Abstract
Mediated by the protein IIAGlc, the phosphoenolpyruvate:sugar phosphotransferase system plays a role in the regulation of activity of other sugar transport systems in Escherichia coli. By using a direct binding assay, a collection of single-Cys replacement mutants in cytoplasmic loops of lactose permease were evaluated for their capacity to bind IIAGlc. Selected Cys replacements in loops IV/V or VI/VII result in loss of binding activity. Analysis of the mutagenesis results together with multiple sequence alignments of a family of proteins that interacts with IIAGlc provides the basis for developing two regions of consensus sequence in those partner proteins necessary for binding to IIAGlc. The requirement for two interaction regions is interpreted in the regulatory framework of a substrate-dependent conformational change that brings those two regions into an orientation optimal for binding IIAGlc.
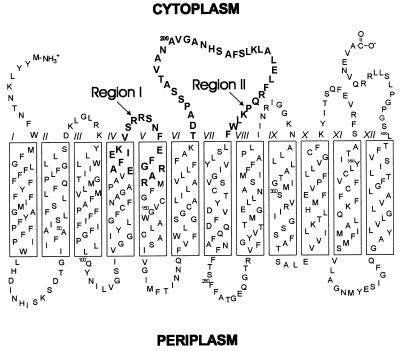
Secondary transport systems use electrochemical gradients to accumulate solutes against a concentration gradient and are found in virtually all biological membranes. The lactose permease (lac permease) from Escherichia coli is the most extensively studied secondary transport system (reviewed in ref. 1). This polytopic, cytoplasmic membrane protein catalyzes the coupled, stoichiometric translocation of β-galactosides such as lactose or melibiose and H+ (2). All evidence suggests that lac permease has 12 transmembrane, α-helical domains that traverse the membrane in a zigzag fashion (Fig. 1), and the tertiary-arrangement of most of the helices has been determined (1, 3, 4). These helical domains are connected by hydrophilic loops with the N and C termini on the cytoplasmic face of the membrane. Substrate binding to lac permease promotes conformational changes in which the helical domains are proposed to undergo sliding motions during substrate turnover (1, 4).
Figure 1.
Secondary-structure model for lactose permease. Two regions of lactose permease (boldface residues) were examined by cysteine-scanning mutagenesis to determine which residues play a role in IIAGlc binding. Region I, cytoplasmic loop IV/V and the flanking residues in helix IV and V. Region II, cytoplasmic loop VI/VII.
Another type of transport system found in bacteria corresponds to the phosphoenolpyruvate:carbohydrate phosphotransferase systems (PTSs) (reviewed in ref. 5). The primary function of this multifunctional, multicomponent system is the concomitant phosphorylation and translocation of sugars across the cytoplasmic membrane (6). For glucose transport, the cytoplasmic proteins Enzyme I, HPr, and IIAGlc transfer a phosphoryl group from phosphoenolpyruvate sequentially from one to the other, and then P-IIAGlc passes the phosphate to the membrane protein IIBCGlc, which both translocates and phosphorylates glucose. The unphosphorylated form of IIAGlc also plays a role in the inhibition of transport of non-PTS sugars such as lactose, maltose, melibiose, and raffinose (termed inducer exclusion) (7–9). In the case of lactose, IIAGlc binds to lac permease, inhibiting the transport of lactose (10, 11). Other allosteric regulatory functions of IIAGlc include inhibition of the phosphorylation of glycerol by binding to glycerol kinase (GK) (12) and either inhibition or activation of adenylyl cyclase (13). Previous work (14) shows that binding of IIAGlc requires lac permease to undergo a substrate-induced conformational change. Polyhistidine insertion into lac permease cytoplasmic loops IV/V or VI/VII or periplasmic loop VII/VIII does not affect substrate binding, as indicated by transport activity, but almost completely abolishes IIAGlc binding.
Site-directed mutagenesis was used to create a version of lactose permease (15) in which all eight Cys residues were replaced with either Ser or Val (Cys-less permease). Cys-less permease is functional with respect to both lactose transport (15) and its ability to bind IIAGlc (14, 16). This construct was used to create a library of mutants in which each individual amino acid residue in the permease was replaced with Cys (reviewed in ref. 1). Using this library, we examined cytoplasmic loop IV/V and the flanking residues in helix IV and V (Region I) and cytoplasmic loop VI/VII (Region II) from lac permease (see Fig. 1) to determine which residues play an important role in binding IIAGlc.
EXPERIMENTAL PROCEDURES
Materials.
Construction of a Cys-less permease and site-directed mutants of lacY encoding Cys-less permease have been described (1, 15). [3H]IIAGlc was purified from E. coli strain HK759 transformed with pRK248 and pEL110 (14, 17).
Membrane Preparations.
Derivatives of E. coli K12 strain T184[lacI+ O+ Z− Y− (A) rpsL met− thr− recA hsdR/F′ lacIq O+ ZD118 (Y+ A+)] (18) harboring plasmid pT7-5/cassette LacY encoding Cys-less lac permease (15) with indicated mutations were used for membrane preparations. Mutations in residues 136 and 221 were unavailable for analysis. Luria–Bertani medium (14) supplemented with 50 μg/ml ampicillin and 20 μg/ml streptomycin was inoculated with an overnight culture and grown at 37°C. When the culture reached an A600 = 0.5, lac permease expression was induced by addition of a final concentration of 1 mM isopropyl 1-thio-β-d-galactopyranoside, and growth was continued for another 4 hours. Cells were harvested by centrifugation for 10 min at 6,000 × g, were washed once in 50 mM sodium phosphate buffer (pH 6.3), and were stored as a pellet at −20°C until use. To prepare membranes, cells were resuspended in phosphate buffer and were passed twice through a French press at 12,000 psi (1 psi = 6.89 kPa); cell debris/unbroken cells were removed by centrifugation for 10 min at 10,000 × g. Membrane vesicles were collected by centrifugation for 90 min at 100,000 × g and were washed once with phosphate buffer, and centrifugation was repeated to pellet the membranes. The final membrane vesicle preparation was resuspended to a protein concentration of ≈20 mg/ml as determined by the bicinchoninic acid protein assay (Pierce).
Binding Assays.
[3H]IIAGlc was purified by previously described methods (14). The binding of [3H]IIAGlc to lac permease was performed by a method modified from Seok et al. (14). The binding assay contained, in 100 μl: 50 mM sodium phosphate (pH 6.3), 2 mM MgCl2, 2 mM DTT, 750 μg of membrane protein from a given membrane vesicle preparation, and 2.4 μg of [3H]IIAGlc (1,080 cpm/μg). Incubation mixtures were set up in Beckman pollyallomer tubes (5 × 20 mm) with and without the addition of 4 mM melibiose. After 5 min of incubation at room temperature, membranes were collected by centrifugation in a Beckman Airfuge for 30 min at 100,000 × g. The supernatant solutions were discarded, and the tubes were rinsed with 100 μl of H2O. The pellets were resuspended in 100 μl Triton X-100 and were transferred to scintillation vials. The tubes were rinsed with 100 μl of H2O and were transferred to the vials containing appropriate resuspended pellets. After adding 10 ml of Aquasol (NEN), membrane-bound IIAGlc was quantitated (cpm) by using a Beckman liquid scintillation counter. All results were reproduced at least two times.
Modification of Lac Permease Mutants.
Single-Cys lac permease mutants were modified with N-ethylmaleimide (NEM). NEM was added to a final concentration of 5 mM to the phosphate buffer, membranes, and H2O described in the binding assay above and was incubated for 5 min at room temperature. The reaction was terminated by adding to this mixture 10 μl of 0.1 M DTT and incubating for 5 min. The remaining binding assay reagents (MgCl2, [3H]IIAGlc, and melibiose) were added, and the procedure was continued as described above.
RESULTS
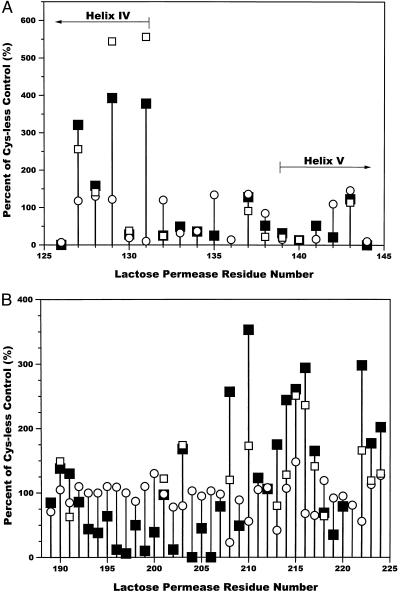
Membrane vesicles were prepared from E. coli expressing Cys-less lac permease (control) or various site-directed mutants of Cys-less permease. The membranes were analyzed for IIAGlc binding with radiolabeled IIAGlc (14). Because IIAGlc binds to lac permease in a sugar-dependent manner, values obtained in the absence of melibiose were subtracted from values in the presence of melibiose to correct for nonspecific binding. The data for the various single-Cys mutants are expressed as the percent of sugar-dependent binding observed with the Cys-less control (230 cpm).‖
In Region I (residues 126–144; see Fig. 1), two Cys replacement mutants, N137C and A143C, exhibited IIAGlc binding comparable to the Cys-less control (Fig. 2A, filled squares). Several mutants in helix IV, A127C, F128C, I129C, and K131C bound a greater amount of IIAGlc than Cys-less permease (321%, 158%, 393%, and 378%, respectively). Mutants E130C, V132C, R135C, E139C, F140C, and R142C exhibited low binding of IIAGlc (13–32% of Cys-less lac permease) whereas mutants S133C, R134C, F138C, and G141C showed moderate binding (36–52% of Cys-less lac permease). Mutants E126C and R144C are unable to bind significant amounts of IIAGlc because they do not bind substrate (19).
Figure 2.
Effect of cysteine-scanning mutagenesis on IIAGlc binding. Lac permease residues from Region I (A) and Region II (B) (Fig. 1) were mutated to cysteine. The percent of IIAGlc bound to the various cysteine mutants compared with the Cys-less lac permease control was determined (■); for some of the residues, the single-cysteine residues were modified with NEM before the binding assays (□). ○, the previously determined initial rates of [14C]-lactose transport as percent of the Cys-less control (20–22). The average rate for Cys-less lac permease was 55 nmol⋅min−1⋅(mg of protein)−1.
In Region II (residues 189–244; Fig. 2B), nine mutants (T189C, D190C, A191C, P192C, V201C, A207C, K211C, L212C, and P220C) bound approximately the same amount of IIAGlc as the Cys-less control (79–138% of Cys-less lac permease). Many mutants (F208C, L210C, A213C, L214C, E215C, L216C, F217C, L222C, W223C, and F224C) in the portion of the loop adjacent to helix VII along with A203C (168% of Cys-less lac permease) bound a greater amount of IIAGlc than did the control (165–353% of Cys-less lac permease). No significant IIAGlc binding was observed for mutants N204C and S206C whereas mutants T196C, V197C, N199C, and G202C exhibited very weak binding (6–12% of Cys-less permease). Mutants S193C, S194C, A195C, A198C, A200C, H205C, S209C, R218C, and Q219C exhibited intermediate levels of binding of IIAGlc (35–69% of Cys-less permease).
The possibility was considered that replacement of various bulky residues by Cys was responsible for the enhanced binding of IIAGlc observed in portions of regions I and II. This idea was tested by N-ethylmaleimide modification, which should restore a bulky sidechain with a variety of the Cys replacement mutants (shown as open squares in Fig. 2 A and B). All of the residues that exhibit enhanced binding of IIAGlc, as well as a selected number of control residues throughout Regions I and II, were subjected to NEM modification. This modification had no or only a slight effect (a <50% decrease of the percent of control values for the unmodified membranes) on mutants A127C, F128C, E130C, V132C, N137C, F138C, E139C, F140C, and A143C in Region I and mutants D190C, V201C, A203C, E215C, F217C, and R218C in Region II. For other mutants in Region II (A191C, F208C, L210C, A213C, L214C, L216C, L222C, W223C, and F224C), modification caused a significant decrease in the amount of IIAGlc bound. Surprisingly, two mutants from region I, I129C and K131C, bound an even greater amount of IIAGlc (an additional 150–170% increase in the percent of control) when the Cys side chain was alkylated.
DISCUSSION
A major purpose of this study was to determine which side chains in loops IV/V and VI/VII of lac permease are critical for interaction with IIAGlc. The approach used was to compare binding of IIAGlc by lac permease with single-Cys replacements in loops IV/V (as well as some flanking residues) and VI/VII to Cys-less permease. Fig. 2 A and B shows the results of those studies. Because effective binding of IIAGlc requires a substrate-dependent conformational change, it was essential to eliminate from consideration those mutations that result in loss of transport (i.e., binding) activity. Consequently, transport rates as reported previously (20–22) also are presented as percent of the Cys-less control in Fig. 2 (open circles). In Region I, residues 126, 130, 131, 133, 134, 136, 139, 140, 141, and 144 were eliminated from consideration because of decreased transport activity (Fig. 2A).
In Region II, it is interesting that mutation of residues 208, 213, and 222 was reported to result in decreased transport activity (20, 21), but the data in Fig. 2B show that the melibiose-dependent binding of IIAGlc was actually enhanced. The decreased transport activity in the mutants is therefore unlikely to result from a defect in substrate binding.
Cys replacements for A127, I129, and K131 in Region I resulted in significantly enhanced binding of IIAGlc (Fig. 2A). These residues are all located in Helix IV (19). It seems reasonable to conclude that, in wild-type lac permease, the arrangement of Helix IV places a limitation on IIAGlc binding; this inhibition is relieved by any of these Cys replacements that may change the tilt of Helix IV and influence the orientation of loop IV/V with respect to loop VI/VII.
A similar clustering of residues in which Cys replacement resulted in increased IIAGlc binding was observed in the region of loop VI/VII vicinal to Helix VII (Fig. 2B). The Cys replacements showing a substantial stimulation were A203, F208, L210, A213, L214, E215, L216, F217, L222, W223, and F224. Because all of these residues are hydrophobic, it is possible that Helix VII is actually longer, including those residues mentioned above that extend out of the membrane into the cytoplasm (see Fig. 1). In that case, the binding stimulation observed in that region may be explained in the same way as for those seen with the Helix IV changes (see above).
Those cases in which stimulation of binding was observed generally involve replacement of a bulkier side chain by the smaller Cys residue. Experiments were carried out to see whether adding more bulk to the Cys side chain by way of NEM modification results in reversal of stimulation (Fig. 2 A and B). There was essentially no effect of alkylation on any of the residues tested in Region I (Fig. 2A, open squares). However, at some sites in Region II, enhanced binding was reduced by NEM treatment. The positions responding to NEM were A191, F208, L210, A213, L214, L222, W223, and F224.
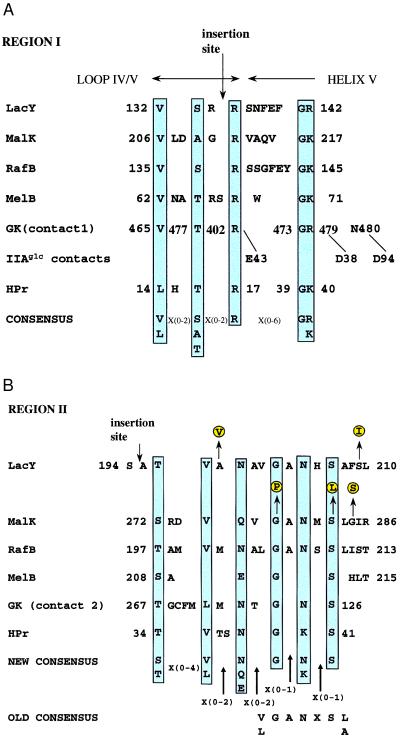
A combination of inspection of the data in Fig. 2A and sequence alignments was used to determine which residues are essential for IIAGlc binding in Region I (Fig. 3A). Mutation of either V132, R135, or R142 in lac permease resulted in decreased IIAGlc binding with no effect on transport. Alignment of this region with the corresponding sequences of the permeases for maltose (MalK, the peripheral membrane component), raffinose (RafB), and melibiose (MelB), showed conservation of the Val and two Arg residues (the second Arg residue was replaced with Lys in MalK, RafB, and MelB). In addition, S133 aligned well with conserved Ser, Ala, or Thr residues in the other permeases. Replacement of S133 with Cys caused decreases in both binding of IIAGlc and transport. Because of the apparent conservation of this residue, we suggest that it may be important for IIAGlc binding. In the alignment, a Gly residue always appeared in the position preceding that corresponding to R142 in lac permease. Although replacement of this residue with Cys is associated with decreased transport, as well as decreased IIAGlc binding, it appears likely that the conserved Gly residue is important for IIAGlc binding. This analysis suggests the five-residue consensus for IIAGlc binding associated with Region I shown in Fig. 3A [(V, L)(X){0,2}(S, A, T)(X){0,2}(R)(X){0,6}(G)(K, R)].
Figure 3.
Multiple sequence alignment of various E. coli proteins that interact with IIAGlc. Accession numbers for the various proteins are as follows: LacY, p02920; RafB, p16522; MelB, K01991; GK, M18393; and HPr, J02796. The corrected sequence of MalK is from ref. 28. Residues proposed to be essential for binding to IIAGlc are shown in blue shaded boxes. The IIAGlc contacts with GK are from ref. 12. (A) Region I alignment. The site of polyhistidine insertion (labeled insertion site) in loop IV/V of lac permease is from ref. 14. (B) Region II alignment. The site of polyhistidine insertion (labeled insertion site) in loop VI/VII is from refs. 14 and 16. Mutants in LacY resulting in loss of inducer exclusion (shown as yellow shaded circles) are from ref. 11. Mutations in MalK resulting in loss of inducer exclusion (yellow shaded circles) are from refs. 8 and 24. The old consensus is from ref. 9.
The crystal structure of the complex of IIAGlc with glycerol kinase has been elucidated. One of the most striking features of the interface is the absence of intermolecular hydrogen bonds (23). The region referred to as contact 1 includes residues in the proposed consensus (Fig. 3A). Therefore, the nature of interaction of IIAGlc with the soluble protein glycerol kinase and membrane-bound permeases may be similar.
It previously was shown that a polyhistidine insertion between R134 and R135 (see Fig. 3A) of loop IV/V results in decreased IIAGlc binding (14). This is precisely the region in which three of the key residues of the proposed consensus are localized. The other two residues of the consensus (G141 and R142) are located in Helix V proximal to loop IV/V (Figs. 1 and 3A). This is consistent with the idea that slight disturbances in the orientation of Helix V with respect to the other helices have substantial effects on IIAGlc binding. In this context, it is worth noting that the interface between Helices IV and V has been identified as an important element in the substrate binding site (19).
A similar approach of multiple sequence alignment and analysis of the data of Fig. 2B was used to deduce residues in region II important for binding of IIAGlc. Mutation of either T196, V197, N199, G202, N204, or S206 of lac permease resulted in essentially complete inhibition of IIAGlc binding with no effect on transport. Alignment of this region with the corresponding sequences of the permeases for maltose (MalK, the peripheral membrane component), raffinose (RafB), and melibiose (MelB) showed absolute conservation of the Gly202 and Ser206 residues. The Thr196 residue was conservatively substituted by Ser in MalK and MelB. The Val197 residue was not found in MelB but was conserved in MalK and RafB. The Asn residue was conservatively substituted by Gln in MalK and by Glu in MelB. The second conserved Asn residue (204) was absent in MelB. Residues associated with contact 2 of glycerol kinase also aligned well with the conserved residues of lac permease. On the basis of this alignment, it is possible to propose a new consensus sequence for the interaction with IIAGlc that consists of six residues [(S, T)(X){0,4}(V, L)(X){0,2}(N, Q, E)(X){0,2}(G)(X){0,1}(N, K)(X){0,1)(S)]. This differs from the previously proposed consensus (9) (labeled old consensus), which was based only on sequence comparisons of LacY, MalK, and RafB.
Further evidence that the region between residues 194 and 210 of lac permease is important derives from the previous finding (14, 16) that a polyhistidine insertion in this region decreases IIAGlc binding. This decrease is probably caused by a steric interference that deters binding. A similar mechanism for binding inhibition may explain the replacement of A198 by Val and S209 (11) by Ile; in both of those cases, smaller residues are replaced by bulkier side chains.
Mutations that result in resistance to inhibition by IIAGlc have been reported for MalK (Fig. 3B). Two of these mutations [G278P (8) and S282L (24)] correspond to residues in the suggested consensus. Another site of mutagenesis is residue Gly284 (8), flanking the consensus. In this case, a likely explanation for the inhibitory effect is imposition of a bulky group that interferes with the protein–protein interaction. Some mutations in the carboxyl-terminal tail of MelB result in resistance to PTS-dependent repression (25). Possibly, these mutations lead to long-range conformational changes that affect the orientation of the conserved regions described here.
An important catalytic interaction of IIAGlc in the PTS pathway is with the phosphocarrier HPr. A model for this interaction has been proposed (26). Chen et al. (27) used NMR to demonstrate chemical shift changes on formation of the IIAGlc-HPr complex that correspond to hydrophobic surfaces at the active sites. Of interest, scanning of the sequence of HPr revealed regions that correspond very well to the proposed consensus patterns for both Regions I and II (see Fig. 3 A and B). The active site region of HPr corresponding to Leu14-Arg17 constitutes a loop connecting the first β-strand to the first α-helix and the amino terminus of that helix. That region, together with residues Gly39-Lys40, provides a perfect match to the Region I consensus. Similarly, a β-sheet region (Thr34-Ser41) shows an excellent match to the Region II consensus. The analysis supports the idea that both catalytic and regulatory interactions involving IIAGlc use similar regions.
The proposed consensus sequences for Regions I and II (see Fig. 3) contain various gaps that might result in a lack of specificity such that these sequences would be found in many proteins. To investigate this possibility, the findpatterns program (see Table 1) was used to search sequences from Fig. 3 for the frequency of occurrence of the Region I and II consensus sequences. Consistent with the layout of Fig. 3, LacY, RafB, and MalK contained only one occurrence each of consensus I and II; MelB contained one instance of the Region I consensus and none of the Region II consensus; and HPr contained one copy of the Region II consensus and none of the Region I pattern. The results of this analysis substantiate the specificity and uniqueness of the two proposed consensus sequences.
Table 1.
E. coli proteins containing regions I and II consensus patterns
| Accession number† | Description of protein |
|---|---|
| Transport proteins | |
| p02920 | Lactose permease |
| p02914 | malK; maltose permease |
| p16552 | rafB; raffinose permease |
| p45535 | ABC transporter |
| Kinases | |
| p14377 | Phosphorylates hydrogenase in response to environmental signals |
| p23837 | Phosphorylates acid phosphatase in response to environmental signals |
| DNA proteins | |
| p25239 | Restriction enzyme |
| p14565 | DNA helicase I |
| p22706 | DNA helicase I |
| q46802 | Transcriptional regulator |
| Membrane proteins | |
| p53517 | Outer membrane protein involved in assembly and export of afimbrial adhesin subunits |
| p39180 | Controls colony form variation and autoaggregation |
| p46009 | Outer membrane protein involved in assembly of fimbriae subunits |
| p07110 | Member of fimbrial export usher family |
| p09745 | Expressed by a shufflon (biological switch) |
| p75857 | Fimbriae export usher |
| p25744 | Integral membrane protein |
| p77804 | Hypothetical transmembrane protein |
| p77315 | Integral membrane protein; part of transport system |
| p52143 | Hypothetical outer membrane protein |
| Cytosolic proteins | |
| p11648 | Aminopeptidase |
| p31459 | 2-keto-3-deoxy-galactonokinase |
| p18775 | Sulfoxide terminal reductase |
| p80668 | Phenylacetaldehyde dehydrogenase |
| p52073 | Glycolate oxidase subunit |
| p03827 | Required for transposition of insertion element |
| p19767 | Insertion element |
| p06959 | Pyruvate dehydrogenase E2 |
| p03067 | Replication initiation protein |
| p16553 | Glycosyl hydrolase |
| p11585 | GTP pyrophosphokinase |
| p25741 | Lipopolysaccharide core biosynthesis protein |
| p02359 | Ribosomal protein S7 |
| p76253 | Member of ring-hydroxylating dioxygenase family |
| p42594 | Hypothetical protein |
| p77689 | Hypothetical protein |
| p32051 | Hypothetical lipoprotein |
The findpatterns program (Wisconsin Package Version 9.1, Genetics Computer Group, Madison, WI) was used to search the entire E. coli genome in the SwissProt database (sw:*_ecoli) for the consensus sequence patterns denoted Regions I [(V,L)(X){0,2}(S,A,T)(X){0,2}R(X){0,6}G(K,R)] and II [(S,T)(X){0,4}(V,L)(X){0,2}(N,Q,E)(X){0,2}G(X){0,1}(N,K) (X){0,1}S] in Fig. 3. The sequences in the table contain at least one copy of both sequence patterns.
The accession numbers are from the SwissProt database.
A further test of the specificity of the proposed consensus sequences was carried out. The complete E. coli genome was surveyed for the occurrence of the Region I and II sequence patterns by using the findpatterns program. Although there are 1,456 finds of the individual patterns in the 4,413 protein sequences examined, there are only 37 cases in which a sequence contains both patterns (see Table 1). This analysis provides further evidence for the specific and limited occurrence of the proposed recognition sequences.
For the purposes of discussion, the 37 hits listed in Table 1 are divided into five arbitrary groups. There are only four transport proteins identified in the search. The permeases for lactose, maltose, and raffinose are cited in Fig. 3. An additional transporter (p45535), a member of the ABC transporter family for which the substrate has not yet been established, may be regulated by IIAglc. There are two related kinases (p14377 and p23837) that are involved in environmental signaling. It is an attractive idea that the PTS, via IIAglc, may be in a communication link with that process. Four proteins that interact with DNA were found to contain the pair of consensus sequences. If IIAglc regulates any of these proteins, this might provide a link between the PTS and transcription. A group of membrane proteins was identified in the search. It is unlikely that the outer membrane proteins would be in the same compartment as IIAglc. However, some of these proteins may be transmembrane and part of transport systems (e.g., p77315). When these proteins are more fully characterized, it would be of interest to search for a regulatory role of IIAglc in their function.
Finally, there is a heterogeneous group of cytosolic proteins identified by the search. There are two noteworthy candidates for regulation by IIAglc in this group. The first, pyruvate dehydrogenase E2 (p06959), is of obvious interest because it is involved in the utilization of pyruvate, one of the products of the PTS pathway. The second, GTP pyrophosphokinase (p11585), is involved in the synthesis of the global regulator ppGpp. It would be of obvious fundamental interest to tie this compound to the PTS pathway via IIAglc.
As indicated above (see Fig. 3), the melibiose permease that is known to be regulated by IIAglc contains only the Region I consensus but does not give a precise match for the Region II pattern. It is therefore possible that some sugar transporters and perhaps other proteins regulated by IIAglc were not detected by the search. In this vein, it is noteworthy that several proteins involved with arabinose transport (p03021, regulatory protein for the arabinose operon; p08531, the inner membrane-associated transporter for L-arabinose; p23910, a protein involved in the transport or processing of arabinose polymers) contain the Region I consensus. Furthermore, two proteins involved in sucrose metabolism (p40715, repressor for Csc operon; p40714, sucrose hydrolase) also contain the Region I pattern. It is also of interest that the protein involved in the carboxylation of pyruvate to form oxaloacetate (p00864) contains the Region II consensus. Exploration of the possible regulation of these proteins by IIAglc may be a fruitful area for future research.
The proposed consensus sequences in regions I and II are consistent with a model in which interaction of IIAGlc with its partners occurs by a common mechanism, whether the partner is membrane-bound (LacY, RafB, MelB) or soluble (MalK, GK, HPr). Moreover, the idea that there are two regions for interaction with IIAGlc is supported by the previous findings that polyhistidine insertions in either of two cytoplasmic loops of lac permease result in loss of binding activity (14, 16). The model that substrate binding to a regulated protein brings the two binding regions into the appropriate alignment is an attractive interpretation of the data presented here.
Acknowledgments
We thank Lynn Miller of the Genetics Computer Group (Madison, WI) and Jonathan Reizer (Department of Biology, University of California at San Diego, La Jolla, CA) for helpful discussions dealing with searches for patterns in protein databases.
ABBREVIATIONS
- PTS
phosphoenolpyruvate:sugar phosphotransferase system
- GK
glycerol kinase
- NEM
N-ethylmaleimide.
Footnotes
The substrate-dependent binding obtained with membranes containing Cys-less lac permease is ≈30% of that observed with wild-type lac permease.
References
- 1.Frillingos S, Sahin-Tóth H, Wu J, Kaback H R. FASEB J. 1998;12:1281–1299. doi: 10.1096/fasebj.12.13.1281. [DOI] [PubMed] [Google Scholar]
- 2.Kaback H R. J Membr Biol. 1983;76:95–112. doi: 10.1007/BF02000610. [DOI] [PubMed] [Google Scholar]
- 3.Foster D L, Boublik M, Kaback H R. J Biol Chem. 1983;258:31–34. [PubMed] [Google Scholar]
- 4.Kaback H R, Voss J, Wu J. Curr Opin Struct Biol. 1997;7:537–542. doi: 10.1016/s0959-440x(97)80119-4. [DOI] [PubMed] [Google Scholar]
- 5.Postma P W, Lengeler J W, Jacobson G R. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1149–1174. [Google Scholar]
- 6.Kaback H R. J Biol Chem. 1968;243:3711–3724. [PubMed] [Google Scholar]
- 7.Saier M H, Jr, Novotny M J, Comeau-Fuhrman D, Osumi T, Desai J D. J Bacteriol. 1983;155:1351–1357. doi: 10.1128/jb.155.3.1351-1357.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dean D A, Reizer J, Nikaido H, Saier M H., Jr J Biol Chem. 1990;265:21005–21010. [PubMed] [Google Scholar]
- 9.Titgemeyer F, Mason R E, Saier M H., Jr J Bacteriol. 1994;176:543–546. doi: 10.1128/jb.176.2.543-546.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osumi T, Saier M H., Jr Proc Natl Acad Sci USA. 1982;79:1457–1461. doi: 10.1073/pnas.79.5.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson T H, Yunker P L, Hansen C L. Biochim Biophys Acta. 1990;1029:113–116. doi: 10.1016/0005-2736(90)90443-r. [DOI] [PubMed] [Google Scholar]
- 12.Hurley J H, Faber H R, Worthylake D, Meadow N D, Roseman S, Pettigrew D W, Remington S J. Science. 1993;259:673–677. [PubMed] [Google Scholar]
- 13.Peterkofsky A, Reizer A, Reizer J, Gollop N, Zhu P, Amin N. Prog Nucleic Acid Res Mol Biol. 1993;44:31–65. doi: 10.1016/s0079-6603(08)60216-0. [DOI] [PubMed] [Google Scholar]
- 14.Seok Y-J, Sun J, Kaback H R, Peterkofsky A. Proc Natl Acad Sci USA. 1997;94:13515–13519. doi: 10.1073/pnas.94.25.13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Iwaarden P R, Pastore J C, Konings W N, Kaback H R. Biochemistry. 1991;30:9595–9600. doi: 10.1021/bi00104a005. [DOI] [PubMed] [Google Scholar]
- 16.Hoischen C, Levin J, Pitaknarongphorn S, Reizer J, Saier M H., Jr J Bacteriol. 1996;178:6082–6086. doi: 10.1128/jb.178.20.6082-6086.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddy P, Fredd-Kuldell N, Liberman E, Peterkofsky A. Protein Expression Purif. 1991;2:179–187. doi: 10.1016/1046-5928(91)90069-u. [DOI] [PubMed] [Google Scholar]
- 18.Teather R M, Bramhall J, Riede I, Wright J K, Furst M, Aichele G, Wilhelm U, Overath P. Eur J Biochem. 1980;108:223–231. doi: 10.1111/j.1432-1033.1980.tb04715.x. [DOI] [PubMed] [Google Scholar]
- 19.Venkatesan P, Kaback H R. Proc Natl Acad Sci USA. 1998;95:9802–9807. doi: 10.1073/pnas.95.17.9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frillingos S, Sahin-Tóth M, Persson B, Kaback H R. Biochemistry. 1994;33:8074–8081. doi: 10.1021/bi00192a012. [DOI] [PubMed] [Google Scholar]
- 21.Frillingos S, Kaback H R. Biochemistry. 1996;35:5333–5338. doi: 10.1021/bi953068d. [DOI] [PubMed] [Google Scholar]
- 22.Frillingos S, Gonzalez A, Kaback H R. Biochemistry. 1997;36:14284–14290. doi: 10.1021/bi972314d. [DOI] [PubMed] [Google Scholar]
- 23.Feese M D, Comolli L, Meadow N D, Roseman S, Remington S J. Biochemistry. 1997;36:16087–16096. doi: 10.1021/bi971999e. [DOI] [PubMed] [Google Scholar]
- 24.Kuhnau S, Reyes M, Sievertsen A, Shuman H A, Boos W. J Bacteriol. 1991;173:2180–2186. doi: 10.1128/jb.173.7.2180-2186.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuroda M, de Waard S, Mizushima K, Tsuda M, Postma P, Tsuchiya T. J Biol Chem. 1992;267:18336–18341. [PubMed] [Google Scholar]
- 26.Herzberg O. J Biol Chem. 1992;267:24819–24823. [PubMed] [Google Scholar]
- 27.Chen Y, Reizer J, Saier M H, Jr, Fairbrother W J, Wright P E. Biochemistry. 1993;32:32–37. doi: 10.1021/bi00052a006. [DOI] [PubMed] [Google Scholar]
- 28.Dahl M K, Francoz E, Saurin W, Boos W, Manson M D, Hofnung M. Mol Gen Genet. 1989;218:199–207. doi: 10.1007/BF00331269. [DOI] [PubMed] [Google Scholar]