage P2 (head width, 60 nm; tail length, 135 nm; tail width, 18 nm; ref. 39). The lysates also contained numerous SopEΦ fragments (data not shown). This phenomenon frequently is observed with phage lysates of the P2 family. Relatives of phage ES18 have a morphology similar to P2 and are isolated occasionally from Salmonella spp. (40). However, in contrast to ES18, neither the absence of TonB (WR1174A36) nor the absence of FhuA (WR1173A36) had any effect on SopEΦ infection.
The similarity of SopEΦ to phages of the P2 family also was examined by Southern blot analysis of the SopEΦ-lysogenic S. typhimurium strain 3351/78DT204;E+ by using probes derived from cloned fragments of S. typhimurium DNA (22) or from a similar region from S. typhi (Fig. 2; Materials and Methods). The region comprising sopE, orfR, orfJ, orfI, orfH, and a part of orf45 was more than 98% identical between S. typhi and S. typhimurium (Fig. 2). The deduced amino acid sequences of both sopE genes were identical except for a N238Y substitution in S. typhi. Because these two proteins are only 89% identical with SopE from S. dublin (accession no. L78932; ref. 17), this suggests a recent event of horizontal transfer of sopE between the serovars Typhi and Typhimurium. The similarity of the S. typhi sopE region with P2-like phages extends over a contiguous sequence of 33 kb (Fig. 2). We used the S. typhi sequence to design probes I, II, III, and IV for Southern blot analysis of SopEΦ (Fig. 2; Materials and Methods). All four probes, as well as probes V, VI, and VII, yielded specific hybridization signals when used to probe chromosomal DNA from the SopEΦ lysogen S. typhimurium 3351/78DT204;E+ (Table 2; see also Fig. 3). These signals were absent when we probed chromosomal DNA from A36DT36;E−. The deduced restriction map indicates that the size of the SopEΦ-pro-phage is approximately 32 kb (Fig. 2), which is very similar to the sizes reported for members of the P2 family (41). Though similar, the P2-like sequences in S. typhi and SopEΦ are not identical: orfG has only 25% sequence identity (Fig. 2) and probe IV did not hybridize with the “overlapping” EcoRV22.5–29 fragment of the SopEΦ-pro-phage (Fig. 2; Table 2), indicating that the 5′ regions of both orfK genes must differ significantly. Furthermore, probe I hybridized only to the 3.5-kb EcoRV fragment, but not to the adjacent “overlapping” 11-kb fragment (V3.5–14.5; Fig. 2 and Table 2), indicating that sequences in this region also may deviate significantly. In conclusion, these data establish SopEΦ as a new member of the P2 family of bacteriophages and suggest that S. typhi CT18 may harbor a similar but distinct phage.
Table 2.
Southern blot analysis of the SopEΦ-pro-phage in S. typhimurium 3351/78
| Probe (source/hybridization temperature)* | Fragments detected, kb† | ||
|---|---|---|---|
| I‡ | (S. typhi/55°C)§ | V0/3.5 | H0/8 |
| II‡ | (S. typhi/55°C) | V3.5/14.5V14.5/20 | H8/11.5H11.5/14H14/27.4 |
| III‡ | (S. typhi/55°C) | V14.5/20V20/22.5 | H14/27.4 |
| IV‡ | (S. typhi/55°C)¶ | V20/22.5 | H14/27.4 |
| V‡ | (S. typhimurium/64°C) | V22.5/29 | H14/27.4 |
| VI‡ | (S. typhimurium/64°C) | V22.5/29 | H27.4/45 |
| VII‡ | (S. typhimurium/64°C) | V29/34.5 | H27.4/45 |
| VIII‖ | (S. typhimurium/64°C) | V29/34.5 | H27.4/45 |
Probes derived from S. typhi or S. typhimurium (Materials and Methods; Fig. 2).
See Fig. 2 for a restriction map.
No signals were obtained with A36DT36;E−.
Probe 1 did not detect fragment V3.5/14.5.
Probe IV did not detect fragment V22.5/29.
Also detects a 2.7-kb EcoRV and a >20-kb HindIII fragment from A36DT36;E−.
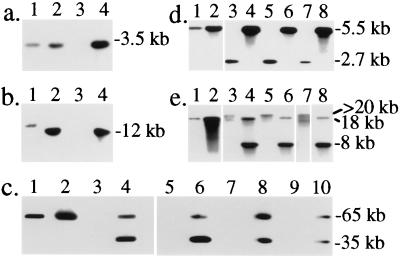
Figure 3.
Southern blot analysis of S. typhimurium M4A36; aphT-Φ. (a) Southern blot analysis of EcoRV-digested chromosomal DNA using probe I (see Fig. 2). (b) Southern blot analysis of HindIII-digested chromosomal DNA using probe IV. Bands in lanes 2 and 4 were shifted because of the presence of the aphT cassette. DNA was from 1, 3351/78DT204;E+; 2, M1063351/78;aphT-Φ; 3, A36DT36;E−; and 4, M4A36;aphT-Φ. (c) Chromosomal DNA was digested with XbaI and analyzed by PFGE and Southern blot hybridization by using probe VI (see Fig. 2). The S. typhimurium strains were: 1, 3351/78DT204;E+; 2, M1063351/78;aphT-Φ; 3, A36DT36;E−; 4, M4A36;aphT-Φ; 5, 3805/96DT186;E−; 6, M63805/96;aphT-Φ; 7, 3739/96DT193;E−; 8, M93739/96;aphT-Φ; 9, 2138/96DT120;E−; and 10, M102138/96;aphT-Φ. (d and e) Chromosomal DNA from strains 1, 3351/78DT204;E+; 2, M1063351/78;aphT-Φ; 3, A36DT36;E−; 4, M4A36;aphT-Φ; 5, 3805/96DT186;E−; 6, M63805/96;aphT-Φ; 7, 2138/96DT120;E−; 8, M102138/96;aphT-Φ was digested with EcoRV (d) or HindIII (e) and analyzed by using probe VIII (see Fig. 2).
Construction of sopE∷aphT-Φ.
To study lysogenic conversion as a mechanism for horizontal transfer of sopE, we constructed a derivative of the SopEΦ-pro-phage residing in the chromosome of 3351/78DT204;E+ carrying a kanamycin resistance marker (see Materials and Methods). The recombinant phage (sopE∷aphT-Φ) was induced from the resulting strain (M1063351/78; aphT-Φ; Materials and Methods). sopE∷aphT-Φ displayed the same host range as wild-type SopEΦ: strains A36DT36;E−, 2138/96DT120;E−, 3805/96DT186;E−, and 1509/96DT193;E− were sensitive whereas strains 3351/78DT204;E+, 839/88DT49;E+, 11635/98DT68;E+, 1/82DT175;E+, and 646/96DT204c;E+ were resistant. Phage morphology also remained unchanged (data not shown). Thus, insertion of the aphT cassette did not alter the properties of SopEΦ.
Lysogenic Conversion Using sopE∷aphT-Φ.
When A36DT36;E− was infected with sopE∷aphT-Φ (see Materials and Methods), kanamycin-resistant lysogens were obtained at a frequency of 10−4. Southern blot and PCR analyses (Fig. 3 a and b; data not shown) verified that sopE∷aphT-Φ was indeed present in the resulting strain (M4A36;aphT-Φ). PFGE analysis followed by Southern hybridization with probe VI demonstrated that sopE∷aphT-Φ in M4A36;aphT-Φ and M1063351/78;aphT-Φ was located on the same 60-kb chromosomal XbaI fragment as SopEΦ in strain 3351/78DT204;E+ (Fig. 3c, compare lanes 1, 2, and 4). The additional band of ca. 35 kb (Fig. 3c, lane 4) may be attributable to the circular, replicative form of sopE∷aphT-Φ or to immature or mature phage particles present in the bacterial cultures. Southern blot analysis lent further support to this hypothesis. The chromosomal EcoRV (2.7-kb) and HindIII (>20-kb) fragments of A36DT36;E− detected by probe VIII were absent in M4A36;aphT-Φ (Fig. 3 d and e; compare lanes 3 and 4). Instead, we detected a new band of the same size as observed in 3351/78DT204;E+ or M1063351/78; aphT-Φ (Fig. 3 d and e, lanes 4; 5.5-kb EcoRV and 18-kb HindIII), corresponding to the right end of the integrated phage. In M4A36; aphT-Φ we also detected a second band (Fig. 3 e, lane 4, 8-kb HindIII, and d, lane 4, the second EcoRV band was of almost identical size; not resolved here) that might be attributable to an extrachromosomal form of sopE∷aphT-Φ. Indeed, in contrast to supernatants from 3351/78DT204;E+ or M1063351/78;aphT-Φ, supernatants from M4A36;aphT-Φ overnight cultures had sopE∷aphT-Φ titers of 104 pfu/ml even without mitomycin C treatment.
In analogous fashion, we prepared sopE∷aphT-Φ lysogens from S. typhimurium strains 2138/96DT120;E−, 3805/96DT186;E−, and 1509/96DT193;E−. PFGE and Southern blot analyses revealed that sopE∷aphT-Φ had integrated into the same chromosomal region as in M4A36;aphT-Φ (Fig. 3 c, lanes 4, 6, 8, and 10, and d and e, lanes 4, 6, and 8). These data suggest that SopEΦ has one preferred attachment site and that it is capable of introducing genes into various S. typhimurium strains by lysogenic conversion.
Distribution of SopEΦ Among S. typhimurium Isolates.
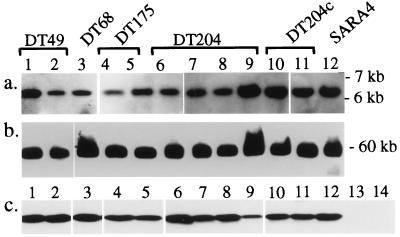
To determine the distribution of SopEΦ among sopE+ S. typhimurium isolates, we have compared the sopE loci of a representative set of isolates by Southern blot analysis. Bacterial DNA was digested with HindIII or EcoRV and analyzed by hybridization with probes II, V, and VII. The bands detected by each of the probes were of identical size in all S. typhimurium isolates tested (Fig. 4a; data not shown). PFGE analysis and Southern hybridization indicated that sopE resides on a 60-kb XbaI fragment in all S. typhimurium strains analyzed (Fig. 4b). Western blot analysis of Salmonella culture supernatants verified that the sopE genes were expressed in all sopE+ isolates tested (Fig. 4c). These data confirm that the sopE loci are functionally and structurally highly conserved among sopE+ wild-type strains of S. typhimurium and suggest that these strains are SopEΦ lysogens.
Figure 4.
Structural and functional conservation of sopE loci from S. typhimurium isolates. (a) Southern blot analysis of EcoRV-digested chromosomal DNA using probe V (see Materials and Methods). (b) PFGE–Southern analysis of XbaI-digested chromosomal DNA using probe VI. (c) Western blot analysis of SopE secretion using an α-SopE antiserum (IM1; Materials and Methods). Isolates shown in a– c: 1, 839/88DT49;E+; 2, 408/88DT49;E+; 3, 11635/98DT68;E+; 4, 1/82DT175;E+; 5, 1646/82DT175;E+; 6, 3351/78DT204;E+; 7, 93/80DT204;E+; 8, 1690/75DT204;E+; 9, 74/80DT204;E+; 10, 646/96DT204c;E+; 11, 6203/97DT204c;E+; 12, SARA4DT20;E+; 13, M1063351/78;aphT-Φ; and 14, 2728/96DT104;E−.
DISCUSSION
Acquisition of the SPI1 type III secretion apparatus was a quantum leap in the evolution of Salmonella spp. into an animal pathogen (20). Presumably, the virulence functions of this type III system are mediated by a set of highly conserved effector proteins (9, 19), which also have been acquired by horizontal gene transfer. However, the distribution of sopE suggests that, even today, at least some effector proteins may be transferred horizontally at appreciable frequency (22). We have used sopE from S. typhimurium to study the mechanisms of horizontal transfer of effector proteins in more detail.
First, we analyzed the distribution of sopE in natural S. typhimurium isolates. Screening of the SARA collection (24) and the collection of the German Salmonella Reference Center has revealed that sopE is present in only a small number of S. typhimurium isolates (Table 1). These included some isolates of phage types DT68 and DT175 and all isolates of phage types DT49, DT204, and DT204c (Table 1). From one of these natural sopE+ S. typhimurium isolates (3351/78DT204;E+) we recovered SopEΦ, a phage carrying the sopE gene within its genome. Because of its insensitivity to mutations in the Salmonella genes fhuA or tonB and its morphological features and based on similarities in DNA-sequence and genome organization, this phage has been assigned to the P2 family of bacteriophages.
Southern blot analyses suggest that all natural sopE+ isolates of S. typhimurium are carrying SopEΦ-pro-phage sequences. Indeed, SopEΦ could be induced from several of these strains. However, we cannot rule out that some of the pro-phages may have become inactivated. This might explain why we were unable to recover functional SopEΦ from several sopE+ S. typhimurium strains tested and why earlier attempts to induce SopEΦ from S. typhimurium SL1344RDNC;E+ had been unsuccessful (22). A derivative of SopEΦ carrying an aphT cassette is able to transfer the resistance marker into various S. typhimurium strains (Fig. 3) and also into S. enterica isolates from several other serovars by lysogenic conversion (S.M., W.R., and W.-D.H., unpublished data). These data suggest strongly that lysogenic conversion with SopEΦ has been the principal mechanism for horizontal transfer of the sopE gene into the serovar Typhimurium and/or horizontal transfer between different Salmonella strains.
Phage P2 originally had been isolated from E. coli (42). It is capable of infecting a wide range of Gram-negative bacteria including Shigella spp., Serratia marcescens, S. typhimurium, Klebsiella pneumoniae, and Yersinia spp. (39). Interestingly, the ΦCTX cytotoxin-converting phage of Pseudomonas aeruginosa is also a member of the P2 family (41). Together with our findings, this implicates effector- or toxin-converting P2 phages as versatile vehicles for horizontal gene transfer not only among Salmonella spp. or among E. coli strains, but also between distantly related Gram-negative bacteria.
Salmonella spp. are known to harbor a multitude of phages, a majority of which are capable of horizontally transferring bacterial genes by transduction (e.g., refs. 8 and 43). It will be of interest to explore whether some of these phages may encode factors modulating the virulence of Salmonella spp. So far, changes in lipopolysaccharide expression mediated by ɛ-phages represent the only well documented example (44). In addition, several reports have described the close association of genes implicated in virulence with phage-like sequences (45–47).
Does lysogenic conversion with SopEΦ modulate virulence? Compared with other well known, toxin-converting phages (41, 48, 49) that have a strong impact on the virulence of the host bacterium, the effects of SopEΦ seem rather subtle. Remarkably, the small group of natural SopEΦ+ strains included the S. typhimurium strains of phage types DT49, DT204, and DT204c, which had caused major epidemics in the United Kingdom and the former East Germany during the 1970s and 1980s (3, 4, 32, 33). In contrast to other strains that were widely spread among cattle in the beginning of the 1970s, strains of these phage types persisted over a long period of time and accounted for a large percentage of bovine and human infection (refs. 3, 4, 32, and 33; W.R., unpublished observation). Bovine infection with S. typhimurium DT204 was associated with severe diarrhea and dehydration that often proved fatal (4). Based on these observations it is tempting to speculate that lysogenic conversion with SopEΦ may have been one of the factors contributing to the epidemic success of S. typhimurium strains of phage types DT49, DT204, and DT204c.
Our data present direct evidence for a role of converting phages in the evolution of Salmonella spp. by providing an efficient mechanism to change the repertoire of translocated effector proteins. According to this model, effector proteins that have been evolved by other bacterial pathogens may become integrated into the genome of a temperate phage. Lysogenic conversion with such a phage could provide any susceptible bacterial pathogen bearing a type III system such as Salmonella spp. with these effector proteins. In this context, the evolution of the virulence functions mediated by the Salmonella SPI1 type III system may be considered as a two-step process. First, SPI1 was acquired via some unknown event of horizontal gene transfer, allowing the bacterium for the first time to interfere directly with signal-transduction pathways within host cells. Second, however, was the subsequent fine-tuning of the cross-talk with the host by acquisition of an optimal set of translocated effector proteins, which probably has been of equal importance. Therefore, the presence of efficient vehicles for horizontal gene transfer, including converting phages, may explain the extreme adaptability of Salmonella spp. and the wide range of hosts that can be infected by this bacterial species.
Acknowledgments
We thank H. Schmieger, M. Hensel, A. Rakin, C. Pelludat, and R. Cladwell for critical review of the manuscript and H. Gattermann and B. Tannert for skillful phage typing. We are grateful to J. E. Galán and J. Heesemann for scientific advice and for generous support. Financial support from the Friedrich Baur Stiftung, the Deutsche Forschungsgemeinschaft, and the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie (to W.-D.H.) is acknowledged.
ABBREVIATIONS
- PFGE
pulsed-field gel electrophoresis
- pfu
plaque-forming unit
- SARA
Salmonella reference collection A
Footnotes
References
- 1.McDonough P L, Timoney J F, Jacobson R H, Khakhria R. J Clin Microbiol. 1989;27:622–627. doi: 10.1128/jcm.27.4.622-627.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacob W K, Kühn H, Kurschner H, Rabsch W. Berl Münch Tierärztl Wochenschr. 1993;106:265–269. [PubMed] [Google Scholar]
- 3.Kühn H, Rabsch W, Tschäpe H, Tietze E. Z Ärztl Fortbild (Jena) 1982;76:607–610. [PubMed] [Google Scholar]
- 4.Threlfall E J, Ward L R, Rowe B. Vet Rec. 1978;103:438–440. doi: 10.1136/vr.103.20.438. [DOI] [PubMed] [Google Scholar]
- 5.van Leeuwen W J, Guinee P A. Zentralbl Bakteriol. 1975;230:320–335. [PubMed] [Google Scholar]
- 6.Bäumler A J. Trends Microbiol. 1997;5:318–322. doi: 10.1016/S0966-842X(97)01082-2. [DOI] [PubMed] [Google Scholar]
- 7.Groisman E A, Ochman H. Trends Microbiol. 1997;5:343–349. doi: 10.1016/S0966-842X(97)01099-8. [DOI] [PubMed] [Google Scholar]
- 8.Schicklmaier P, Schmieger H. Appl Environ Microbiol. 1995;61:1637–1640. doi: 10.1128/aem.61.4.1637-1640.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hueck C J. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galán J E, Curtiss R., III Proc Natl Acad Sci USA. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watson P R, Galyov E E, Paulin S M, Jones P W, Wallis T S. Infect Immunol. 1998;66:1432–1438. doi: 10.1128/iai.66.4.1432-1438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collazo C M, Galán J E. Mol Microbiol. 1997;24:747–756. doi: 10.1046/j.1365-2958.1997.3781740.x. [DOI] [PubMed] [Google Scholar]
- 13.Fu Y, Galán J E. Mol Microbiol. 1998;27:359–368. doi: 10.1046/j.1365-2958.1998.00684.x. [DOI] [PubMed] [Google Scholar]
- 14.Hardt W-D, Galán J E. Proc Natl Acad Sci USA. 1997;94:9887–9892. doi: 10.1073/pnas.94.18.9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galyov E E, Wood M W, Rosqvist R, Mullan P B, Watson P R, Hedges S, Wallis T S. Mol Microbiol. 1997;25:903–912. doi: 10.1111/j.1365-2958.1997.mmi525.x. [DOI] [PubMed] [Google Scholar]
- 16.Jones M A, Wood M W, Mullan P B, Watson P R, Wallis T S, Galyov E E. Infect Immunol. 1998;66:5799–5804. doi: 10.1128/iai.66.12.5799-5804.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wood M W, Rosqvist R, Mullan P B, Edwards M H, Galyov E E. Mol Microbiol. 1996;22:327–338. doi: 10.1046/j.1365-2958.1996.00116.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhou D, Mooseker M S, Galán J E. Science. 1999;283:2092–2095. doi: 10.1126/science.283.5410.2092. [DOI] [PubMed] [Google Scholar]
- 19.Galán J E. Curr Opin Microbiol. 1999;2:46–50. doi: 10.1016/s1369-5274(99)80008-3. [DOI] [PubMed] [Google Scholar]
- 20.Groisman E A, Ochman H. Cell. 1996;87:791–794. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 21.Hardt W-D, Chen L M, Schuebel K E, Bustelo X R, Galán J E. Cell. 1998;93:815–826. doi: 10.1016/s0092-8674(00)81442-7. [DOI] [PubMed] [Google Scholar]
- 22.Hardt W-D, Urlaub H, Galán J E. Proc Natl Acad Sci USA. 1998a;95:2574–2579. doi: 10.1073/pnas.95.5.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 24.Beltran P, Plock S A, Smith N H, Whittam T S, Old D C, Selander R K. J Gen Microbiol. 1991;137:601–606. doi: 10.1099/00221287-137-3-601. [DOI] [PubMed] [Google Scholar]
- 25.Anderson E S, Ward L R, De Saxe J D H. J Hyg. 1977;78:94–127. doi: 10.1017/s0022172400056187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Callow B R. J Hyg. 1959;57:346–359. doi: 10.1017/s0022172400020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lilleengen K. Acta Pathol Microbiol Scand. 1948;77,Suppl.:7–127. [Google Scholar]
- 28.Miller V L, Mekalanos J J. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsolis R M, Bäumler A J, Stojiljkovic I, Heffron F. J Bacteriol. 1995;177:4628–4637. doi: 10.1128/jb.177.16.4628-4637.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsolis R M, Bäumler A J, Heffron F, Stojiljkovic I. Infect Immunol. 1996;64:4549–4556. doi: 10.1128/iai.64.11.4549-4556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stent G. Molecular Biology of Bacterial Virus. New York: Freeman; 1963. pp. 74–77. [Google Scholar]
- 32.Threlfall E J, Ward L R, Rowe B. Brit Med J. 1978;2:426–428. doi: 10.1136/bmj.2.6143.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wray C, McLaren I M, Jones Y E. J Med Microbiol. 1998;47:483–487. doi: 10.1099/00222615-47-6-483. [DOI] [PubMed] [Google Scholar]
- 34.Nutter R L, Bullas L R, Schultz R L. J Virol. 1970;5:754–764. doi: 10.1128/jvi.5.6.754-764.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmieger H. Mol Gen Genet. 1972;119:75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 36.Botstein D. Ann N Y Acad Sci. 1980;354:484–490. doi: 10.1111/j.1749-6632.1980.tb27987.x. [DOI] [PubMed] [Google Scholar]
- 37.Hendrix R W, Smith M C, Burns R N, Ford M E, Hatfull G F. Proc Natl Acad Sci USA. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campbell A. In: The Bacteriophages. Calendar R, editor. Vol. 1. New York: Plenum; 1988. pp. 1–17. [Google Scholar]
- 39.Bertani L E, Six E. In: The Bacteriophages. Calendar R, editor. Vol. 1. New York: Plenum; 1988. pp. 73–143. [Google Scholar]
- 40.Kuo T-T, Stocker B A D. Virology. 1970;42:621–632. doi: 10.1016/0042-6822(70)90308-9. [DOI] [PubMed] [Google Scholar]
- 41.Nakayama K, Kanaya S, Ohnishi M, Terawaki Y, Hayashi T. Mol Microbiol. 1999;31:399–419. doi: 10.1046/j.1365-2958.1999.01158.x. [DOI] [PubMed] [Google Scholar]
- 42.Bertani G. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schicklmaier P, Moser E, Wieland T, Rabsch W, Schmieger H. Antonie Van Leeuwenhoek. 1998;73:49–54. doi: 10.1023/a:1000748505550. [DOI] [PubMed] [Google Scholar]
- 44.Popoff M Y, LeMinor L. Antigenic Formulas of the Salmonella serovars. 7th Ed. Paris: Institut Pasteur; 1997. [Google Scholar]
- 45.Farrant J L, Sansone A, Canvin J R, Pallen M J, Langford P R, Wallis T S, Dougan G, Kroll J S. Mol Microbiol. 1997;25:785–796. doi: 10.1046/j.1365-2958.1997.5151877.x. [DOI] [PubMed] [Google Scholar]
- 46.De Groote M A, Ochsner U A, Shiloh M U, Nathan C, McCord J M, Dinauer M C, Libby S J, Vazquez-Torres A, Xu Y, Fang F C. Proc Natl Acad Sci USA. 1997;94:13997–14001. doi: 10.1073/pnas.94.25.13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conner C P, Heithoff D M, Julio S M, Sinsheimer R L, Mahan M J. Proc Natl Acad Sci USA. 1998;95:4641–4645. doi: 10.1073/pnas.95.8.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheetham B F, Katz M E. Mol Microbiol. 1995;18:201–208. doi: 10.1111/j.1365-2958.1995.mmi_18020201.x. [DOI] [PubMed] [Google Scholar]
- 49.Waldor M K, Mekalanos J J. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]