Abstract
This study examines the impact of a series of mutations at position 553 on the kinetic and structural properties of soybean lipoxygenase-1 (SLO-1). The previously uncharacterized mutants reported herein are I553L, I553V, and I553G. High-resolution x-ray studies of these mutants, together with the earlier studied I553A, show almost no structural change in relation to the WT-enzyme. By contrast, a progression in kinetic behavior occurs in which the decrease in the size of the side chain at position 553 leads to an increased importance of donor–acceptor distance sampling in the course of the hydrogen transfer process. These dynamical changes in behavior are interpreted in the context of two general classes of protein motions, preorganization and reorganization, with the latter including the distance sampling modes [Klinman JP (2006) Philos Trans R Soc London Ser B 361:1323–1331; Nagel Z, Klinman JP (2006) Chem Rev 106:3095–3118]. The aggregate data for SLO-1 show how judicious placement of hydrophobic side chains can influence enzyme catalysis via enhanced donor–acceptor hydrogenic wave function overlap.
Keywords: protein dynamics, hydrogenic wave function overlap
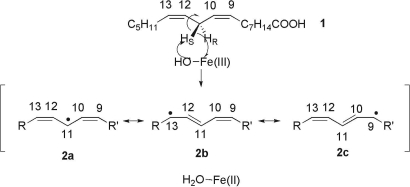
The properties of the C–H bond cleavage reaction, catalyzed by soybean lipoxygenase-1 (SLO-1), have played a major role in shifting the formalisms that describe enzyme function (1–6). As shown in Scheme 1, SLO-1 catalyzes a proton-coupled electron transfer from the C-11 of bound substrate to the active site ferric-hydroxide, producing a substrate-derived free radical and ferrous-water (note ref. 7 and references therein).
Scheme. 1.
Consensus mechanism for SLO-1 in which the active site Fe(III)-OH abstracts a proton and electron from the C-11 position of substrate. The structures 2a, 2b, and 2c indicate the three extreme resonance forms in which the single electron density resides at positions 11, 13, and 9, respectively.
This reaction exhibits one of the largest primary kinetic isotope effects seen in any enzyme reaction (kH/kD ≈80 at room temperature), together with small values for both the energy of activation (Ea = 2.1 ± 0.2 kcal/mol) and the difference in Ea for deuterium vs. protium transfer (Ea(D) − Ea(H) = 0.9 ± 0.2 kcal/mol) (8). The composite properties of the WT-SLO-1 constitute some of the strongest evidence in support of a hydrogen tunneling mechanism in an enzyme reaction, with SLO-1 serving as a “gold standard” for computational chemists attempting to model and understand the role of quantum mechanical hydrogen transfer in enzyme function (e.g., refs. 9–14).
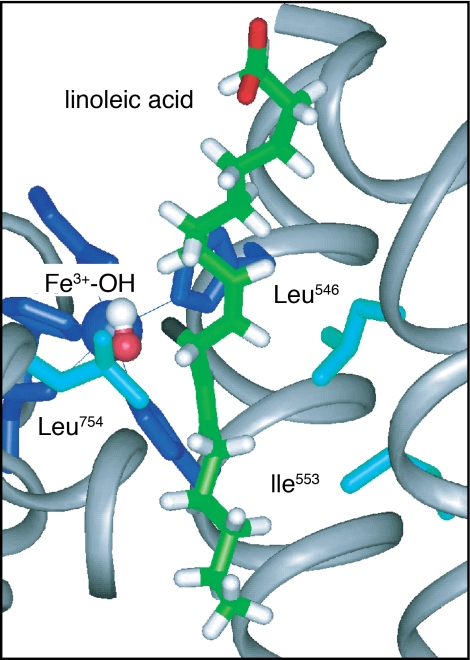
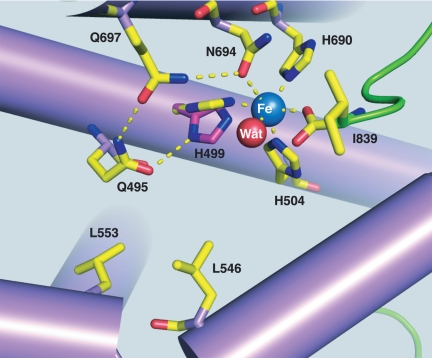
In addition to the detailed kinetic characterizations that have been carried out for WT-SLO-1 (1, 2), site-specific mutagenesis has been pursued for a number of hydrophobic residues that reside at or near the bound substrate; these residues include Ile-553, Leu-546, and Leu-754 (8, 15), illustrated in Fig. 1. In the case of Leu-546 and Leu-754 the side chains seem to play a role in “sandwiching” the bound substrate into its correct position for reaction with the active site iron, whereas Ile-553 is more distal, residing a helix turn away from Leu-546. Previously published results, in which each of these residues was converted to an alanine, indicated a number of distinctive properties (8, 15). In all instances, the observed kinetic isotope effects remained large (≈80–100), whereas L546A and L754A produced a reduction in rate of 62-fold and 950-fold, respectively. These changes in rate were accompanied by an increase in Ea from 2.1 to 4.1 kcal/mol, as well as a noticeable increase in ΔEa = Ea(D) − Ea(H) to 1.9 ± 0.6 and 2.0 ± 0.5 kcal/mol, respectively (8, 15).
Fig. 1.
Active site structure of SLO-1, illustrating the placement of the reactive carbon of substrate in relation to Leu-546, Leu-754, and Ile-553 (hydrogen that is cleaved is labeled in black). The bound substrate, linoleic acid, has been modeled into the active site (15).
It is the latter property that has received the greatest attention. As discussed, the observed constancy of the kinetic isotope effects, together with the increase in ΔEa, is ascribed to mutant-derived defects in the active site that lead to an increased initial distance between the reacting C-11 of substrate and oxygen of Fe(III)-OH in the mutants. The deletion of hydrophobic side chains also produces a lower frequency (softer potential) for motion within the active site, such that an increase in distance sampling between the reacting atoms can compensate for the longer initial distance (15, 16). It is the increased distance sampling (or gating) that increases the value for ΔEa. The reason for this increased ΔEa is straightforward: The deuterium, because of its shorter wavelength, undergoes transfer at a shorter distance than for protium with a concomitant increase in Ea(D) in relation to Ea(H) (17).
The previously reported behavior of I553A is also of considerable interest, given the absence of a significant impact of I553A on rate for the protium substrate, together with the largest increase seen in ΔEa = 4.0 ± 0.3 kcal/mol (8, 15). This finding implied a number of features regarding the role of distance sampling in catalysis. First, there was the possibility that the contribution of distance sampling to the observed energy of activation for protium transfer was small in this case, with the barrier to reaction coming primarily from local changes in active site electrostatics and bonding (the lambda term) that must occur for efficient tunneling to take place (note ref. 18). Second, the results indicated that a hydrophobic residue distal from the reactive bond(s) could influence the transmission of motion to the active site that is necessary to increase the probability of wave function overlap for deuterium transfer between the donor and acceptor atoms. The original studies of I553A were criticized (14), with the claim that experimental differences between this mutant and WT-SLO-1 were not statistically significant and that a better way to examine the data was to average the observed behavior for the two enzymes. It should be noted that the result of such averaging was to make the data conform more closely to previously existing models for the temperature dependences of kinetic isotope effects and their relationship to the H-tunneling process (19).
In the present study, results are provided that extend the mutations at position 553 to include all naturally occurring aliphatic side chains (Leu, Ile, Val, Ala, and Gly). As the data indicate, there is a trend in the direction of an ever increasing requirement for distance sampling as the bulk of the side chain is reduced. To verify that these effects are, in fact, dynamical in nature, x-ray crystallographic analyses were carried out for I553A, I553V, and I553G at close to atomic resolution. The structural data indicate virtually identical active sites with only small differences in the position of either one of the ligands to the active site iron or the side chain of Leu-546. These data both dispel previous claims that the large ΔEa for I553A could have been due to “incorrect” experimental analysis and reinforce the concept of dynamical differences between the WT-SLO-1 and its mutants as the source of the observed trends in kinetic parameters.
Results
Kinetic Parameters.
To extend the earlier investigation of WT-SLO-1 and I553A, a series of mutants at position 553 was created: I553L, I553V, and I553G, in which there is a progressive reduction in bulk relative to the naturally occurring amino acid side chain:
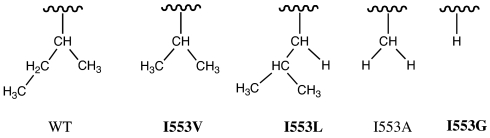 |
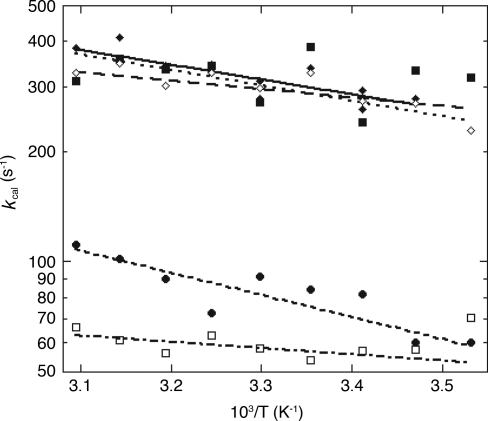
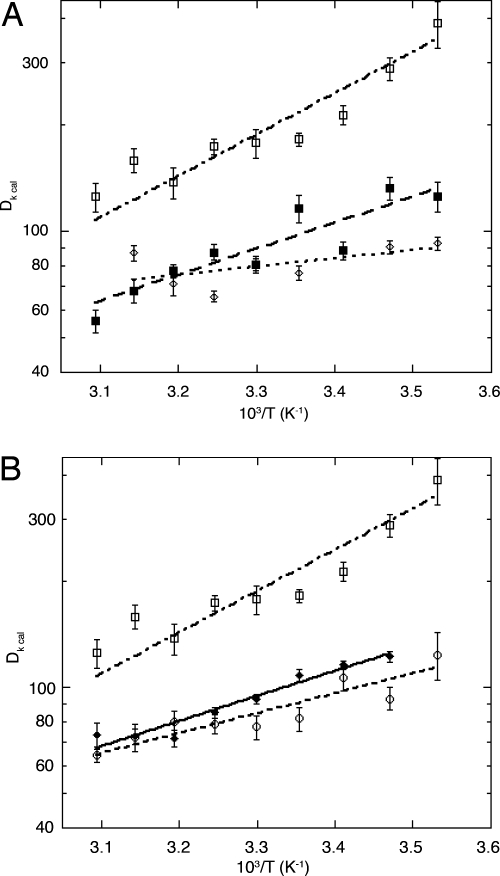
We have ordered the mutants based on the degree of substitution at the β-carbon because this ordering correlates with the observed experimental parameters (see Table 1). A similar correlation between substitution at the β-carbon and tunneling efficiency has already been seen in the reaction catalyzed by horse liver alcohol dehydrogenase (20). For each of the previously uncharacterized mutants (in bold above), steady state kinetic data were collected by using protio-linoleic acid and 11,11-dideuterio-linoleic acid between 10°C and 50°C. At the outset, the rate of product formation was measured by following the appearance of product hydroperoxide at 234 nm under the ambient oxygen concentration. Subsequent experiments tested whether ambient oxygen levels were saturating at each temperature by varying the oxygen tension between ≈10% to 100% O2. Small corrections were then applied as needed [see supporting information (SI) Materials and Methods] to yield the final values for kcat, Km(S), and their respective isotope effects, SI Tables 2–4. The kcat value for I553L of 322 ± 33 s−1, averaged over all temperatures, is either equal to or slightly increased in relation to WT-SLO-1 at 30°C (kcat = 297 ± 12 s−1), whereas the kcat values for I553V and I553G are decreased ≈3- and 5-fold, respectively. Data fitted to the full experimental temperature range for kcat and D(kcat) are illustrated in Figs. 2 and 3, respectively. One notable feature for all of the mutants studied herein is the fact that the values for D(kcat) remain very large, with the magnitude of D(kcat) at 25°C similar to WT-SLO for I553V and I553L, and the value for I553G exceeding WT at all temperatures.
Table 1.
Summary of empirical and computed Arrhenius parameters
| 1SLO mutant | Experimental Arrhenius parameters |
Calculated Arrhenius parameters* |
Input parameters† |
||||
|---|---|---|---|---|---|---|---|
| Ea(H), kcal/mol | Ea(D) − Ea(H), kcal/mol | AH/AD | Ea(D) − Ea(H), kcal/mol | AH/AD | r0, Å | ΩGating, cm−1 | |
| WT | 2.1 ± 0.2‡ | 0.9 ± 0.2‡ | 18 ± 5‡ | 0.9§ | 20§ | 1.02§ | 167§ |
| Ile553 → Val | 2.4 ± 0.5 | 2.6 ± 0.5 | 0.3 ± 0.2 | 2.6 | 1.1 | 1.46 | 68 |
| Ile553 → Leu | 0.4 ± 0.7 | 3.4 ± 0.6 | 0.3 ± 0.4 | 3.4 | 0.33 | 1.85 | 52 |
| Ile553 → Ala | 1.9 ± 0.2‡ | 4.0 ± 0.3‡ | 0.12 ± 0.06‡ | 4.0§ | 0.11§ | 2.34§ | 42§ |
| Ile553 → Gly | 0.03 ± 0.04 | 5.3 ± 0.7 | 0.027 ± 0.034 | 5.3 | 0.026 | 3.14 | 35 |
*Calculated by treating the hydrogen wave functions as Morse oscillators. The significance of these calculations is the trends in parameters, not their absolute magnitude.
† r0 is the initial distance that hydrogen would have to transfer between the donor and acceptor (prior to gating). Ωgating is the frequency of the gating mode.
‡Data from ref. 15.
§Data from ref. 16.
Fig. 2.
Plot of ln kcat vs. 1/T for WT-SLO-1 (◇◇), I553A (♦), I553L (■), I553V (●), and I553G (□). Data for WT and I553A are from ref. 15.
Fig. 3.
Plots of ln D(kcat) vs. 1/T. (A) WT-SLO-1 (◇), I553L (■), I553G (□). (B) I553V (○), I553A (♦), I553G (□). The data are presented in two frames, to avoid crowding with I553G used as the frame of reference. Data for WT and I553A are from ref. 15.
In Table 1 the experimental parameters Ea, Ea(D) − Ea(H), and AH/AD are summarized. The enthalpies of activation using protio-linoleic acid have either stayed the same as for WT (I553V) or decreased (I553L and I553G). The latter may explain the slight increase in rate for I553L, but the decreases in rate for I553V and I553G indicate a corresponding decrease in the TΔS‡ term. Notably, for all three mutants, there is an increase in the temperature dependence of the kinetic isotope effects (KIEs) that is in the same direction seen for I553A and distinct from the behavior for the WT-SLO-1. Of considerable importance for our understanding of the parameters that control H-tunneling in SLO-1, the trend in increasing ΔEa/decreasing AH/AD values mirrors the decreasing presence of space filling side chains at the β-carbon: AH/AD (WT) = 18 ± 5 > AH/AD (I553L and I553V) = 0.3 ± 0.30 > AH/AD I553A = 0.12 ± 0.06 > AH/AD I553G = 0.027 ± 0.034.
X-Ray Structures for Mutants at Position 553.
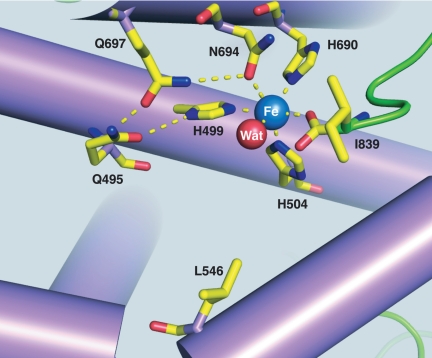
The structures for all four I553 mutant proteins have now been determined at close to atomic resolution (1.45 Å, 1.60 Å, 1.40 Å and 1.65 Å for I553L, I553V, I553A and I553G, respectively), SI Table 5. These structures are found to be virtually superimposable onto the structure for the WT-protein with some subtle differences at the active site in the region of the mutation. As shown in Fig. 4 for I553L, one of the histidine ligands to the active site iron is modeled in two conformations. In the high-resolution structure for the WT, the H499 is seen as a mixture of conformations that includes the alternate position observed for I553L, indicating inherent mobility for this side chain and its partner Q495. Another difference seen among the I553 mutants is the orientation of the hydrophobic side chain of L546 that resides one helix turn away and on the same face as the I553 side chain, Fig. 5 for I553G. It is clear the L546 can exist in (at least) two conformations that place the iso-butyl side chain either facing or rotated away from the side chain at position 553, and in the I553A structure the side chain is modeled as a statistical mixture of both rotamers. There does not seem to be a relationship between the size of the substitution at position 553 and the position of the side chain at 546, with I553A and I553L indicating one conformation and I553G and I553V showing the alternate conformation. The final difference observed from WT and I553L is a change in the rotamer for Q495 in the I553V, I553A and I553G mutants. The rotation of this side chain does not fundamentally disturb the hydrogen-bonding interactions of the Q495 carboxamide with Q697 and H499, and probably reflects an inherent flexibility in this residue.
Fig. 4.
Configuration of ligands to the active site iron atom in I553L. The leucine side chain at position 546 resides one helix turn away, in the direction of the catalytically active iron center. The shadow at position 553 traces out the region occupied by isoleucine in the WT-SLO.
Fig. 5.
Configuration of ligands to the active site iron atom in I553G. The leucine side chain at position 546 resides one helix turn away, in the direction of the catalytically active iron center. The shadow at position 553 traces out the region occupied by isoleucine in the WT-SLO.
Perhaps the most important conclusion from the new structures for SLO is that complete deletion of the side chain at I553 (e.g., I553G) leads neither to a collapse of the native structure nor to the presence of an ordered network of water molecules. The structural studies imply, instead, the increasing creation of a cavity as a result of mutation that may be expected to alter either the conformation of bound substrate and/or the resulting parameters that affect the efficiency of wave function overlap from the substrate donor to Fe(III)-OH acceptor. With regard to the issue of substrate binding, modeling of linoleic acid into the structure for the WT-SLO-1 indicates a possible van der Waals contact between the Ile side chain and the position 14 of bound substrate. An examination of the impact of mutation on the binding of substrate to the mutant proteins is possible from the kinetically measured Km values for the deuterated substrate, where rate-determining chemistry means that kcat/Km = kcat/KD. From SI Tables 2–4, KD values are estimated to fall between 3 to 4 μM, with KD = 10 ± 4 and 3.5 ± 0.2 for the WT-SLO-1 and I553A (15). The data thus indicate no impairment of substrate binding, which, together with the structural similarities among the mutants (Figs. 4 and 5), implicates a dynamical role for I553 in the hydrogen tunneling process.
Discussion
Distinguishing the Types of Heavy Atom Motions That Impact H-Tunneling.
The data presented herein allow a careful analysis of the impact of an amino acid side chain, I553, on the properties for hydrogen tunneling in the SLO-1 reaction. Because this side chain is ≈15 Å from the active site iron center it offers an excellent opportunity to understand the role of distal residues on the tunneling process, in particular, the impact of an imposed packing defect on the catalytic features of the active site. It can be seen that the impact of mutation on rate is fairly small and, in the two instances where rates are decreased, the result cannot be ascribed to an increase in Ea as this parameter is either unchanged or significantly reduced. Based on studies of a thermophilic alcohol dehydrogenase (21, 22) it is conceptually beneficial to parse the protein motions that control H-tunneling into two categories (23). The first class of motions has been labeled preorganization and reflects an equilibrium sampling of multiple protein conformations that may involve large segments of protein on time scales anywhere from seconds to nanoseconds. In WT protein, a subset of these conformations provides the requisite interactions between the protein backbone and bound substrate for tunneling to take place. Because the rate of reaction is thus related to the probability of achieving the optimal subset of protein conformations, the TΔS‡ contribution to ΔG‡ (= ΔH‡ − TΔS‡) may be expected to play a dominant role in protein preorganization. From a functional viewpoint, a small value of Ea for protein preorganization guarantees that a very large number of protein conformations can be readily accessed and interconverted while avoiding the trapping of protein into nonproductive, local minima. The expectation of a small change in Ea for distal mutations is in fact realized for the I553L and I553G mutants, where the observed decreases in rate correlate with decreases in TΔS‡, attributed to a smaller probability of achieving the conformers that promote the precise placement of substrate and other side chains necessary for efficient H-tunneling. In a recent study of a double mutant involving two distal side chains in the Escherichia coli dihydrofolate reductase (EcDHFR), Kohen and coworkers (24) observed a similar trend in which Ea was unchanged or reduced, with the large change in rate arising from a decrease in TΔS‡.
Once the family of properly preorganized conformations has been achieved, fine-tuning of the local environment controls the barrier crossing; this fine-tuning is expected to be more proximal to the reacting bonds and to occur on a faster time scale (nanoseconds to picoseconds) than preorganization. This second type of motion has been labeled as the reorganization and can be contextualized within a modified Marcus equation that takes into account the difference between electron and hydrogen tunneling reactions (8, 15, 25, 26). As shown in Eq. 1, three major factors determine the efficiency of H-transfer: (i) a Marcus term that contains lambda (λ), the sum of the contribution of inner and local outer reorganization to the reaction barrier and ΔG°, the reaction driving force; (ii) the Franck Condon overlap that defines the probability of wave function overlap between the H-donor and acceptor as a function of the mass (mH), frequency (ωH), and distance (rH) traveled by the transferred hydrogen; and (iii) a distance sampling term that reflects the barrier encountered, Ex, in reducing the distance between the H-donor and acceptor:
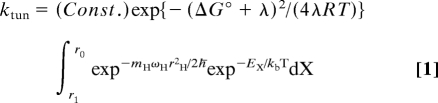 |
where h is Planck's constant divided by 2π, ωx is the frequency of the gating oscillation, rx is the distance over which the gating unit moves, and mx is the mass of the gating unit. The expression given in Eq. 1 is for a reaction in which tunneling occurs exclusively between ground state vibrational levels (an expanded equation that includes tunneling between excited vibrational levels is available in ref. 15). Inspection of the three exponentials in Eq. 1 shows that the first is temperature-dependent and mass-independent. As discussed in an earlier work (15), the majority of tunneling in SLO-1 is expected to take place from the zero point energy level, although a small contribution from excited state vibrational modes arises, especially for deuterium transfer. Modeling shows that the latter will produce a small temperature dependence to the KIE, with values for AH/AD that remain above unity. The second exponential in Eq. 1 contains the Franck Condon term for wave function overlap, which is temperature independent, although strongly mass dependent. However, when this Franck Condon term is integrated over a range of donor–acceptor distances, as shown, the full term within the integral sign becomes both mass- and temperature-dependent. Although the expression for reorganization has historically (in the case of electron transfer) been confined to the lambda parameter in the first exponential of Eq. 1 (26), it is useful to expand the definition of reorganization for hydrogen transfer to include the distance sampling term that arises in the third exponential of Eq. 1. As has been discussed in the original derivation of the Marcus theory for electron transfer (26, 27), the reorganization factors that control hydrogen tunneling may be expected to be primarily manifest in the enthalpic barrier to catalysis.
Physical Origin of the Behavior of the I553 Mutants.
The results provided herein lead to a large number of experimental parameters that include the structures for the mutant proteins, together with rate parameters, their temperature dependences (Ea), the experimental KIEs and their temperature dependences (ΔEa). It is likely that any dynamical behavior produced by the mutations at position 553 will be manifested through a network of residues whose motions are correlated with I553. As discussed above, the impact of mutation on rate seen for I553V and I553G, with no attendant increase in Ea for reaction of the protio-substrate, is ascribed to a decreased probability of these proteins to reach the family of reactive conformers, i.e., a hindered preorganization, together with relatively little change in its reorganization barrier. The lack of an impact on lambda (as defined above) may not be unexpected in light of the nature of the mutations at position 553 (all uncharged aliphatic side chains or hydrogen) and their distance from the active site. The impact on the distance sampling term for the protio-substrate is a little more difficult to assess. In an earlier modeling study, it was concluded that the introduction of the mutations at position 553 leads to an increased initial distance between the donor and acceptor atoms with the trends in increased distance (rather than their actual values) of primary importance to our understanding of the SLO-1 behavior (16). Note that an increase in distance, without compensation via gating, would increase the size of the observed kinetic isotope effects (21), which has not been observed with the exception of the I553G mutant. Thus, it is likely that the transfer of the protium in all of the mutant proteins requires more gating than with the WT-SLO-1. Given the lack of a significant perturbation of Ea, however, the observed properties for the mutant proteins almost certainly result from a combination of decreasing force constants for the protein gating mode coupled to the need for a relatively small increase in the degree of distance sampling between the donor and acceptor atoms when protium is undergoing transfer.
The most significant result to emerge from this study is the regular trend in Ea(D) − Ea(H) and AH/AD, toward increasingly larger and smaller values, respectively, as the size of the cavity caused by deletion of the side chain at position 553 is increased. The trend in AH/AD is a direct consequence of the progressive inflation of Ea(D) in relation to Ea(H) in proceeding from the WT-enzyme to I553G. The increasing elevation for Ea(D) indicates the extreme difficulty deuterium encounters in moving from donor to acceptor when the distance between these heavy atoms deviates from the optimal configuration of the WT-enzyme. Basically, because of its heavier mass and resulting smaller wavelength, deuterium finds itself unable to move between the donor and acceptor wells without substantial distance sampling (17), elevating the value of Ex in Eq. 1, which is then reflected in an elevated Ea(D) term. The need for closer approach between the donor and acceptor atoms becomes increasingly acute as the extent of the “defect” in the active site reaches the limit of an absent side chain, in which case the experimental KIE has actually become elevated. The latter is consistent with the notion that in I553G the closest approach (after gating) between the donor and acceptor atoms cannot approach that of the WT-protein. According to the above picture, the wavelength of the protium is long enough, within the confines of the SLO-1 active site, such that some increase in the initial donor–acceptor distance can be tolerated without a significant compromise in reactivity. In the case of the transfer of the heavier deuterium, however, the increase in donor–acceptor distance becomes increasingly unacceptable: the dominant factor that produces the trends in ΔEa is, thus, the smaller wave length for deuterium (a consequence of its larger mass), necessitating a closer approach between the donor and acceptor atoms before effective wave function overlap can occur (17).
It can be seen, from the above discussion, that there is an interplay between the preorganization and reorganization terms that control H-tunneling, such that mutation of a protein can produce a complex result. In the case of the I553 mutants of SLO-1, mutation is concluded to both compromise the ability of the enzyme to achieve its family of optimal catalytic conformers and to impair the ability of deuterium to move between the donor and acceptor wells within the achievable family of preorganized states. The increase in the gating requirement during D-transfer is also complex, reflecting a concomitant change in the donor acceptor distance and the frequency that controls the distance sampling mode. In fact, the decrease in the frequency for the gating mode(s) that arises from the mutations makes gating more accessible, enabling the mutant enzymes to “compensate” for an initial imperfect placement of the hydrogen donor in relation to its acceptor via enhanced access to the distance sampling modes. The participation of two compensating features is likely the origin of a fortuitous agreement in kcat values for protio-substrate with WT-SLO and the I553A mutant (15).
Implicit in the above discussion is the assumption that mutations at position 553 have no impact on the inner and outer sphere reorganization represented in the λ term of Eq. 1. The hydrogen bonding network among N694, Q697, and Q495 has been proposed to control the flexibility of N694, which in turn, controls the reactivity and coordination number of the active site iron. The observation that the orientation of Q495 is seen to change in some of the I553 mutants (compare Figs. 4 and 5) raised the issue of whether this structural change could also be impacting the experimental rate parameters. We consider this explanation highly unlikely for a number of reasons that include: (i) the observation of two configurations for the H499/Q495 hydrogen bonding pair in WT-SLO; (ii) the fact that the full H-bonding network is retained, along with the liganding of H499 to the iron, among all of the mutants; and (iii) the published finding that the Q495E retained full enzyme activity (28).
Link Between Enzyme Efficiency and Tunneling.
The aggregate data for SLO-1 that include huge primary deuterium kinetic isotope effects for WT-enzyme and all mutants studied thus far, an unexpectedly small temperature dependence of the size of the isotope effects with WT-enzyme, and highly temperature dependent KIEs for the distal mutations at position 553 provide some of the strongest evidence in support of a full tunneling process in an enzyme reaction. Whereas the importance of hydrogen tunneling has increasingly achieved wide acceptance, a discussion has ensued regarding the extent to which enzymes catalyze the tunneling event, i.e., whether it may be expected that the extent of tunneling will be the same or different in relation to a comparable reaction in solution (29–31). One approach to address this question is to study the properties of a model reaction in solution. In the context of SLO-1, Stack and coworkers have previously reported a relevant model, observing a KIE of only 2.7 (32). This comparison, at face value, may lead to the conclusion that tunneling is far more significant for SLO-1 than for the model reaction in solution. However, a range of other explanations are possible, beginning with the difficulty of designing a system that mimics the properties of the enzyme active site, followed by the possibility of different rate-limiting steps in the two systems. It is equally likely that extensive tunneling is also occurring in the model reaction but with properties that are different from those in the enzyme reaction. In this instance it is no longer valid to ask the question: To what extent does the enzyme increase the percentage of reaction that occurs via tunneling? The relevant question becomes: How does the enzyme promote the tunneling process?
Site-specific mutagenesis offers a means of addressing the latter. In particular, the combination of site-specific mutagenesis and the investigation of temperature dependent isotope effects afford us a unique view into the origins of catalytic efficiency in SLO-1. From the very small temperature dependence of the KIE for the WT-SLO-1, we propose that the enzyme is able to sample a family of ground state conformers that promote a very close approach of the donor to acceptor atoms and efficient wave function overlap. In fact, this distance is small enough that deuterium as well as protium is able to undergo transfer without the requirement for a great deal of distance sampling. By contrast, the introduction of a single mutation ≈15 Å from the active site iron impacts deuterium in a fairly dramatic manner, as evidenced by both the large values for ΔEa and the exalted KIE for I553G. In this manner, it may be expected that changes in side chains closer to the bound substrate will have quite dramatic effects on the rate and Ea for protium transfer as well. This property has already been reported in the case of the more proximal L546A and L754A mutants of SLO-1, which produce reduced values for kcat and elevated values for Ea in the case of protium transfer, together with values for ΔEa well below that for the WT-enzyme, while retaining the very large KIE of close to 100 (15). These results for L546A and L754A indicate a significant impact of mutagenesis on the efficiency of protium as well as deuterium transfer, illustrating how the judicious placement of proximal hydrophobic side chains within the binding pocket of SLO-1 has created an active site whose properties are optimized for the hydrogen tunneling process. That is, the evolution of the active site has not necessarily made tunneling more prominent. It has, however, made the rate of transfer of hydrogen via tunneling significantly more efficient. This property may well be the case for all enzymes that catalyze C–H activation.
Materials and Methods
Mutagenesis and Protein Purification.
Each mutant lipoxygenase was prepared, expressed, and purified according to procedures described in refs. 1, 2, and 6) (see SI Materials and Methods).
Enzyme Kinetics.
Steady-state kinetics were performed on a Cary 300 UV/vis spectrophotometer in the single wavelength mode. The reaction progress was monitored by following the production of 13-(S)-hydroperoxy-9,11-(Z,E)-octadecadienoic acid [13-(S)-HPOD] formation (ε234 = 23,600 M−1·cm−1) (1, 2). Rate constants were corrected for the iron content of each preparation and for oxygen levels below saturation. Noncompetitive isotope effects were obtained by comparing rate constants for protio-linoleic acid to 11,11-dideuterio-LA. Details of the enzymatic assays and synthesis of substrates are found in SI Materials and Methods.
Crystallization and Data Collection.
Crystals of mutant proteins were grown and cryocooled as described in ref. 33. Diffraction data from all mutants were collected at 100 K at the Structural Biology Center 19-ID beamline of the Advanced Photon Source. The details regarding data collection and crystallographic refinement are in SI Materials and Methods.
Supplementary Material
ACKNOWLEDGMENTS.
Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357. This work was supported by National Institutes of Health Grant GM25765 and National Science Foundation Grant MCB0446395 (to J.P.K.). M.P.M. was supported in part by National Institutes of Health–Ruth L. Kirschstein Postdoctoral Fellowship GM64218.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710643105/DC1.
References
- 1.Glickman MH, Klinman JP. Nature of rate-limiting steps in the soybean lipoxygenase-1 reaction. Biochemistry. 1995;34:14077–14092. doi: 10.1021/bi00043a013. [DOI] [PubMed] [Google Scholar]
- 2.Glickman MH, Klinman JP. Lipoxygenase reaction mechanism: Demonstration that hydrogen abstraction from substrate precedes dioxygen binding during catalytic turnover. Biochemistry. 1996;35:12882–12892. doi: 10.1021/bi960985q. [DOI] [PubMed] [Google Scholar]
- 3.Glickman MH, Wiseman JS, Klinman JP. Extremely large isotope effects in the soybean lipoxygenase-linoleic acid reaction. J Am Chem Soc. 1994;116:793–794. [Google Scholar]
- 4.Hwang CC, Grissom CB. Unusually large deuterium isotope effects in soybean lipoxygenase are not caused by a magnetic isotope effect. J Am Chem Soc. 1994;116:795–796. [Google Scholar]
- 5.Jonsson T, Glickman MH, Sun S, Klinman JP. Experimental evidence for extensive tunneling of hydrogen in the lipoxygenase reaction implications for enzyme catalysis. J Am Chem Soc. 1996;118:10319–10320. [Google Scholar]
- 6.Rickert K, Klinman JP. The nature of hydrogen transfer in soybean lipoxygenase-1: Separation of primary and secondary isotope effects. Biochemistry. 1999;38:12218–12228. doi: 10.1021/bi990834y. [DOI] [PubMed] [Google Scholar]
- 7.Knapp MJ, Seebeck FP, Klinman JP. Steric control of oxygenation regiochemistry in soybean lipoxygenase-1. J Am Chem Soc. 2001;123:2931–2932. doi: 10.1021/ja003855k. [DOI] [PubMed] [Google Scholar]
- 8.Knapp MJ, Klinman JP. Environmentally coupled hydrogen tunneling: Linking catalysis to dynamics. Eur J Biochem. 2002;269:3113–3121. doi: 10.1046/j.1432-1033.2002.03022.x. [DOI] [PubMed] [Google Scholar]
- 9.Hatcher E, Soudackov V, Hammes-Schiffer S. Proton-coupled electron transfer in soybean lipoxygenase. J Am Chem Soc. 2004;126:5763–5775. doi: 10.1021/ja039606o. [DOI] [PubMed] [Google Scholar]
- 10.Hatcher E, Soudackov V, Hammes-Schiffer S. Proton-coupled electron transfer in soybean lipoxygenase: Dynamical behavior and temperature dependence of kinetic isotope effects. J Am Chem Soc. 2007;129:187–196. doi: 10.1021/ja0667211. [DOI] [PubMed] [Google Scholar]
- 11.Lehnert N, Solomon EI. Density-functional investigation on the mechanism of H-atom abstraction by lipoxygenase. J Biol Inorg Chem. 2003;8:294–305. doi: 10.1007/s00775-002-0415-6. [DOI] [PubMed] [Google Scholar]
- 12.Tejero I, Eriksson LA, Gonzalez-Lafont A, Marquet J, Lluch JM. Hydrogen abstraction by soybean lipoxygenase-1: Density-functional theory study on active site models in terms of Gibbs free energies. J Phys Chem B. 2004;108:13831–13838. [Google Scholar]
- 13.Olsson MHM, Siegbahn PEM, Warshel A. Manganese(II) Zero-field interaction in cambialistic and manganese superoxide dismutases and its relationship to the structure of the metal binding site. J Am Chem Soc. 2004;126:2820–2828. doi: 10.1021/ja036503x. [DOI] [PubMed] [Google Scholar]
- 14.Siebrand W, Smedarchina Z. Temperature dependence of kinetic isotope effects for enzymatic carbon-hydrogen bond cleavage. J Phys Chem B. 2004;108:4185–4195. doi: 10.1021/jp2022117. [DOI] [PubMed] [Google Scholar]
- 15.Knapp MJ, Rickert K, Klinman JP. Temperature-dependent isotope effects in soybean lipoxygenase-1: Correlating hydrogen tunneling with protein dynamics. J Am Chem Soc. 2002;124:3865–3874. doi: 10.1021/ja012205t. [DOI] [PubMed] [Google Scholar]
- 16.Meyer M, Klinman JP. Modeling temperature dependent kinetic isotope effects for hydrogen transfer in a series of soybean lipoxygenase mutants: The effect of anharmonicity upon transfer distance. Chem Phys. 2005;319:283–296. doi: 10.1016/j.chemphys.2005.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruno WJ, Bialek W. Vibrationally enhanced tunneling as a mechanism for enzymatic hydrogen transfer. Biophys J. 1992;63:689–699. doi: 10.1016/S0006-3495(92)81654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klinman JP. The role of tunneling in enzyme catalysis of C-H activation. Biochim Biophys Acta. 2006;1757:981–987. doi: 10.1016/j.bbabio.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 19.Kohen A, Klinman JP. Enzyme catalysis: Beyond classical paradigms. Acc Chem Res. 1998;31:397–404. [Google Scholar]
- 20.Bahnson BJ, Colby TD, Chin JK, Goldstein BM, Klinman JP. A link between protein structure and enzyme catalyzed hydrogen tunneling. Proc Natl Acad Sci USA. 1997;94:12797–12802. doi: 10.1073/pnas.94.24.12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohen A, Cannio R, Bartolucci S, Klinman JP. (1999) Enzyme dynamics and hydrogen tunneling in a thermophilic alcohol dehydrogenase. Nature. 1999;399:496–499. doi: 10.1038/20981. [DOI] [PubMed] [Google Scholar]
- 22.Liang Z-X, Lee T, Resing KA, Ahn NG, Klinman JP. Thermal-activated protein mobility and its correlation with catalysis in thermophilic alcohol dehydrogenase. Proc Natl Acad Sci USA. 2004;101:9556–9561. doi: 10.1073/pnas.0403337101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagel Z, Klinman JP. Tunneling and dynamics in enzymatic hydride transfer. Chem Rev. 2006;106:3095–3118. doi: 10.1021/cr050301x. [DOI] [PubMed] [Google Scholar]
- 24.Wang L, Goodey NM, Benkovic SJ, Kohen A. Coordinated effects of distal mutations on environmentally coupled tunneling in dihydrofolate reductase. Proc Natl Acad Sci USA. 2006;103:15753–15758. doi: 10.1073/pnas.0606976103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcus RA, Sutin N. (1985) Electron transfers in chemistry and biology. Biochim Biophys Acta. 1985;811:265–322. [Google Scholar]
- 26.Kuznetsov AM, Ulstrup J. Proton and hydrogen atom tunneling in hydrolytic and redox enzyme catalysis. Can J Chem. 1999;77:1085–1096. [Google Scholar]
- 27.Marcus RA. Electron-transfer reactions in chemistry: Theory and experiment (Nobel lecture). Angew Chem Int Ed Eng. 1993;32:1111–1121. [Google Scholar]
- 28.Schenk G, Neidig ML, Zhow J, Holman TR, Solomon EI. Spectroscopic characterization of soybean lipoxygenase-1 mutants: The role of second coordination sphere residues in the regulation of enzyme activity. Biochemistry. 2003;42:7294–7302. doi: 10.1021/bi027380g. [DOI] [PubMed] [Google Scholar]
- 29.Doll KM, Bender BR, Lin Y, Finke RG. The first experimental test of the hypothesis that enzymes have evolved to enhance hydrogen tunneling. J Am Chem Soc. 2003;125:10877–10884. doi: 10.1021/ja030120h. [DOI] [PubMed] [Google Scholar]
- 30.Doll KM, Finke RG. A compelling experimental test of the hypothesis that enzymes have evolved to enhance quantum mechanical tunneling in hydrogen transfer reactions: The beta-neopentylcobalamin system combined with prior adocobalamin data. Inorg Chem. 2003;42:4849–4856. doi: 10.1021/ic0300722. [DOI] [PubMed] [Google Scholar]
- 31.Kemsley J. Enzyme tunneling idea questioned: Comparison of H-transfer reactions with and without enzyme shows no enhanced tunneling. Chem Eng News. 2003;81:29–30. [Google Scholar]
- 32.Goldsmith CR, Jonas RT, Stack DP. C-H bond activation by a ferric methoxide complex: Modeling the rate-determining step in the mechanism of lipoxygenase. J Am Chem Soc. 2002;124:83–96. doi: 10.1021/ja016451g. [DOI] [PubMed] [Google Scholar]
- 33.Tomchick DR, Phan P, Cymborowski M, Minor W, Holman TR. Structural and functional characterization of second-coordination sphere mutants of soybean lipoxygenase-1. Biochemistry. 2001;40:7509–7517. doi: 10.1021/bi002893d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.