Abstract
The widely observed dispensability of duplicate genes is typically interpreted to suggest that a proportion of the duplicate pairs are at least partially redundant in their functions, thus allowing for compensatory affects. However, because redundancy is expected to be evolutionarily short lived, there is currently debate on both the proportion of redundant duplicates and their functional importance. Here, we examined these compensatory interactions by relying on a genome wide data analysis, followed by experiments and literature mining in yeast. Our data, thus, strongly suggest that compensated duplicates are not randomly distributed within the protein interaction network but are rather strategically allocated to the most highly connected proteins. This design is appealing because it suggests that many of the potentially vulnerable nodes that would otherwise be highly sensitive to mutations are often protected by redundancy. Furthermore, divergence analyses show that this association between redundancy and protein connectivity becomes even more significant among the ancient duplicates, suggesting that these functional overlaps have undergone purifying selection. Our results suggest an intriguing conclusion—although redundancy is typically transient on evolutionary time scales, it tends to be preserved among some of the central proteins in the cellular interaction network.
Keywords: evolution, systems biology
Gene duplications have long been perceived as a source of genetic redundancy that contributes to the robustness of phenotypes (1–3). The assumption is that for a portion of the duplicate pairs, there exists a functional overlap, which enables one gene copy to compensate for mutations in its partner. Examples of such compensation by duplicates have frequently been observed in a wide variety of organisms and systems (cf. ref. 4).
From an evolutionary perspective, functional overlaps of gene duplicates may serve to increase the evolvability of organisms (5) but are also expected to be unstable (6, 7). Specifically, if a gene's function can be compensated for by a redundant duplicate, mutations in that gene would have no effect on the phenotype. As a result, such mutations could not be selected against, and redundancy would be gradually lost (8).
Because of the inherently unstable nature of functional overlaps, it is thought that they are rapidly eliminated on evolutionary time scales (8–10). In line with this assumption, recent estimates suggest that the proportion of duplicate pairs that can effectively compensate for each other's loss is low [10% (3, 11)], compared with the majority of duplicates with little or no compensation (or “backup”) capacity. These considerations have recently sparked controversy as to whether functionally overlapping duplicates play any significant biological role, other than accelerating evolutionary rates (8, 11, 12).
Notably, although evidence suggests that a rapid loss of functional overlap indeed describes the fate of most duplicated genes, this hypothesis is also violated by numerous well documented examples (13, 14). In one such case, recent knockdown experiments in Caenorhabditis elegans have revealed duplicate genes that have been conserved in a functionally redundant state for >80 million years of evolution (15). Furthermore, it was demonstrated in both S. cerevisiae and in C. elegans that duplicate genes evolve more slowly than singletons, despite an initial increased evolutionary rate (16, 17), indicating that some essential functions are more likely endowed with redundancies. More recently, a combined proteomic and phenotypic analysis in yeast suggested that a preponderance of redundancy could also exist between alternative pathways (18). Taken together, these pieces of evidence suggest that, in particular types of systems, genetic redundancy may play an as-yet-unidentified role that could provide a basis for its extended conservation. Although it is unlikely that functional overlaps have been conserved solely for the sake of buffering the mutations (8, 19, 20), the possibility that they could be advantageously used for a range of different functionalities is intriguing (4, 6). If such functionalities do exist, they pose two evolutionary questions. One is how these functional overlaps have initially been fixated in the population after the duplication event. The second is how the system has evolved to use these functional overlaps. Models have been proposed that may explain the first stage, namely fixation of the duplicated state (6, 7). These models are based on differential properties of the redundant duplicates with respect to their functional efficiency and/or mutation rates.
In the present study, we used the yeast protein interaction network to search for functional characteristics rendering redundant gene duplicates unique compared with the majority of nonredundant duplicates. We examined whether redundancies are randomly distributed within the protein interaction network or are strategically allocated to certain nodes, assuming that deviation for randomness should indicate selection. Our results indicated that redundant partners are significantly more frequently associated with the so-called protein network “hubs” (i.e., genes whose protein products bind a particularly large number of protein partners). Notably, when inspecting the entire genome, which is dominated by proteins that lack redundant partners, Jeong et al. (21) found a strong connection between “centrality” (i.e., tendency to interact with multiple partners), and lethality; i.e., they found increased essentiality of the highly connected nodes. In contrast to this entire genome survey, we focused here exclusively on duplicated genes that are more likely to have preserved partially redundancies. We found that highly connected nodes are more likely than lowly connected ones to have preserved partially redundant paralogs. We conclude that although “centrality” does imply “lethality” (21), the proportion of essential hubs would have been even higher if it were not for the preferential allocation of redundant duplicates to some of the hubs. We then provide extensive corroboration of these conclusions from single- and double-knockout experiments and from literature mining.
Results
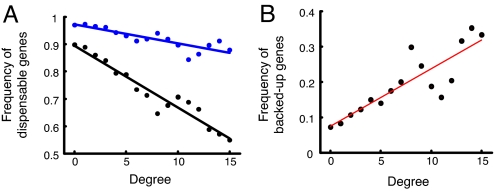
To characterize redundancy, we analyzed the extent to which connectivity correlates with higher proportions of essential genes but separately for singletons and duplicates (Fig. 1A). Although a general association between connectivity and dispensabily has been previously shown (21), we show here that adding the distinction between duplicates to singletons distinction provides new and unexpected insights. In both singletons and duplicates, we found that the highly connected proteins are typically more essential (Fig. 1A). Yet, strikingly, this association is characterized by very different slopes among the two sets of genes. Although the singletons show the familiar sharp decline in dispensability as a function of their degree (21), the duplicates show only a faint correlation. Moreover, we observed that the difference between the proportion of essential singletons and the proportion of essential duplicates increases with connectivity. In other words, although it was long been known that duplicates are more dispensable than singletons (1, 3), we show that this difference is far more pronounced among the protein network hubs.
Fig. 1.
Proportion of redundant duplicates as a function of connectivity in the protein interaction network. (A) Proportion of duplicates with a viable knockout phenotype is shown as a function of the number of their physical association partners in the protein interaction network. Plots were calculated separately for genes with duplicates (blue) and singletons (black). For drawing the curve for the duplicate genes, all duplicated genes at each value of degree connectivity were pooled. Then, the proportion of dispensable genes in each pool was computed and shown on the y axis. P values for the two slopes, calculated by means of logistic regression, were 1.4 × 10−35 for singletons and 5 × 10−5 for duplicates. (B) Estimated proportion of redundant duplicates as a function of their connectivity in the protein interaction network (for calculation details, see SI Appendix 2). The P value on the slope calculated by means of logistic regression was 1.5 × 10−10.
To quantify this statement, we estimated the proportion of redundant duplicates, frd(k), for any given degree of connectivity, k, through
where NkObs is the number of observed essential duplicates at degree k, and NkExp is the number of duplicates with degree k that would have been expected to be essential if there were no redundancy among duplicates. We then calculated NkExp by NkExp = Nktotalfs(k), where fs(k) is the fraction of singletons at degree k that are essential for viability, and Nktotal is the total number of duplicate genes with degree k. The estimated proportion of redundant duplicate pairs as a function of the duplicates' connectivity is plotted in Fig. 1B. These results demonstrate that highly connected proteins are more likely than lowly connected proteins to have retained a potentially compensating duplicate.
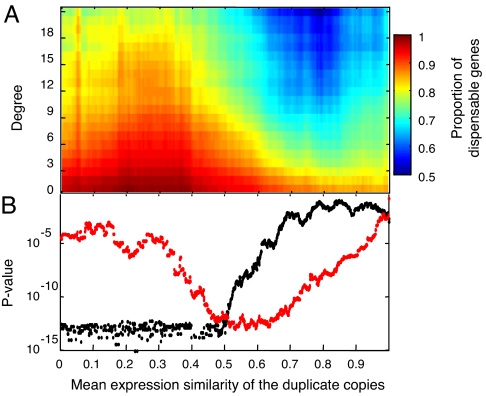
We next turned to examine how another feature of duplicates—the extent of their coregulation interacts with their degree of connectivity in affecting essentiality. It was suggested that gene duplicates that are consistently coexpressed are unlikely to have redundant functions (4, 22). The rationale is that systematically coregulated duplicate genes may be simultaneously required for a given functionality and therefore cannot substitute for each other's absence. Fig. 2 shows the proportion of nonessential duplicates as a function of both the expression similarity of the duplicate pairs and their connectivity within the protein network. Duplicates that physically interact with only a few partners (Fig. 2A; connectivity values <3) appear to be nonessential, almost regardless of their expression similarity. The dispensability of these genes may attest to the dispensability of their biochemical functions.
Fig. 2.
Joint dependence of gene dispensability on connectivity within the protein interaction network and on expression similarity among paralogs. (A) Proportion of dispensable genes from the total set of paralogs is shown (blue, low proportion of dispensable genes and red, high proportion of dispensable genes) as a function of their degree of connectivity in the protein interaction network and the expression similarity between the paralogous pair members. A version including also the relatively few negatively correlated duplicate pairs is qualitatively similar, although with less statistical power (see SI Fig. 7). (B) P values (plotted in red) illustrating the association between degree connectivity and dispensability were tested for paralogous pair populations and stratified according to expression similarity. These are compared with the P values (plotted in black) illustrating the enrichment of functionally redundant paralogs. The plot was generated by sliding a window of width 0.3, along the expression similarity axis. For statistical details on P value calculations, see Materials and Methods.
Intriguingly, however, as we examine gene duplicates with higher connectivity values (Fig. 2A), the question of whether they are essential or dispensable becomes highly dependent on whether or not the duplicate copies are coexpressed (Fig. 2). Specifically, if a protein has many interaction partners and its expression is tightly coregulated with that of its duplicate copy, it will, in most cases, appear to be essential in knockout experiments. In contrast, proteins that have equally high interaction partners but whose expression is not coregulated with that of their duplicate copies are typically dispensable, implying functional redundancy (Fig. 2A). Thus, by analyzing the dependency between essentiality, expression similarity, and connectivity [see also supporting information (SI) Appendix 1], we demonstrate here that dispensability of gene duplicates is strongly associated with how these duplicates are regulated and the number of different binding partners with which they interact. Specifically, we found that, especially among the noncoregulated duplicates, backup capacity is distributed nonrandomly, demonstrating preferential tendency to concentrate in the network's hubs (see SI Appendix 2).
To experimentally validate our predictions, we performed double-knockout experiments involving dozens of duplicate gene pairs. We deleted protein network hubs and, as a control, sparsely connected proteins, each with their respective paralogs. In the case of protein hubs, we excluded from our analysis all hubs that are tightly coexpressed with their duplicate copies, because these are unlikely to be redundant (Fig. 2; also refs. 4 and 22). For sparsely connected proteins, we considered dispensable genes with a single duplicate copy (see Materials and Methods).
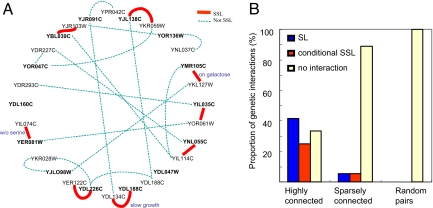
To generate the double-deletion strains, we crossed haploid cells deleted for a gene of interest with another haploids deleted for the corresponding duplicate. This procedure resulted in a collection of diploids that were heterozygous for both mutations. We then sporulated these diploids, obtaining haploid spores with varying combinations of the two mutations, and then assessed the fitness of the double-knockout strains. As shown in Fig. 3 and SI Appendix 3, in 8 of the 12 tested protein hubs, we found a significant reduction in fitness when genes were codeleted with their duplicate copies. These effects ranged from complete loss of viability (synthetic lethality; 41.6% of these 12 cases) to slow growth (synthetic sick; 8.3% of these 12 cases) or lethality under certain growth conditions (16.6% of these 12 cases). These results strongly contrast with those we obtained among the sparsely connected proteins (nonhubs), where only 11% (2/18) showed any impact on phenotype when codeleted with their duplicates. Reassuringly, for 11 of the 14 hubs in our dataset, there have been previous indications in the literature suggesting some type of compensation (see SI Table 1 and SI Appendix 3). This is in contrast to our control set of sparsely connected duplicates, in which such evidence was reported for only 2 of 18 duplicate pairs.
Fig. 3.
Results of the synthetic sick and lethal double-knockout experiments. (A) Pairs of dispensable genes for which genetic interaction was tested are connected by a solid red line in cases where SSL interaction was found and by a dashed blue line in cases where no interaction was observed. The hub-paralog pairs are arranged clockwise, starting from 12:00 (hub YJL138C, followed by its paralog YKR059W); all hubs are designated in boldface type. As a negative control, we codeleted hubs and randomly picked paralogs of other hubs. In instances of double knockout of the following hubs (YER081W and YMR105C) and their respective paralogs (YIL074C and YKL127W, respectively), SSL interactions were obtained only in specific growth conditions (lack of serine and galactose as a carbon source, respectively). Four hubs (YER081W, YDL226C, YJL098W, and YOR136W) were found to have two or three paralogs. For these cases, we searched for SSL interactions with all paralogs, yet we never found additional interactions (data not shown). For two hubs, we were unable to examine genetic interactions, either because of the essential nature of the hub itself (YDL047W, which in the database appears as viable, yet in our experiments, with specific genetic background, is extremely sick) or because of very low spore viability (YDL160C). (B) Proportions of the different genetic interactions obtained in all three double-knockout experiments are shown. Highly connected, double-knockout experiments in which both the highly connected gene and its duplicate were deleted; sparsely connected, double-knockout experiments in which both the sparsely connected gene and its duplicate were deleted; random pairs, double-knockout experiments in which both the highly connected gene and a randomly chosen paralog of another hub were deleted; SL, synthetic lethality; conditional SSL, lethality under specific conditions and slow growth; no interaction, no detectable fitness effect under the conditions tested.
Because several of the hubs in the set we examined contained more than one duplicate gene copy, we investigated whether all given duplicates are equally likely to compensate for the loss originating from the deletion. Accordingly, we selected all hubs in our collection that had three or fewer duplicate copies (constituting a total of four or fewer genes). We then separately codeleted these hubs with each of their different duplicates, generating alternative double-knockouts. The results from this experiment (see SI Appendix 3) suggested that, for any given hub, there is only one gene partner whose absence generates synthetic interaction with the deletion of the hub. That said, we cannot exclude the possibility that functional redundancy exists, even in the gene pairs that did not yield a synthetic interaction; but this redundancy was not revealed by the double-knockout, e.g., due to a third redundant partner (23).
In addition, we asked whether paralogs of hubs that are compensated for by their duplicates are also highly connected. Indeed, we found that they have a significantly higher number of protein partners compared with the average gene in the genome [P value for difference in connectivity = 2.6 × 10−4 (t test)]. Furthermore, we found that in seven of the eight cases of synthetic sick or lethal phenotypes, the hub and its paralog share a significant portion of their protein interaction partners (P < 0.01 for each of the seven pairs, using a hypergeometric test).
To firmly associate these synthetic interactions with compensations, an alternative interpretation of these experimental results must have been examined. Specifically, it could be argued that the deletion of the discussed hubs could destabilize the cellular network to such an extent that many random additional deletions, on the background of the hub's deletion will also produce lethality. To rule out this possibility, we performed another set of negative control double-knockout experiments, in which we paired the hubs previously analyzed, with duplicates of other randomly selected hubs. Strikingly, none of the 12 double knockouts we performed showed any effect on cell viability (see Fig. 3 and SI Appendix 3). These results suggest that there is genuine information in the identity of the codeleted gene and that only the true paralogs may generate genetic interactions, arguably because of functional compensation to the hubs.
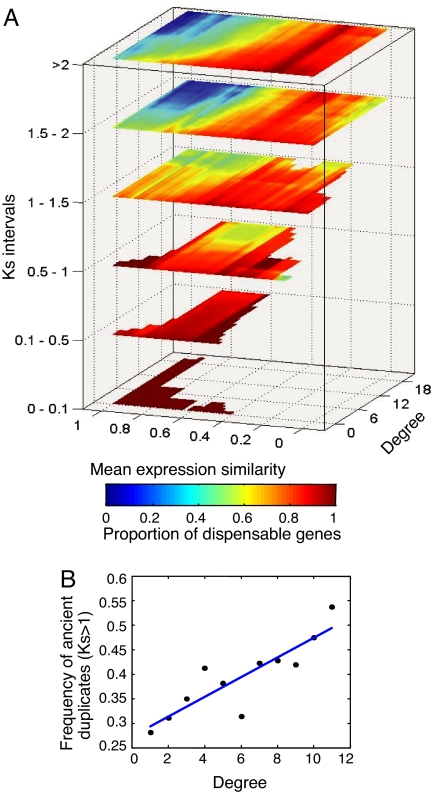
One possible interpretation of our results is that functional overlaps of gene duplicates have been evolutionarily conserved more frequently, among protein network hubs. To examine the evolutionary processes responsible for the association between redundancy and connectivity, we tested how the approximated age of duplication affects the correlation between the proportion of dispensable duplicates to both (i) the connectivity of duplicates in the protein network and (ii) the expression similarity of the duplicate copies (Fig. 4). [Age of duplication was roughly estimated by the extent of synonymous substitutions (Ks) (8)]. We roughly discern three separate evolutionary regimes. In the first phase, immediately after the duplication event (0 < Ks < 0.1), duplicate pairs are both tightly coexpressed and highly dispensable. This result may reflect either compensation due to the functional similarity of duplicated genes before divergence or a dispensability of the biochemical function of the duplicated gene (24). In the second phase (0.1 < Ks < 1), we observe, in line with studies reported in refs. 9, 10, 25, and 26, a decline in the expression similarity of the duplicates, concomitant with a gradual loss of their dispensability. Notably, during these first two evolutionary stages, the dependency of knockout phenotypes on both protein connectivity and expression similarity of duplicate genes is very weak. In fact, such dependency only becomes significant during what we consider to be the third evolutionary phase, corresponding to highly ancient duplication events with divergence levels of Ks > 1. Remarkably, it thus becomes evident that the correlation shown in Fig. 2 primarily reflects an association between redundancy and connectivity in ancient, rather than in recent, duplicates. This is further substantiated by a 3-way ANOVA test (P = 0.009), demonstrating the interaction between Ks and degree in affecting duplicate dispensability (Table 6 in SI Appendix 1). This finding may suggest that compensations of protein network hubs by their duplicates is not a simple epiphenomenon of gene duplication but rather represent a functionality that has evolved through purifying selection. We have further examined the proportion of remote paralogs (Ks > 1) among pairs with increasing degree connectivity (Fig. 4B). Interestingly, the proportion of remote (presumably more ancient) pairs increases with degree connectivity, consistent, although not exclusively, with a prolonged retention of duplications in involving highly connected proteins.
Fig. 4.
Relationships among gene dispensability, connectivity, expression similarity, and evolutionary divergence. (A) Dispensability as a function of degree and expression similarity among paralogs (as in Fig. 3A), tested separately for pairs with different Ks values. (B) The proportion of remote (Ks > 1) pairs in each window of degree connectivity. Similarity to data in Fig. 2, all duplicated genes at each value of degree connectivity were pooled. Then, the proportion of genes in each pool that have a remote paralog was computed and shown on the y axis.
In an attempt to at least partially understand the additional value gained from such redundancies, we manually searched the literature for all references of duplicate gene pairs in yeast that were experimentally demonstrated to be redundant (see Materials and Methods for a description of the literature search). Specifically, we labeled genes “redundant” if literature indicates that they meet two criteria: first, clear findings in non high-throughput studies documenting their functional overlap; and second, experimental validation of compensatory interactions between the pair members. To limit the size of the dataset to one that is reasonable for a manual search of the National Center for Biotechnology Information PubMed database, we defined a sequence similarity threshold (see Materials and Methods) and only examined duplicate pairs meeting this criterion. The resulting analysis yielded 112 carefully validated redundant paralogous pairs (for a full list, see SI Table 1). Plotting the frequency of redundant genes within the total curated set as a function of their degree of connectivity, we again observed that the proportion of redundancies significantly increased, with increasing connectivity (Fig. 5) (P = 1.7 × 10−6; logistic regression).
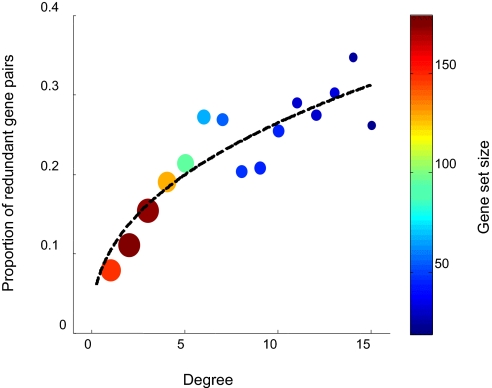
Fig. 5.
Proportion of functionally redundant duplicate pairs in a literature curated dataset as a function of their connectivity in the protein interaction network. The data for the analysis consisted of a list of 766 duplicate-gene pairs selected by a sequence similarity criterion (BLAST e value <3 × 10−108). Each of these pairs was subjected to a manual literature examination in search of evidence for functional redundancy. This procedure resulted in 112 redundant pairs. At each degree connectivity, the value at the y axis denotes the fraction of genes with that degree that have an annotated redundant paralog in the set of 112 pairs. Proportions were calculated by normalizing to the total set of curated paralogs, thus avoiding potential biases associated with literature over-representation of highly connected proteins. Both color and size of the data points represent the number of genes in a given category (colors specified by the color bar at Right). Analysis was performed by applying a sliding window of width = 2 on the degree axis.
Despite incompleteness and potential bias (e.g., because certain functional categories of genes are more likely to be represented in the literature), we reasoned that our list could at least partially assist in clarifying the roles performed by such redundant duplicates. Relying on the curated list we found that the biological functions of hubs that are “backed-up” by redundant partners represent a variety of categories associated with different hierarchies of gene regulation. These range from transcriptional regulators (e.g., the pair Fkh1 and Fkh2) to posttranslational protein modifiers such as kinases (e.g., Mrk1 and Rim11, which are homologs of the mammalian Gks-3 involved in Wnt pathway regulation), phosphotases (e.g., Ppz2 and Ppz1), and ubiquitin ligases (e.g., Bul1 and Bul2). Furthermore, we find a fair representation of components of signaling pathways (e.g., Sro7 and Sro77); isozymes (e.g., Cit1 and Cit2); and membrane transporters (e.g., Trk1 and Trk2).
Discussion
By combining bioinformatics, experiments, and literature mining, we demonstrate here that proteins with a large number of physically interacting protein partners are more frequently associated with functionally redundant gene duplicates. An alternative interpretation to our bioinformatics results (Fig. 1) could be that the dispensability of even the most highly connected duplicates does not result from compensations and redundancy but rather simply because these genes carry out less-essential functions (24). Nevertheless, such an interpretation could explain the data only if the frequency of nonessential functions increased with increasing degree among duplicates more than among singletons. Because we cannot support this interpretation, we conclude that the increased difference between dispensable duplicates to dispensable singletons among the protein network hubs most likely reflects compensatory interactions.
Previously, a classification was suggested, distinguishing between hubs whose partners are coexpressed (party hubs) and hubs whose partners are differentially expressed (date hubs) (27). By examining duplicate dispensability according to these criteria, we found no significant difference in the representation of these two gene types in the data (data not shown).
It was convincingly shown that hubs are more likely than lowly connected genes to be essential (21). Not only do our results not contradict these early findings, they are in good agreement with them, because we show too increased proportion in essential functionalities among the highly connected proteins. Essentiality of the functions carried out by the hubs either manifest themselves by increased rate of essential genes among the singletons or enhanced rate of compensations by redundancies among the duplicates. Thus, we hypothesize that without redundancy, the fraction of hubs with lethal single-gene knockout phenotypes would have been even higher than is actually the case. In line with this possibility, examples of essential functions performed by pairs of redundant, and consequently dispensable, gene duplicates have been reported (4, 14, 28).
Several points of caution regarding our assumption that hubs represent proteins with essential function should be taken. These include the possibility that some essential genes have more annotated interaction partners simply because they were studied more extensively and the valid possibility that essentiality of hubs may owe itself to the high probability that at least one of their many interactions will be essential (29). Another point of caution relates to the observation that variations on experimental and modeling methodology may affect the interpreted network topology (30). Indeed, any interpretation of our results is subject to the possibility that the protein interaction data used in this study represents only a fraction of the total underlying interaction network and that some of the annotated interactions represent false positives. Together with that, because the experimental methods used for collecting the protein–protein interactions were mostly high-throughput (affinity tag, yeast two-hybrid, etc.), they are likely not biased against detecting protein associations among particular gene sets, e.g., essential genes.
Our findings raise an intriguing question: Are redundant duplicates associated with biological roles that differ from the roles played by the majority of duplicate pairs that do not functionally overlap? In principle, high connectivity in protein networks is suggestive of one of two possibilities: (i) involvement in protein complexes [party hubs (27)] or (ii) labile interactions [date hubs (27)] typically played by posttranscriptional regulators. From examination of our curated list, it is clearly apparent that most compensated hubs fall into the second category with functions varying from posttranscriptional regulators, signaling scaffolds, or isozymes. This is also consistent with the dissimilarity in the expression of redundant duplicates (see Fig. 2 and ref. 22). It is, thus, tempting to suggest that redundant duplicates tend to be associated with regulatory functionalities, such as posttranscriptional or metabolic regulation.
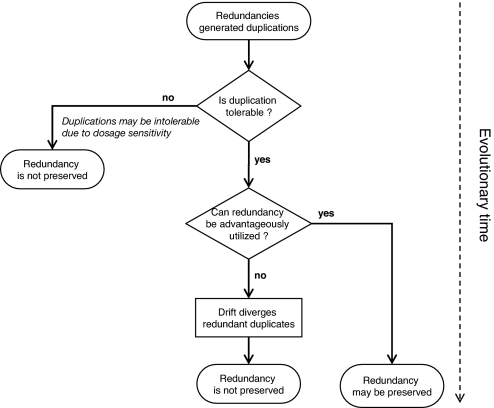
Why some of the hubs have retained a redundant gene duplicate whereas others have not remains an open question. We propose that the answer involves two separate criteria pertaining to two different evolutionary time scales as depicted in Fig. 6. Briefly, we estimate that redundancy has been conserved where (i) the immediate dosage doubling of the duplication event was not deleterious and (ii) the functional overlap offered an evolutionary advantage in wild type. Plausible evolutionary advantages of redundancy is discussed in refs. 4 and 7.
Fig. 6.
Schematic drawing of a proposed evolutionary time flow chart, describing duplicate retention in the genome.
Materials and Methods
Duplicate Gene Dataset and Protein–Protein Physical Interaction Data.
A total of 2,216 duplicate genes were collected based on PBLAST as described in ref. 22. The list of paralog pairs used in this study, along with the paralogs' corresponding values of mean expression similarity and degree connectivity, are provided in SI Table 2. The degree of connectivity of each of the genes in the protein interaction network was retrieved from the GRID database (40) (http://biodata.mshri.on.ca/yeast_grid/servlet/SearchPage), which combines literature-derived and high-throughput physical protein–protein interactions. (See further details in SI Appendix 2.)
Single Gene Mutant Phenotype Data.
Viable vs. nonviable phenotypes of all gene deletions were downloaded from www-sequence.stanford.edu/group/yeast_deletion_project/Essential_ORFs.txt.
Hypotheses Testing and Computation of P Values.
The hypothesis of whether or not backup prevails in a particular set of paralogs was tested by comparing the proportion of genes with a viable knockout phenotype contained within that set, with the proportion of genes with viable phenotypes among the singletons, a population of genes that is assumed not to have backup. The P values for this hypothesis were computed based on the c2 test for comparing proportions. To test the significance of the association between degree connectivity and percentage of dispensable genes, we used the logistic regression model (41), which enabled us to test both the existence of a negative association between degree connectivity and dispensability and compute a P value for its statistical significance.
Synthetic Sick and Synthetic Lethal Experiments: Strains, Media, Growth Conditions, and Tetrad Analysis.
The following criteria were used when choosing genes for the double-knockout experiments: For highly connected proteins, we examined all nonessential dispensable hubs (with >10 physically interacting partners) that had a nonsimilarly expressed paralog (0 < mean expression similarity <0.3). Based on the June 2005 version of the GRID database. For sparsely connected proteins, we examined all dispensable nonhubs (0–1 physically interacting partners for both paralogs) that had only one duplicate (based on the June 2005 version of the GRID database).
All S. cerevisiae disruption strains used in the present work are based on the following genetic backgrounds: BY4741: MATa, his3Δ1, leu2Δ0, met15Δ0, and ura3Δ0 and BY4742: MATα, his3Δ1, leu2Δ0, lys2Δ0, and ura3Δ0. All disruptions were marked by kanMX4 (42).
Yeast cells were grown in YEPD (1% yeast extract, 2% Bacto peptone, 2% dextrose). Sporulation was carried out in SPO medium (1% potassium acetate, 0.1% yeast extract, and 0.05% dextrose) by incubating cells for 72h at 25°C.
Diploid selection and tetrad analysis were carried out by using the Singer MSM Manual Micromanipulator, according to the manufacturer's instructions. Genetic interactions were scored by conventional tetrad analysis. (See further details in SI Appendix 2.)
Literature Curation of Redundant Gene Pairs.
All paralogous gene pairs corresponding to a BLASTP e value threshold <3 × 108 were identified by using the default BLASTP parameters. We then applied a Perl script that, for each such pair, collected all references in PubMed for which both pair members were concomitantly cited in the same reference. We then manually inspected the resulting list of >2,000 abstracts and publications. In a typical search, we first attempted to infer from the abstract and, with the aid of the SGD database, the functional relationship between the duplicate pair members. In particular, we searched for sentences clearly stating that functional overlap and compensatory interactions were established for the two paralogs. This is in contrast to sentences clearly describing functional divergence (distinct functions for each of the duplicate pair members). In some cases, we resorted to reading entire manuscripts to arrive at final conclusions. We classified genes as “redundant” if they met the following criteria: (i) clear documentation in the literature, from non high-throughput studies, of their functional overlap and (ii) experimental validation of compensatory interactions between the pair members. This search yielded 112 highly validated “redundant” paralogous pairs (for a full list, see SI Table 1).
Supplementary Material
ACKNOWLEDGMENTS.
We thank all members of the Y.P. lab for fruitful discussions and Pedro Bordalo, Alex De-Luna, Roy Kishony, Martin Kupiec, Michael Springer, Itay Tirosh, and Itay Yanay for critical review of the manuscript. We thank the Ben-May Foundation, the W. Strauss Foundation, and the Minerva Foundation for grant support. Y.P. is an incumbent of the Rothstein Career Development Chair in Genetic Diseases.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711043105/DC1.
References
- 1.Ohno S. Evolution by Gene and Genome Duplication. Berlin: Springer; 1970. [Google Scholar]
- 2.Conant GC, Wagner A Duplicate genes and robustness to transient gene knock-downs in Caenorhabditis elegans. Proc R Soc Lond B Biol Sci. 2004;271:89–96. doi: 10.1098/rspb.2003.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu Z, et al. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421:63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- 4.Kafri R, Levy M, Pilpel Y. The regulatory utilization of genetic redundancy through responsive backup circuits. Proc Natl Acad Sci USA. 2006;103:11653–11658. doi: 10.1073/pnas.0604883103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirschner M, Gerhart J. Evolvability. Proc Natl Acad Sci USA. 1998;95:8420–8427. doi: 10.1073/pnas.95.15.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nowak MA, Boerlijst MC, Cooke J, Smith JM. Evolution of genetic redundancy. Nature. 1997;388:167–171. doi: 10.1038/40618. [DOI] [PubMed] [Google Scholar]
- 7.Krakauer DC, Nowak MA. Evolutionary preservation of redundant duplicated genes. Semin Cell Dev Biol. 1999;10:555–559. doi: 10.1006/scdb.1999.0337. [DOI] [PubMed] [Google Scholar]
- 8.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 9.Gu Z, Nicolae D, Lu HH, Li WH. Rapid divergence in expression between duplicate genes inferred from microarray data. Trends Genet. 2002;18:609–613. doi: 10.1016/s0168-9525(02)02837-8. [DOI] [PubMed] [Google Scholar]
- 10.Makova KD, Li WH. Divergence in the spatial pattern of gene expression between human duplicate genes. Genome Res. 2003;13:1638–1645. doi: 10.1101/gr.1133803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin YS, Hwang JK, Li WH. Protein complexity, gene duplicability and gene dispensability in the yeast genome. Gene. 2007;387:109–117. doi: 10.1016/j.gene.2006.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lynch M, Conery JS. The evolutionary demography of duplicate genes. J Struct Funct Genomics. 2003;3:35–44. [PubMed] [Google Scholar]
- 13.Steingrimsson E, et al. Mitf and Tfe3, two members of the Mitf-Tfe family of bHLH-Zip transcription factors, have important but functionally redundant roles in osteoclast development. Proc Natl Acad Sci USA. 2002;99:4477–4482. doi: 10.1073/pnas.072071099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearce AC, et al. Vav1 and vav3 have critical but redundant roles in mediating platelet activation by collagen. J Biol Chem. 2004;279:53955–53962. doi: 10.1074/jbc.M410355200. [DOI] [PubMed] [Google Scholar]
- 15.Combinatorial. Tischler J, Lehner B, Chen N, Fraser AG. RNA interference in C. elegans reveals that redundancy between gene duplicates can be maintained for >80 million years of evolution. Genome Biol. 2006;7:R69. doi: 10.1186/gb-2006-7-8-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jordan IK, Wolf YI, Koonin EV. Duplicated genes evolve slower than singletons despite the initial rate increase. BMC Evol Biol. 2004;4:22. doi: 10.1186/1471-2148-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis JC, Petrov DA. Preferential duplication of conserved proteins in eukaryotic genomes. PLoS Biol. 2004;2:e55. doi: 10.1371/journal.pbio.0020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelley R, Ideker T. Systematic interpretation of genetic interactions using protein networks. Nat Biotechnol. 2005;23:561–566. doi: 10.1038/nbt1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lynch M, O'Hely M, Walsh B, Force A. The probability of preservation of a newly arisen gene duplicate. Genetics. 2001;159:1789–1804. doi: 10.1093/genetics/159.4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagner A. Birth and death of duplicated genes in completely sequenced eukaryotes. Trends Genet. 2001;17:237–239. doi: 10.1016/s0168-9525(01)02243-0. [DOI] [PubMed] [Google Scholar]
- 21.Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 22.Kafri R, Bar-Even A, Pilpel Y. Transcription control reprogramming in genetic backup circuits. Nat Genet. 2005;37:295–299. doi: 10.1038/ng1523. [DOI] [PubMed] [Google Scholar]
- 23.Borenstein E, Ruppin E. Direct evolution of genetic robustness in microRNA. Proc Natl Acad Sci USA. 2006;103:6593–6598. doi: 10.1073/pnas.0510600103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He X, Zhang J. Higher Duplicability of Less Important Genes in Yeast Genomes. Mol Biol Evol. 2005;23(1):144–151. doi: 10.1093/molbev/msj015. [DOI] [PubMed] [Google Scholar]
- 25.Gu Z, Rifkin SA, White KP, Li WH. Duplicate genes increase gene expression diversity within and between species. Nat Genet. 2004;36:577–579. doi: 10.1038/ng1355. [DOI] [PubMed] [Google Scholar]
- 26.Papp B, Pal C, Hurst LD. Evolution of cis-regulatory elements in duplicated genes of yeast. Trends Genet. 2003;19:417–422. doi: 10.1016/S0168-9525(03)00174-4. [DOI] [PubMed] [Google Scholar]
- 27.Han JD, et al. Evidence for dynamically organized modularity in the yeast protein–protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 28.Enns LC, et al. Two callose synthases, GSL1 and GSL5, play an essential and redundant role in plant and pollen development and in fertility. Plant Mol Biol. 2005;58:333–349. doi: 10.1007/s11103-005-4526-7. [DOI] [PubMed] [Google Scholar]
- 29.He X, Zhang J. Why do hubs tend to be essential in protein networks? PLoS Genet. 2006;2:e88. doi: 10.1371/journal.pgen.0020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hakes L, Robertson DL, Oliver SG. Effect of dataset selection on the topological interpretation of protein interaction networks. BMC Genomics. 2005;6:131. doi: 10.1186/1471-2164-6-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu X, Zhu L, Guo J, Zhang DY, Lin K. Prediction of yeast protein–protein interaction network: insights from the Gene Ontology and annotations. Nucleic Acids Res. 2006;34:2137–2150. doi: 10.1093/nar/gkl219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ekman D, Light S, Bjorklund AK, Elofsson A. What properties characterize the hub proteins of the protein–protein interaction network of Saccharomyces cerevisiae? Genome Biol. 2006;7:R45. doi: 10.1186/gb-2006-7-6-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolch W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat Rev Mol Cell Biol. 2005;6:827–837. doi: 10.1038/nrm1743. [DOI] [PubMed] [Google Scholar]
- 34.Dard N, Peter M. Scaffold proteins in MAP kinase signaling: More than simple passive activating platforms. Bioessays. 2006;28:146–156. doi: 10.1002/bies.20351. [DOI] [PubMed] [Google Scholar]
- 35.Kim PM, Lu LJ, Xia Y, Gerstein MB. Relating three-dimensional structures to protein networks provides evolutionary insights. Science. 2006;314:1938–1941. doi: 10.1126/science.1136174. [DOI] [PubMed] [Google Scholar]
- 36.Gasch AP, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cutler S, McCourt P. Dude, where's my phenotype? Dealing with redundancy in signaling networks. Plant Physiol. 2005;138:558–559. doi: 10.1104/pp.104.900152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papp B, Pal C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- 39.Taylor JS, Raes J. Duplication and Divergence: The Evolution of New Genes and Old Ideas. Annu Rev Genet. 2004;38:615–643. doi: 10.1146/annurev.genet.38.072902.092831. [DOI] [PubMed] [Google Scholar]
- 40.Breitkreutz BJ, Stark C, Tyers M. The GRID: The General Repository for Interaction Datasets. Genome Biol. 2003;4:R23. doi: 10.1186/gb-2003-4-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sokal RR, Rohlf FJ. Biometry: the Principles and Practice of Statistics in Biological Research. New York: W. H. Freeman; 1995. [Google Scholar]
- 42.Brachmann CB, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 43.Dwight SS, et al. Saccharomyces Genome Database (SGD) provides secondary gene annotation using the Gene Ontology (GO). Nucleic Acids Res. 2002;30:69–72. doi: 10.1093/nar/30.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.