Abstract
The generation of enzymes to catalyze specific reactions is one of the more challenging problems facing protein engineers. Structural similarities between the enzyme scytalone dehydratase with nuclear transport factor 2 (NTF2) suggested the potential for NTF2 to be re-engineered into a scytalone dehydratase-like enzyme. We introduced four key catalytic residues into NTF2 to create a scytalone dehydratase-like active site. A C-terminal helix found in scytalone dehydratase but absent in NTF2 also was added. Mutant NTF2 proteins were tested for catalytic activity by using a spectroscopic assay. One of the engineered enzymes exhibited catalytic activity with minimal kcat and Km values of 0.125 min−1 and 800 μM, respectively. This level of catalytic activity represents minimally a 150-fold improvement in activity over the background rate for substrate dehydration and a dramatic step forward from the catalytically inert parent NTF2. This work represents one of the few examples of converting a protein scaffold into an enzyme, outside those arising from the induction of catalytic activity into antibodies.
One of the most alluring promises of protein engineering is the ability to create a biocatalyst for any given chemical reaction. Attempts using a combination of site-directed mutagenesis guided by structural and biochemical information have been somewhat successful (1). Even so, the creation of novel biocatalysts remains a challenging problem (2).
Given that the current understanding of how one might create a potent biocatalyst is limited, we have proposed a classification that ranks the enzyme engineering efforts in terms of three levels of increasing difficulty (1). The first category includes the modification of the substrate or cofactor specificity of a given enzyme while retaining the catalytic activity of the enzyme scaffold. Examples of successful applications of this approach are: the modification of the substrate specificity of subtilisin (3–5), the generation of chymotrypsin-like specificity within trypsin (6), and the change in the cofactor specificity of glutathione reductase (7) and lipoamide dehydrogenase (8). The second level envisions the introduction of residues that will confer catalytic activity on a binding protein. This level recently has been accomplished by the re-engineering of cyclophilin into a proline-specific endopeptidase (9). A catalytic triad similar to that found in serine proteases was grafted into the peptidyl-prolyl binding cleft. The resultant protein had an efficiency (kcat/Km) of 0.7 × 104 M−1⋅s−1 toward X-Pro dipeptide substrates and a rate enhancement of (kcat/kuncat) 8 × 108 fold. The third, and most challenging group, imagines taking a protein scaffold, devoid of catalytic and substrate binding activities, and introducing residues that convert the scaffold into a catalyst.
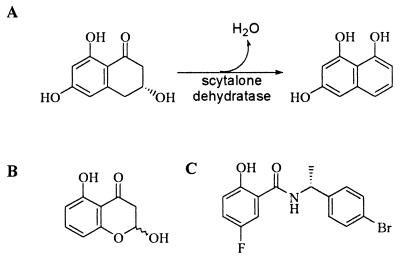
Previously, we focused our efforts on the creation of hybrid enzymes through the fusion of protein modules joining a substrate binding domain and a catalytic domain (10). Although this approach holds promise, we wanted to explore the other levels of our enzyme engineering classification, namely the challenge of converting a protein scaffold into an enzyme. The choice of a suitable scaffold protein was limited by the types of reactions we had chosen to emulate. We focused on reactions that are simple, require no cofactor, and had previously been well characterized. The enzyme scytalone dehydratase fulfills these requirements. Found only in fungi, and as the major determinant of rice blast disease, scytalone dehydratase catalyses two steps in melanin biosynthesis (Fig. 1A). The reaction involves elimination of water via α-proton abstraction to form a conjugated product. From analysis of crystal structures of scytalone dehydratase bound to two different tight binding inhibitors (11, 12), and coupled with site-specific mutagenesis of key residues, a catalytic mechanism was proposed (13).
Figure 1.
(A) Natural reaction catalyzed by scytalone dehydratase. (B) Structure of DDBO. (C) Structure of tight binding inhibitor.
The tertiary structural similarity of scytalone dehydratase to the nuclear transport factor 2 (NTF2 or p10) recently was highlighted (14), although the sequence identity of the two proteins is less than 20%. NTF2 is involved in protein translocation across the nuclear membrane and possesses no catalytic function. We became interested in whether NTF2 could be engineered to express scytalone dehydratase activity. The work described herein details how we mapped the active site residues of scytalone dehydratase onto the structure of NTF2, made the appropriate mutations by site-directed mutagenesis, and characterized the modified NTF2 mutants.
MATERIALS AND METHODS
Materials.
Restriction enzymes, T4 DNA ligase, Taq polymerase, and Wizard DNA preparation systems were obtained from Promega. Some restriction enzymes also were obtained from New England Biolabs. Ultrapure dNTPs were obtained from Boehringer Mannheim. Agarose for analytical gel electrophoresis was obtained from Kodak. Agarose for preparative gel electrophoresis was obtained from FMC. Purified scytalone dehydratase, 2,3-dihydro-2,5-dihydroxy-4H-benzopyran-4-one (DDBO), and scytalone dehydratase inhibitors were obtained from Douglas Jordan and Gregory Basarab (DuPont Agricultural Products). All other materials were of the highest quality available from commercial sources.
Plasmids.
pDB1, pDB2, pDB3, and pET21 expression vectors containing the wild-type and modified scytalone dehydratase gene were obtained from Jim Steffens (DuPont Agricultural Products). An expression vector (pRSETB) containing the NTF2 gene was obtained from Mary Moore (Baylor College of Medicine, Houston).
Molecular Biology.
Site-directed mutagenesis was performed by using the single overlap extension method (15, 16). Small-scale (5 ml) DNA preparations were performed by using Wizard DNA preps (Promega) in accordance with the manufacturer’s procedure. Large-scale (100–500 ml) DNA preparations were performed by using Qiagen Midi-Prep Tip 100 in accordance with the manufacturer’s procedure. Extraction of DNA from low melting point agarose used the Wizard PCR DNA Prep resin.
Protein Expression.
NTF2 constructs were transformed into electrocompetent BL21 (DE3) pLysS (Novagen) and then grown in LB broth containing 100 mg/ml of ampicillin either at 37°C or 25°C until an OD600 of 0.4–0.6 was reached. Protein expression was induced by addition of isopropyl β-d-thiogalactoside (IPTG) to a final concentration of 1 mM. Cells were allowed to grow in the presence of IPTG for either a further 4 hr (37°C) or 16 hr (25°C). After this time the cells were harvested by centrifugation at 8,000 rpm using a GS-3 rotor (Beckman). Cell paste was stored at −80°C until required.
Protein Purification.
All constructs were His-tagged at the amino terminus, which allowed purification using Ni-nitrilotriacetic acid (NTA) resin. A 1-ml Ni-NTA column was poured in a 5-ml syringe containing a frit made of glass wool, washed in accordance with manufacturer’s instructions, and then equilibrated with buffer A (50 mM Tris/300 mM NaCl/50 mM imidazole, pH 8.0). Harvested cells were thawed, resuspended in 30 ml of buffer A, and sonicated by using a Branson Sonifier 450 sonicator, 1/2-inch tip 3 × 2 min at 50% duty cycle. Cell debris was pelleted by centrifugation at 14,000 rpm in an SS-34 rotor. The supernatant was loaded onto the column and washed with buffer A until the flow-through had an A280 <0.01 (≈ 50 ml). Bound protein was eluted by using a stepwise gradient of imidazole from 0.1 to 0.5 M in buffer A.
Enzyme Assays.
Ligand binding assays were performed by measuring quenching of intrinsic protein fluorescence at 340 nm upon excitation at 290 nm using an SLM–Aminco (Urbana, IL) 8000 spectrofluorometer. Continuous assays of enzymatic activity were performed essentially as described by using DDBO as substrate (17). Product formation was followed by determining the change of absorbance at 320 nm (Δɛ = 1.5 mM−1⋅cm−1) with time. Discontinuous assays also were performed. At timed intervals 100-μl aliquots were removed from a 1-ml reaction, held at 25°C, containing 12 mM substrate and up to 3 μM engineered enzyme. The aliquots were stored at −80°C until required. From each aliquot 30 μl was diluted to a final volume of 1 ml, and the remaining substrate was converted to product by using an excess of the wild-type scytalone dehydratase. The difference in absorbance at 320 nm then can be used to calculate the total amount of product formed and hence the concentration of substrate that was present. Spectrophotometric assays were performed by using a Cary-1 (Varian) spectrophotometer.
Structure visualization was done by using the program quanta (Molecular Simulations, San Diego), with Protein Databank files 1STD and 1OUN of scytalone dehydratase and NTF2, respectively.
RESULTS
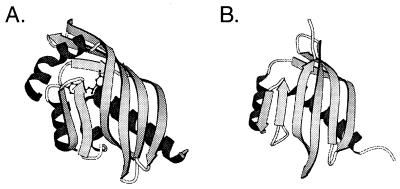
The structural similarity between the scytalone dehydratase and NTF2 is quite remarkable considering that the two proteins have low sequence homology and do not share a common function. The two main structural differences between scytalone dehydratase and NTF2 are a C-terminal α-helix, which although it doesn’t appear to contact the substrate directly is thought to be important in excluding water from the active site and a region of β-sheet that is extended in scytalone dehydratase (Fig. 2).
Figure 2.
Structural comparison of (A) scytalone dehydratase with (B) NTF2 (after ref. 14).
The proposed catalytic mechanism of scytalone dehydratase implicates residues important for substrate binding and catalysis and follows the general mechanism for α-proton abstraction proposed by Gerlt and Gassman (18, 19). In the active site, Asp-31 and His-85 appear to act as a catalytic dyad with His-85 functioning as the general base responsible for abstraction of a proton from the substrate. Tyr-30 and Tyr-50 bind and activate a water molecule that is hydrogen-bonded to the carbonyl of the substrate, lowering the pKa of the proton to be abstracted by His-85. His-110 acts to stabilize the intermediate. In addition to the residues implicitly involved in catalysis, Ser-129 and Asn-131 are thought to aid in correct binding and orientation of the substrate.
By using quanta it was possible to superimpose the structures of scytalone dehydratase and NTF2 and identify the residues in NTF2 that are equivalent to the residues described above. Table 1 shows the result of this comparison. Somewhat surprising was the finding that NTF2 had equivalents to the two key catalytic residues, His-85 and Asp-31. A conservative change was found in Gln for Asn-131 in the enzyme. No equivalent of His-110 was found in NTF2; this residue is part of the β-sheet extension present in scytalone dehydratase but not in NTF2.
Table 1.
Identification of residues of NTF2 equivalent to the active site residues of scytalone dehydratase
| Scytalone dehydratase residue | Proposed function | NTF2 equivalent |
|---|---|---|
| Tyr-30 | Assists protonation of substrates’ carbonyl | Phe-22 |
| Asp-31 | Activation of His-85 | Asp-23 |
| Tyr-50 | Assists protonation of substrates’ carbonyl | Trp-41 |
| His-85 | Proton abstraction | His-66 |
| Val-108 | Involved in positioning of His-110 | Leu-89 |
| His-110 | Assists in hydroxide elimination | None |
| Ser-129 | Orientation of substrate | Phe-99 |
| Asn-131 | Orientation of substrate | Gln-101 |
Residues in NTF2 identified as being in equivalent positions to, but differing from, the mechanistically important residues of scytalone dehydratase were changed by using the overlap extension method of site-directed mutagenesis. Mutations W41Y and F22Y were introduced followed by F99S and Q101N to yield the quadruple (quad) mutant. The C-terminal α-helix of scytalone dehydratase also was introduced by using overlap extension. Residues 152–172 of scytalone dehydratase were fused to residue 123 of NTF2 to generate the quad mutant plus flap. Introduction of the His-110 mutation was difficult because of the lack of corresponding structure in NTF2. To avoid unnecessarily large disruptions in NTF2 we chose to introduce a four-residue pair consisting of a Pro-His and its partner residue pair, Gly-His, from the opposing side of the β-sheet. This addition was sufficient to extend the β-sheet and theoretically to place the histidine residue in an appropriate position to play a role in catalysis. Mutants were expressed and purified to apparent homogeneity.
Investigation of the NTF2 mutants began with an assessment of ligand binding. Because it was likely that the binding of scytalone to NTF2 and the engineered mutants was likely to be very much weaker than to the wild-type scytalone dehydratase, we chose to use a tight-binding inhibitor of scytalone dehydratase (Fig. 1C). Inhibitor binding was monitored by using quenching of intrinsic protein fluorescence, with the results shown in Table 2. From previously published work, this inhibitor has a Ki of 0.2 nM. The quad mutant plus flap bound this inhibitor with a Kd of 35 μM, and the quad mutant plus flap plus His bound with a Kd of 40 μM. Interestingly, the mutant that contained the flap, His, and the Tyr residues involved in coordinating the water bound the inhibitor with a Kd of 14 μM. This finding suggests a steric effect in the quad mutant plus flap plus His may prevent tighter binding of the inhibitor. Assuming that scytalone dehydratase and NTF2 bind the inhibitor and substrate in a similar fashion, the Kd for substrate for the quad mutant plus flap plus His would be 35 × 105 μM.
Table 2.
Comparison of kinetic parameters of the engineered NTF2 mutants with those of the wild-type scytalone dehydratase
| Construct | Kd (inhibitor), μM | kcat (DDBO), s−1 | Km (DDBO), μM | kcat/Km, μM−1⋅s−1 | kcat/kuncat |
|---|---|---|---|---|---|
| Wild-type scytalone dehydratase* | 0.0001 | 400 | 15 | 27 | 2.8 × 107 |
| Wild-type NTF2 | 185 | None detected | N.A. | N.A. | N.A. |
| Quad mutant + flap† | 42 | (a) 1.67 | 3.5 × 106 | 0.48 × 10−6 | 1.2 × 105 |
| (b) 2.1 × 10−3 | 800 | 2.6 × 10−6 | 150 | ||
| Double mutant + flap + His | 14 | None detected | N.A. | N.A. | N.A. |
| Quad mutant + flap + His | 35 | None detected | N.A. | N.A. | N.A. |
With the improved binding of the inhibitor by the mutants, we next addressed the catalytic activity of the NTF2 mutants. Activity was determined by monitoring the dehydration of a scytalone analogue, DDBO (17) (Fig. 1B). For the wild-type enzyme this assay can be done in a continuous fashion by following the increase in absorbance at 320 nm. Initial screening of mutant NTF2 proteins for catalytic activity was performed in a continuous manner by using 150 μM DDBO. The quad mutant plus flap was the only NTF2 mutant to show significant activity above the spontaneous rate of dehydration. The continuous assay gave an apparent kcat of 0.0018 min−1, ≈ twice that of the value for the rate of spontaneous dehydration measured in this study (8.67 × 10−4 min). However, from the results of the inhibitor binding study we expect that this mutant enzyme would have a Km for DDBO significantly higher than 150 μM. We would have to use much higher DDBO concentrations to saturate the enzyme and obtain the true value of kcat. Because of technical considerations, measurement of catalytic activity using concentrations of DDBO above 1 mM required the use of a discontinuous assay. For this assay, aliquots were removed from a reaction mix at timed intervals, and remaining substrate was converted to product by using excess wild-type scytalone dehydratase. The amount of substrate present in each aliquot was determined by measuring the difference in absorbance at 320 nm of the aliquot before and after addition of the wild-type enzyme. In the absence of enzyme, a plot of substrate vs. time shows an exponential decay caused by the spontaneous dehydration of substrate. The rate constant of this process (kuncat) was determined to be 8.67 × 10−4⋅min−1, which compares to the previously measured value of 3.0 × 10−4⋅min−1 (D. B. Jordan and G. S. Basarab, personal communication). The experiment then was repeated in the presence of enzyme, and the resulting data were fit, by using kinsim, to a kinetic model that included a term to allow for the spontaneous dehydration of substrate. The kinsim simulation allows for a range of values for kcat and Km (Table 2). Although we cannot accurately report values of the specific parameters, we can set limits on the specificity of our engineered enzyme toward DDBO. By using the kinetic values obtained from the kinsim simulation the engineered NTF2 has an efficiency (kcat/Km) between 0.47 × 10−6⋅μM−1⋅min−1 and 2.6 × 10−6⋅μM−1⋅min−1 toward DDBO. This result compares to the efficiency of the wild-type scytalone dehydratase of 27 μM−1⋅s−1.
DISCUSSION
That nature has created a unique biocatalyst de novo for every chemical reaction seems unfeasible. Levinthal (21) has pointed out that for a 100-aa protein to sample every possible conformation it would take 1027 years for the protein to fold into the correct conformation. Similarly, for nature to explore all the sequence space available to a 100-aa protein would require production of 20100 different proteins. If only 1 μg of each variant was produced, starting materials with a mass of 1.27 × 10124 g, greater than the mass of the earth (5.98 × 1027 g), would be required. It seems likely therefore, that in the evolution of proteins and specifically enzymes, nature has recruited motifs and domains from other functions and retooled them to change specificity and chemistry. The α/β barrel is one such example of a protein scaffold that serves as the framework for chemically diverse enzymatic reactions that seem to have evolved through changes in key amino acids within the active site (22).
By using two proteins that belong to the α + β fold group, NTF2 and scytalone dehydratase, we have sought to create a unique biocatalyst by mimicking processes that we believe take place in nature, namely the retooling of active sites for different catalytic functions. Our goal is to minimally reconfigure the protein scaffold to confer both substrate binding and enzymatic activity on this fragment.
Analysis of the crystal structure of NTF2 reveals the presence of a hydrophobic pocket in the same region as the hydrophobic active site of scytalone dehydratase and binds to the small molecular weight G-protein Ran (23). A phenylalanine residue of Ran binds into the hydrophobic pocket of NTF2. Because of the similar overall structure and presence of an appropriately placed hydrophobic pocket in NTF2, we reasoned that it should be possible to decorate the hydrophobic pocket of NTF2 with residues from scytalone dehydratase that confer substrate binding and catalysis.
Surprisingly, the wild-type NTF2 was found to be capable of binding the tight-binding inhibitor of scytalone dehydratase, although the difference in Kd values is a factor of 106, disfavoring binding to NTF2. The interaction of the inhibitor with NTF2 is most likely a consequence of its hydrophobic nature rather than a specific interaction with the protein. By decorating the hydrophobic pocket of NTF2 with residues that should confer substrate binding and catalysis we were able to observe a value for kcat/Km between 0.47 × 10−6⋅μM−1⋅min−1 and 2.6 × 10−6⋅μM−1⋅min−1 toward DDBO. If we presume the binding of inhibitor and DDBO to our construct parallels their affinity for scytalone dehydratase, the Km for DDBO would be greater than mM. We suspect that the Km for DDBO is indeed in that range, because simulations of the kinetic assay converge to a span of Km values from 0.8 to 3,500 mM. Consequently, kcat is minimally a value of 150 over background and most likely higher. Given that wild-type NTF2 possess no scytalone dehydratase activity and appears to be unable to bind the tight-binding inhibitor with high affinity, it is remarkable that we have been able to convert the NTF2 scaffold into an enzyme. This result clearly indicates the applicability of retooling protein scaffolds into catalytically active proteins able to act on substrates previously not associated with that scaffold.
Mutagenesis studies of the serine protease, substilisin, parallel the work described here (24). Replacement of the three key catalytic residues with alanine lowered kcat, by a factor of 107, with little effect on Km. However, this protein was still capable of a rate acceleration of 2.7 × 103 above the background rate of hydrolysis. The most dramatic loss of activity was with the first mutation, a drop in kcat of 10−5 for Asp and 10−7 for His and Ser. This finding suggests that there could well be some synergy between the residues in and around the active site. The implication is that even with several elements important to catalysis already in place, the introduction of an appropriately positioned final catalytic residue would be sufficient to dramatically raise catalytic activity. This hypothesis is particularly appropriate to consider because the only NTF2 mutant to show activity above the spontaneous rate of dehydration lacks the equivalent of His-110, a residue known to be important in the catalytic mechanism of scytalone dehydratase.
Although the absence of structural information prevents a detailed discussion of the reasons for the limited catalytic activity of the engineered enzyme, it is possible to speculate as to what some of the problems may be. As described above, the introduction of one key residue could be sufficient to dramatically improve catalytic activity. However, one would expect that this concept requires the associated active site residues to be already appropriately positioned for catalysis. We would be very fortunate if the residues that were mutated in NTF2 to mimic the active site of scytalone dehydratase naturally adopted the correct orientation that allowed them to be effective in catalysis. In a naturally evolved enzyme such geometrical arrangement of residues essential for catalytic activity is achieved, in part, through interaction with second shell residues. The rational design of such interactions is something that is likely to be difficult to predict, but could be optimized through the use of random mutagenesis coupled with a suitable biological selection.
Although the current trend in protein engineering seems to be moving away from rational approaches and toward more stochastic methods of producing biocatalysts with novel reactivities it is likely that a combination of the two approaches will yield results. We propose to use the system under discussion to test the feasibility of such an approach. In the first phase of this work we used a rational approach to introduce into a protein scaffold residues that will confer substrate binding and catalysis. This initial rational approach made sense because structural and kinetic information is available that allowed identification of key residues. Moreover, to introduce four residues using random mutagenesis would require screening of a library of greater than 1012 members (25). Having been able to introduce some catalytic activity into a protein scaffold we feel that we are now in a position to move to the second phase of this work, namely improvement of either substrate binding or catalysis or both using random mutagenesis techniques.
Acknowledgments
We thank Dr. J. Steffens (DuPont Agricultural Products) for providing the scytalone dehydratase expression clones; Drs. D. B. Jordan and G. S. Basarab (DuPont Agricultural Products) for providing the substrate and inhibitor used in this work and for communicating results before publication; Prof. M. Moore and Dr. C. Lane (Baylor College of Medicine) for providing the NTF2 expression clone and advice on purification of the NTF2 protein; and Drs. C. P. Scott and M. Ostermeier for their critical reading of the manuscript. S.M.F. is supported by the cancer research fund of the Damon Runyon-Walter Winchell foundation fellowship, DRG-1398.
ABBREVIATIONS
- DDBO
2,3-dihydro-2,5-dihydroxy-4H-benzopyran-4-one
- NTF2
nuclear transport factor 2
- quad mutant
NTF2 containing W41Y, F22Y, F99S, and Q101N mutations
- quad mutant plus flap
quad mutant plus 20 amino acid C-terminal α-helix from scytalone dehydratase
References
- 1.Nixon A E, Ostermeier M, Benkovic S J. Trends Biotechnol. 1998;16:258–264. doi: 10.1016/s0167-7799(98)01204-9. [DOI] [PubMed] [Google Scholar]
- 2.Ballinger M D, Tom J, Wells J A. Biochemistry. 1995;34:13312–13319. doi: 10.1021/bi00041a006. [DOI] [PubMed] [Google Scholar]
- 3.Ballinger M D, Tom J, Wells J A. Biochemistry. 1996;35:13579–13585. doi: 10.1021/bi961543h. [DOI] [PubMed] [Google Scholar]
- 4.Corey M J, Corey E. Proc Natl Acad Sci USA. 1996;93:11428–11434. doi: 10.1073/pnas.93.21.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wells J A, Cunningham A C, Graycar T P, Estell D A. Proc Natl Acad Sci USA. 1987;84:5167–5171. doi: 10.1073/pnas.84.15.5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hedstrom L, Szilagyi L, Rutter W J. Science. 1992;255:1249–1253. doi: 10.1126/science.1546324. [DOI] [PubMed] [Google Scholar]
- 7.Scrutton N S, Berry A, Perham R N. Nature (London) 1990;343:38–43. doi: 10.1038/343038a0. [DOI] [PubMed] [Google Scholar]
- 8.Bocanegra J A, Scrutton N S, Perham R N. Biochemistry. 1993;32:2737–2740. doi: 10.1021/bi00062a001. [DOI] [PubMed] [Google Scholar]
- 9.Quemeneur E, Moutiez M, Charbonnier J-B, Menez A. Nature (London) 1998;391:301–304. doi: 10.1038/34687. [DOI] [PubMed] [Google Scholar]
- 10.Nixon A E, Warren M S, Benkovic S J. Proc Natl Acad Sci USA. 1997;94:1069–1073. doi: 10.1073/pnas.94.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lundqvist T, Rice J, Hodge C N, Basarab G S, Pierce J, Lindqvist Y. Structure (London) 1994;2:937–944. doi: 10.1016/s0969-2126(94)00095-6. [DOI] [PubMed] [Google Scholar]
- 12.Nakasako M, Motoyama T, Kurahashi Y, Yamaguchi I. Biochemistry. 1998;37:9931–9939. doi: 10.1021/bi980321b. [DOI] [PubMed] [Google Scholar]
- 13.Jordan, D. B., Basarab, G. S., Steffens, J. J., Pfrogner, B. R., Schwartz, R. S. & Wawrzak, Z. (1999) Pesticide Sci., in press.
- 14.Murzin A G. Curr Opin Struct Biol. 1996;6:386–394. doi: 10.1016/s0959-440x(96)80059-5. [DOI] [PubMed] [Google Scholar]
- 15.Higuchi R, Krummel B, Saiki R K. Nucleic Acids Res. 1988;15:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 17.Thompson J E, Basarab G S, Pierce J, Hodge C N, Jordan D J. Anal Biochem. 1998;256:1–6. doi: 10.1006/abio.1997.2489. [DOI] [PubMed] [Google Scholar]
- 18.Gerlt J A, Gassman P G. J Am Chem Soc. 1991;113:9667–9669. [Google Scholar]
- 19.Gerlt J A, Gassman P G. J Am Chem Soc. 1992;114:5928–5934. [Google Scholar]
- 20.Chen J M, Xu S L, Wawrzak Z, Basarab G S, Jordan D B. Biochemistry. 1998;37:17735–17744. doi: 10.1021/bi981848r. [DOI] [PubMed] [Google Scholar]
- 21.Levinthal C. J Chim Phys. 1968;65:44–45. [Google Scholar]
- 22.Babbit P C, Gerlt J A. J Biol Chem. 1997;272:30591–30594. doi: 10.1074/jbc.272.49.30591. [DOI] [PubMed] [Google Scholar]
- 23.Stewart M, Kent H M, McCoy A J. J Mol Biol. 1998;277:635–646. doi: 10.1006/jmbi.1997.1602. [DOI] [PubMed] [Google Scholar]
- 24.Carter P, Wells J A. Nature (London) 1988;332:564–568. doi: 10.1038/332564a0. [DOI] [PubMed] [Google Scholar]
- 25.Kuchner O, Arnold F H. Trends Biotechnol. 1997;15:523–530. doi: 10.1016/S0167-7799(97)01138-4. [DOI] [PubMed] [Google Scholar]