Abstract
TMC278 is a diarylpyrimidine (DAPY) nonnucleoside reverse transcriptase inhibitor (NNRTI) that is highly effective in treating wild-type and drug-resistant HIV-1 infections in clinical trials at relatively low doses (∼25–75 mg/day). We have determined the structure of wild-type HIV-1 RT complexed with TMC278 at 1.8 Å resolution, using an RT crystal form engineered by systematic RT mutagenesis. This high-resolution structure reveals that the cyanovinyl group of TMC278 is positioned in a hydrophobic tunnel connecting the NNRTI-binding pocket to the nucleic acid-binding cleft. The crystal structures of TMC278 in complexes with the double mutant K103N/Y181C (2.1 Å) and L100I/K103N HIV-1 RTs (2.9 Å) demonstrated that TMC278 adapts to bind mutant RTs. In the K103N/Y181C RT/TMC278 structure, loss of the aromatic ring interaction caused by the Y181C mutation is counterbalanced by interactions between the cyanovinyl group of TMC278 and the aromatic side chain of Y183, which is facilitated by an ∼1.5 Å shift of the conserved Y183MDD motif. In the L100I/K103N RT/TMC278 structure, the binding mode of TMC278 is significantly altered so that the drug conforms to changes in the binding pocket primarily caused by the L100I mutation. The flexible binding pocket acts as a molecular “shrink wrap” that makes a shape complementary to the optimized TMC278 in wild-type and drug-resistant forms of HIV-1 RT. The crystal structures provide a better understanding of how the flexibility of an inhibitor can compensate for drug-resistance mutations.
Keywords: drug design, drug resistance, nonnucleoside reverse transcriptase inhibitor, polymerase, x-ray crystallography
Combinations of drugs have been used to successfully treat HIV-1 infections, permitting dramatic reduction in viral loads and restoration of the immune system. However, drug treatments do not eliminate the infection and treatment must be life-long. There are problems with drug toxicity, and prolonged drug exposure can lead to the emergence of drug-resistant mutant viruses. Because toxicity and drug resistance complicate treatment strategies, the development of new anti-AIDS drugs remains essential. TMC278 {(4-[[4-[[4-[(1E)-2-cyanoethenyl]-2,6-dimethylphenyl]amino]-2-pyrimidinyl]amino]benzonitrile} (1, 2), also referred to as rilpivirine and R278474, is a diarylpyrimidine (DAPY) nonnucleoside reverse transcriptase inhibitor (NNRTI) that was developed in a multidisciplinary structure-based design effort (3, 4) intended to discover drugs that can inhibit a wide range of the known drug-resistant HIV-1 strains. In cell-based assays, TMC278 inhibits a broad spectrum of HIV-1 variants (Table 1), including strains that are partially or completely resistant to the existing NNRTI drugs (3). Phase I and II clinical trials have shown that TMC278 effectively reduces viral load and maintains low viral load levels with no virus rebound in patients even after 48 weeks of treatment (5, 6).
Table 1.
Crystal structures of HIV-1 RT/NNRTI complexes were solved in the course of developing TMC278 (7, 8) along a path that originated with the first-generation TIBO compounds (9). Most of those structures were obtained at ∼2.6 to 3.0 Å resolution. The concept of strategic flexibility, which allows DAPY NNRTIs to bind to the different NNRTI-binding pockets, caused by various resistance RT mutations, through “wiggling” (torsional flexibility) and “jiggling” (ability to reposition), emerged from this study. This concept led to a proposed general strategy for designing flexible drugs to target an array of resistant mutants (7). Multivariate analyses of biological, chemical, and structural data also indicated that the DAPY NNRTIs have multiple potential binding modes (10). Although the wiggling and jiggling of DAPY compounds help them to retain their potency against mutant HIV-1 viruses, these same characteristics hindered the crystallographic studies of HIV-1 RT in complexes with TMC278 and etravirine (TMC125), another DAPY clinical candidate. The underlying problem is that the flexibility of the drug, and its binding mode, could introduce conformational heterogeneity into the periodic arrangement of RT/drug complexes in the crystal lattice (4). Numerous attempts to obtain a crystal structure of RT/TMC278 (literally thousands of crystallization trials) failed, and the “best” crystals diffracted X-rays to only ∼6.0 Å resolution.
A systematic protein engineering approach was used to obtain a mutant form of RT that yielded better diffracting crystals of the RT/TMC278 complex. Successful protein engineering included (i) truncating the termini of the protein, (ii) removing surface lysine and glutamic acid patches, and (iii) altering amino acid residues to make lattice contacts and/or remove some of the lattice contacts seen in earlier crystal forms. This mutated RT produced crystals of the HIV-1 RT/TMC278 complex in a crystal form that is distinct from the reported crystals of RT/NNRTI complexes. One of the crystal forms of the engineered RT/NNRTI complexes diffracted X-rays to 1.8 Å, significantly better than any of the reported structures of HIV-1 RT. The L100I/K103N and K103N/Y181C double mutant (in the p66 subunit only) RTs were designed based on the above construct, and their structures in complexes with TMC278 were determined at 2.9 and 2.1 Å resolution, respectively.
Results and Discussion
Engineering RT for High-Resolution Diffraction.
Numerous earlier attempts to obtain a crystal structure of the HIV-1 RT/TMC278 complex failed, and the best crystals diffracted X-rays to only 6 Å resolution. In contrast, the engineered RT/TMC278 complex crystallized in a previously uncharacterized form, and the crystals diffracted X-rays to 1.8 Å resolution (Table 2). The structure of wild-type HIV-1 RT/TMC278 was determined by molecular replacement by using the structure of RT/R147681 (PDB ID 1S6Q) as the starting model and refined to 1.8 Å resolution to Rwork and Rfree of 0.221 and 0.248, respectively. This high-resolution structure of HIV-1 RT has excellent stereochemistry (>91% of amino acid residues are in the most favored regions of the Ramachandran plot with no outliers; Procheck G factor = 0.25) and a reliable solvent model. The form of recombinant RT (1B1) (11) used in our previous structural studies of RT/NNRTI complexes crystallized with the symmetry of space group C2 with unit cell volume ∼1.6 × 106 Å3, one molecule per asymmetric unit, approximate solvent content of 64%, and a Matthews coefficient of 3.4. The p66 fingers and thumb subdomains are flexible and not involved in any significant crystal contacts. Earlier structural studies have shown that NNRTI binding is accompanied by repositioning of the thumb and fingers subdomains, resulting in a conformation of RT with a wide cleft between these mobile subdomains (12, 13). Comparing the crystal structures of a number of RT/NNRTI complexes revealed that individual NNRTIs have both short-range and long-range effects on the conformation of RT and affect the precise positioning of the p66 thumb and fingers subdomains. We predicted that several DAPY inhibitors, by virtue of their structural flexibility and compactness, have the ability to bind RT in more than one conformation (4). These different binding modes for a single NNRTI also may lead to differences in the positions of the fingers and thumb. In the context of a crystal lattice, this heterogeneity in the arrangement of RT molecules would reduce the resolution of x-ray diffraction from the crystal.
Table 2.
Diffraction data and refinement statistics
| Wild-type RT/TMC278 | K103N/Y181C mutant HIV-1 RT/TMC278 | L100I/K103N mutant HIV-1 RT/TMC278 | |
|---|---|---|---|
| PDB ID | 2ZD1 | 3BGR | — |
| X-ray source | CHESS F1 | BNL X25 | CHESS F1 |
| Wavelength, Å | 0.9176 | 0.9800 | 0.9176 |
| Space group | C2 | C2 | C2 |
| Cell constants (a, b, c in Å; β in °) | 163.37, 73.26, 110.07 Å, 100.07° | 162.84, 73.18, 108.92 Å, 100.79° | 163.53, 73.40, 109.29 Å, 100.60° |
| Resolution range, Å | 50.0–1.8 | 50–2.1 | 50–2.9 |
| Number of unique reflections (number of observations) | 113,256 (328,451) | 72,851 (244,765) | 28,140 (92,353) |
| Completeness, % | 96.0 | 98.9 | 98.8 |
| Rmerge (in last shell) | 0.091 (0.572) | 0.065 (0.568) | 0.107 (0.655) |
| Average I/σ(I) | 12.4 | 11.3 | 8.6 |
| σ cutoff | −0.5 | −1.0 | −1.0 |
| Refinement statistics | |||
| Total number of atoms (inhibitor/solvent atoms) | 8,582 (28/637) | 8,421 (28/504) | 7,960 (28/0) |
| Resolution, Å | 40.0–1.8 | 40.0–2.1 | 40.0–2.9 |
| Number of reflections (Rfree set) | 113,072 (2,260) | 70,651 (2,189) | 28,135 (1,423) |
| Completeness, % | 95.2 | 98.9 | 98.7 |
| Cutoff criteria | ∣F∣ < 0 | ∣F∣ < 0 | ∣F∣ < 0 |
| Rwork | 0.220 | 0.229 | 0.241 |
| Rfree | 0.248 | 0.269 | 0.299 |
| Root mean square deviations | |||
| Bond lengths, Å | 0.008 | 0.008 | 0.009 |
| Bond angles, ° | 1.4 | 1.43 | 1.49 |
An engineered RT variant (RT52A) crystallized in a previously uncharacterized form with the symmetry of C2 space group and a unit cell volume of ∼1.3 × 106 Å3. The unit cell volume and the solvent content are reduced by ∼18% and 28%, respectively, compared with the C2 crystal form obtained with the parental RT. The lower solvent content of ∼56% with a Matthews coefficient of 2.8, compared with 64% solvent of the old C2 unit cell, with a Matthews coefficient of 3.4, reflects significantly tighter packing of the RT molecules in the crystal lattice. The previously uncharacterized crystal form involves new protein contacts; of the new contacts, a set of back-to-back interactions between the p66 thumb and p66 fingers of symmetry-related RT molecules may be critical in stabilizing the positions of p66 thumb and fingers subdomains in the crystal lattice [supporting information (SI) Fig. 5]. The tighter packing of the engineered RT molecules and the specific intermolecular interactions seen with this form of RT may have contributed to the higher-order and high-resolution (1.8 Å) diffraction. A total of 113,072 unique reflections were used to refine the structure of one RT molecule (Table 2), which is ∼2.3 times the number of observations (49,347 reflections) used in refining the published highest-resolution (2.2 Å) structure (PDB ID 1VRT) of an HIV-1 RT/NNRTI complex (14). The substantial increase in experimental measurements leads to higher accuracy and greater overall reliability of the current structure.
Individual subdomains of the engineered wild-type (RT52A) RT/TCM278 structure and 1B1 RT/TMC120 structure are highly similar. The overall Cα atom superposition of the structures had an rmsd of 1.6 Å, primarily because of small differences in the relative positioning of the subdomains. The Cα superposition of the binding pocket regions of both structures (residues 98–110, 178–190, and 226–240 of the p66 subunit) showed an rmsd of 0.85 Å. The overall similarity in the binding of the two DAPY compounds, as shown in Table 3, also suggests only subtle or modest effects of the crystal-engineered mutations on the inhibitor binding. The engineered RT52A also exhibited both DNA polymerization and RNase H activities similar to 1B1 RT (data not shown).
Table 3.
The variation of torsion angles (τ, in °) reflects the role of strategic flexibility of TMC278 in evading the effect of drug-resistance mutations
| τ1 | τ2 | τ3 | τ4 | τ5 | Total energy, kcal/mol | |
|---|---|---|---|---|---|---|
| Wild-type RT/TMC278 | −97 | 16 | −11 | −12 | −50 | −131.3 |
| K103N/Y181C RT/TMC278 | −98 | 17 | −6 | −10 | −49 | −132.7 |
| L100I/K103N RT/TMC278 | −115 | −2 | −6 | 10 | −16 | −131.3 |
| Free-state TMC278 | −109 | 35 | −13 | −7 | 0 | −133.0 |
| Wild-type RT/TMC120 | −100 | 18 | 0 | −14 | N/A | N/A |
The total energy (in kcal/mol) of the NNRTI only at its specific conformations was calculated by using the MACROMODEL program of the molecular modeling software suite from Schrödinger. The “free-state” conformation represents the lowest-energy “horseshoe” conformation of TMC278 in vacuum. The RT-bound conformations of TMC278, which are almost isoenergetic with the free-state conformation of TMC278, are attained by varying the torsion angles (wiggling).
The 1.8 Å Resolution Structure of the Wild-Type HIV-1 RT/TMC278 Complex.
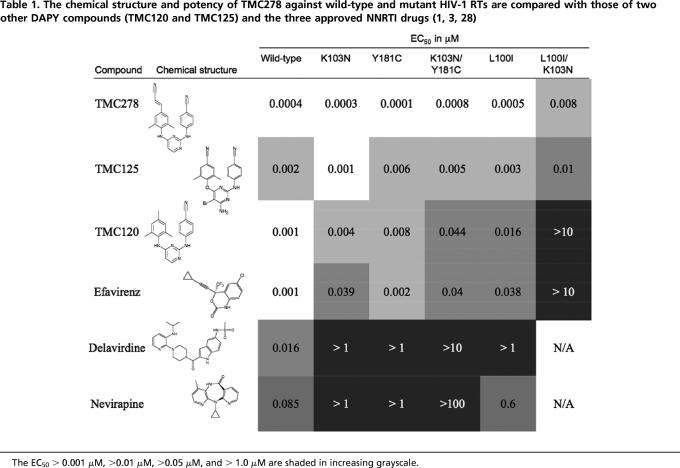
Overall, the structure of the p66/p51 RT heterodimer (Fig. 1A) in the HIV-1 RT/TMC278 complex resembles the open-cleft conformation seen in the previous structures of RT/NNRTI complexes (14–16). The electron-density maps (Fig. 1B) unambiguously defined the position and conformation of TMC278 in the structure of HIV-1 RT/TMC278 complex. TMC278 has a conformation that is similar to the horseshoe conformation seen with other DAPY inhibitors (7), with the three aromatic rings connected by two linking amino groups and a cyanovinyl (acrylonitrile) substituent that is unique to TMC278 (Fig. 1C). The torsion angles of the rotatable bonds (τ1–τ4) of TMC278 (Table 3) have values similar to those of the prototype DAPY analog TMC120 (R147681/dapivirine) bound to RT (7), although the two structures were determined in two different crystal forms by using two different RT constructs.
Fig. 1.
Structure of HIV-1 in complex with TMC278. (A) Overall structure of the wild-type HIV-1 RT/TMC278 complex determined at 1.8 Å resolution. (B) The position and conformation of TMC278 were defined by the difference (∣Fo∣ − ∣Fc∣) electron density calculated at 1.8 Å resolution (3.5σ contours). (C) Chemical structure of TMC278. The τ angles define the torsional flexibility of TMC278 (Table 3).
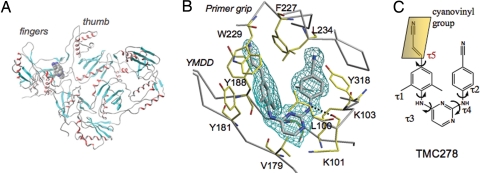
TMC278 makes important contacts with a number of key amino acids in the NNRTI-binding pocket (Fig. 2). The hydrogen bond between a linker nitrogen atom of TMC278 and the main-chain carbonyl oxygen of K101 is conserved in the binding of many NNRTIs. The second linker nitrogen is involved in a water-mediated hydrogen bond network with the main-chain carbonyl group of E138 of the p51 subunit (Fig. 2A). The dimethylphenyl ring and its attached 4-cyanovinyl group interact with the hydrophobic core of the binding pocket (4). The cyanovinyl group is positioned to fit into a hydrophobic tunnel formed by the side chains of amino acid residues Y188, F227, W229, and L234; this tunnel opens toward the nucleic acid-binding cleft (Fig. 2B). A similar tunnel was seen in the binding of a cyanovinyl-containing iodo-pyridinone (IOPY) NNRTI (PDB ID 2B5J) (17). In the free TMC278 molecule, the cyanovinyl group is expected to be coplanar with the dimethylphenyl ring. However, in the RT-bound conformation, the plane of the cyanovinyl group is inclined 50° to the plane of the dimethylphenyl ring. The extensive interactions of the cyanovinyl group with the hydrophobic tunnel may explain why TMC278 is the most potent of the DAPY analogs (3).
Fig. 2.
Binding mode of TMC278 to HIV-1 RT. (A) Interactions of TMC278 (gray) with NNRTI-binding pocket residues (in yellow). (B) The molecular surface (orange) defines the hydrophobic tunnel that accommodates the cyanovinyl group of TMC278.
The high-resolution structure provides a reliable solvent model. The amino acid residues K101 and K103 are solvent exposed (Fig. 2A), and, if mutated, each can confer NNRTI resistance. In the RT/TMC278 structure, the Nζ atom of K103 interacts with two water molecules, whereas the corresponding Nζ of K101 interacts with four oxygen atoms: the carbonyl oxygen of G99, both carboxyl oxygen atoms of E138 (of the p51 subunit), and a water molecule (SI Fig. 6). The location of the K101–Nζ atom in the TMC278 complex is similar to that in the recently published structure of the HIV-1 RT/GW420867X complex (18); however, the identification of the interaction between K101–Nζ and four surrounding oxygen atoms, including one from a solvent water molecule, defines an interesting polar environment for K101. The different environments for and interactions of K101 and K103 may help account for the differences in resistance seen when these two lysines are mutated, even though both of their side chains point toward a common putative entrance to the NNRTI-binding pocket.
Structure of the K103N/Y181C Double Mutant RT/TMC278 Complex.
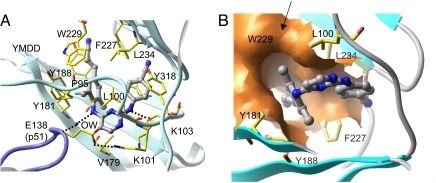
K103N and Y181C are the two resistance mutations most frequently observed in patients treated with NNRTIs, and viruses carrying these mutations show high levels of resistance to existing NNRTIs. However, TMC278 inhibits K103N, Y181C, and K103N/Y181C RT mutants at an EC50 < 1 nM (Table 1) (3). We determined the crystal structure of the K103N/Y181C mutant RT/TMC278 complex at 2.1 Å resolution with Rwork and Rfree of 0.229 and 0.269, respectively (Table 2). Superposition of this structure onto the wild-type RT/TMC278 structure revealed no major conformational changes for the bound TMC278 (Fig. 3). We use the number of distances <4.5 Å between pairs of atoms, one from RT and the other from TMC278, as an indicator of the extent of the hydrophobic interactions between RT and TMC278 (SI Tables 4 and 5). In the K103N/Y181C mutant RT/TMC278 complex, the number of such distances is 51, which is almost the same number of distances, 52, in the wild-type RT/TMC278 complex. A slight tilt (5°) of τ3 (Table 3) results in displacement of the dimethylphenyl-4-cyanovinyl group away from the mutated Y181C side chain. The interaction between the dimethylphenyl ring of TMC278 and the aromatic side chain of Y181 is lost, and a void is created by the mutation. In the structure of the mutant RT, the amino acid residue Y183, which is part of the conserved Y183MDD motif at the polymerase active site, is shifted by ∼1.5 Å toward the NNRTI-binding pocket, permitting it to participate in the binding of TMC278 by binding the cyanovinyl group (Fig. 3).
Fig. 3.
Superposition of K103N/Y181C mutant RT (cyan ribbon, yellow side chains)/TMC278 (orange) complex on the wild-type RT (blue ribbon and gray side chain)/TMC278 (gray) complex. The YMDD motif in the mutant structure is repositioned closer to TMC278, which leads to an important interaction between the cyanovinyl group and the highly conserved Y183 residue. Despite the rearrangements in the inhibitor position and conformation and the binding-pocket residues, the extents of the inhibitor–protein interactions remain almost unchanged (SI Tables 4 and 5).
The ability of the cyanovinyl group of TMC278 to recruit Y183 helps to compensate for the loss of interactions caused by Y181C mutation; the involvement of Y183 in this compensatory interaction is particularly fortuitous and significant because Y183 is completely conserved in all HIV-1 sequences. This mode of compensatory interaction is different from that observed for another NNRTI, HBY 097, which developed a hydrogen bond with the thiol group of the mutated C181 side chain (19); in the K103N/Y181C RT/TMC278 structure, the thiol group of C181 has a hydrogen bond with a water molecule at the equivalent position of the Oη atom of Y181 in the wild-type RT/TMC278 structure. Subtle conformational changes (as reflected by the torsion angles τ; see Table 3) of TMC278 in the K103N/Y181C mutant RT/TMC278 complex enhance the interactions of the cyanovinyl group with the modified hydrophobic tunnel (Fig. 2B), supplementing the contributions of the interactions with Y183. The extent of the interaction between the other mutated amino acid, N103, and TMC278 is comparable to the interaction between K103 of wild-type RT and TMC278: the number of distances <4.5 Å between the atoms of TMC278 and the amino acid K/N103 are 16 and 17 in the wild-type and the mutant structures, respectively (SI Table 5).
Structure of the L100I/K103N Mutant RT/TMC278 Complex.
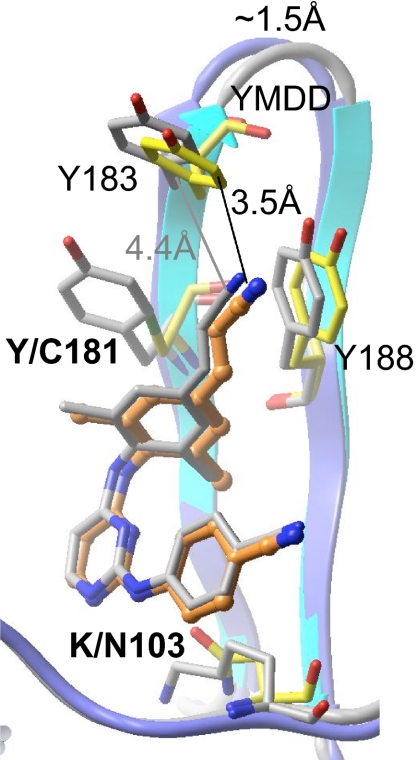
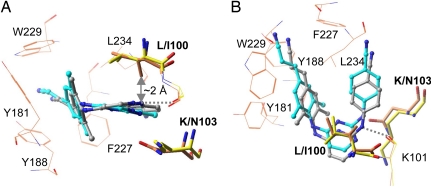
Among the known NNRTI-resistance mutations the L100I/K103N double mutation has the greatest effect on the potency of TMC278. However, TMC278 still inhibits the double mutant at ∼8 nM EC50 (Table 1). We determined the crystal structure of L100I/K103N mutant RT/TMC278 complex at 2.9 Å resolution. The refined structure has R and Rfree of 0.240 and 0.299, respectively (Table 2). In the wild-type RT/TMC278 structure, L100 is near the center of the pocket and primarily interacts with the central pyrimidine ring of TMC278; K103 is located on the other side of the pyrimidine ring. Comparison of the structures of the L100I/K103N mutant RT/TMC278 and wild-type RT/TMC278 complexes (Fig. 4) shows that β-branching of I100 in the L100I mutant would lead to steric conflict with the inhibitor if TMC278 were to bind in a conformation similar to that seen in the wild-type RT/TMC278 complex; in the mutant structure, the Cγ2 atom of I100 would be only ∼2 Å away from the position of the central pyrimidine ring of TMC278 when it is bound to wild-type RT. However, when TMC278 binds to the L100I/K103N mutant RT the drug undergoes significant conformational (wiggling) and positional (jiggling) rearrangements compared with the position in which it binds to wild-type RT (Fig. 4 and SI Tables 4 and 5). To avoid steric conflict with the L100I mutation, TMC278 shifts away from I100 and toward N103 (Fig. 4A); the position of the entire inhibitor molecule is displaced by ∼1.5 Å in the pocket. The number of distances <4.5 Å between TMC278 and I100 is 12 in the complex with the L100I/K103N mutant RT, which is considerably less than the 28 and 30 distances <4.5 Å in the complexes with wild-type RT and K103N/Y181C mutant (SI Table 5); however, in compensation, the number of protein–ligand distances <4.5 Å for residue 103 increases from 16 and 17 in the wild-type and K103N/Y181C mutant structures, respectively, to 27 in the L100I/K103N mutant RT/TMC278 structure.
Fig. 4.
Comparison of L100I/K103N mutant RT (orange side chains)/TMC278 (cyan) structure with the wild-type RT (yellow side chains)/TMC278 (gray) structures reveals wiggling (A) and jiggling (B) of TMC278.
In the L100I/K103N complex, the rotatable torsion angles τ1–τ5 of TMC278 are changed by 18°, 18°, 5°, 22°, and 34°, respectively, with respect to the wild-type RT/TMC278 complex. Unlike its configuration in the wild-type RT/TMC278 and Y181C/K103N mutant RT/TMC278 structures, the cyanovinyl group is almost coplanar with the dimethylphenyl ring in the L100I/K103N mutant RT/TMC278 structure (Table 3). In the L100I/K103N complex structure, the amino acid residues in the NNRTI-binding pocket are rearranged to optimize the inhibitor–protein interactions, which contrasts with an earlier proposal that the basis of the effects of the L100I mutation was a loss of interactions with Y181 and Y188 (20). However, analysis of all of the structural results shows that L100I introduces a significant distortion in the NNRTI-binding pocket. NNRTIs that do not have the ability to wiggle and jiggle and adapt their shape to the various pockets found in the NNRTI-resistant RTs fail against the known mutants because their binding is susceptible to steric hindrance, because they lose key hydrophobic interactions, or because mutations like K103N interfere with entry of the NNRTIs into the pocket.
Role of the Cyanovinyl Group of TMC278.
The cyanovinyl group of TMC278 is not present in the other DAPY analogs. Analysis of the crystal structures suggests that the cyanovinyl group contributes to the enhanced potency of TMC278 relative to the other DAPY analogs and that this moiety helps TMC278 to retain potency against NNRTI-resistance mutations. As has already been discussed, the cyanovinyl group is positioned in a cylindrical tunnel connecting the NNRTI-binding pocket to the nucleic acid-binding cleft that resembles a “piston-and-ring” structure (Fig. 2B). The extent of the interactions between the cyanovinyl group and the hydrophobic tunnel is conserved despite rearrangements in RT and TMC278 that accompany the pocket mutations (SI Table 4). Apparently, the maintenance of cyanovinyl group interactions with RT is critical for retaining the potency of TMC278 against a broad range of NNRTI-resistance mutations. Analysis of the torsional flexibility (Table 3) clearly demonstrates how TMC278 is resilient in overcoming the effects of drug-resistance mutations. A 2D infrared spectroscopic study of TMC278 complexed with the engineered RT52A HIV-1 RT (21) revealed that the conformational distribution of drug–protein complexes is relaxing on the tens of picoseconds time scale; i.e., TMC278 loses structural “memory” of its binding mode within tens of picoseconds. These motions are consistent with the concept that TMC278 is flexible even when bound to HIV-1 RT and can change its conformation to adapt to the elastic NNRTI-binding pocket.
Implications for Drug Design.
High-resolution structures of RT provide opportunities for understanding inhibitor–protein interactions with greater accuracy, more reliable determination of the structural effects of resistance mutations, and systematic structure-based drug design targeting the NNRTI-binding pocket. The opening of the tunnel to the nucleic acid-binding site suggests the possibility of extending NNRTIs so that they could interact directly with the conserved residues involved in dNTP and/or nucleic acid binding, a concept that has been previously proposed (22, 23). The interactions of the TMC278 cyanovinyl group with the hydrophobic tunnel enhances the binding of the inhibitor, and the group also is important for the potency of the inhibitor against drug-resistance mutations. Comparison of structures of TMC278 in complexes with L100I/K103N and wild-type RT clearly demonstrated the importance of strategic flexibility (wiggling) and repositioning (jiggling).
The RT-bound conformations of TMC278 are somewhat different from each other and from its free-state low-energy conformation (Table 3) obtained by using the molecular modeling software Schrödinger (www.schrodinger.com). However, the total energy calculated for the different conformations of TMC278 (Table 3 and SI Fig. 7) are not significantly different from its free-state low-energy conformation. It is expected that a small molecule would bind to a receptor approximately in its low-energy conformation. The fact that TMC278 can achieve near-low-energy conformations when bound to different forms of HIV-1 RT explains why TMC278 maintains its high potency against the mutant RTs.
The HIV-1 RT binding pocket for NNRTIs is flexible (14) and can accommodate a diverse range of small molecule chemotypes. The binding pocket flexibility can be described as a molecular “shrink wrap” phenomenon in which the protein structure adapts and can form a complementary shape to surround the bound inhibitor (SI Movie 1). Analysis of the K103N/Y181C mutant RT/TMC278 structure reveals how TMC278 can take advantage of the structural flexibility of RT, inducing localized changes in the protein that lead to interactions with Y183 that compensate the loss of the hydrophobic interaction caused by the Y181C mutation. The fact that compensatory changes can occur both in the protein and in the drug suggests that optimal drug design strategies should carefully consider and take advantage of the flexibility of both the inhibitor and protein. The potential flexibility of both the protein and the drug should be strategic considerations in early stages of programs to design drugs that are intended to be broadly effective against targets that readily mutate and develop drug resistance.
Methods
Expression, Purification, and Crystallization.
The IB1 RT used in our earlier crystallographic studies of RT/NNRTI complexes (11) produced RT/TMC278 crystals that diffracted (at best) to only 6 Å resolution. To overcome this obstacle and obtain suitable diffraction data for structural studies, a systematic crystal engineering approach that improved resolution was used. The engineered RT (RT52A) used in the current study has amino acid residues 1–555 in the p66 chain containing K172A and K173A mutations and 1–428 in the p51 chain when compared with 1B1 RT. The RT/TMC278 complexes were crystallized from RT52A at 20 mg/ml in 10 mM Tris (pH 8.0) and 75 mM NaCl containing TMC278 at a 5:1 molar ratio of TMC278 to RT. Crystals were obtained in hanging drop-vapor diffusion setups at 4°C. The well solution contained 12% PEG 8000, 100 mM ammonium sulfate, 10 mM MgCl2, 15 mM spermine, and 50 mM imidazole buffer at pH 6.8. The crystals grew to appropriate size for diffraction within 1 week. Crystals of the RT/TMC278 complexes were dipped for 10 s in their respective mother liquors containing 25% ethylene glycol for cryoprotection. The cryoprotected crystals were flash-cooled in liquid N2 and transported to synchrotron sources.
Structure Solution.
Diffraction data were collected from one crystal of each type of RT/NNRTI complex at the Cornell High Energy Synchrotron Source (CHESS) F1 and Brookhaven National Laboratory (BNL) X25 beamlines. The data were processed by using HKL2000 (24). The engineered RT/TMC278 complexes crystallized in a previously uncharacterized crystal form (Table 2). The previously reported structure of the RT/R147681 complex (7) was used as a starting model for obtaining molecular replacement solutions for the structure of the wild-type RT/TMC278 complex. The 1.8 Å resolution structure of the wild-type RT/TMC278 complex was used as the starting model for obtaining the structures of I100L/K103N mutant and K103N/Y181C mutant RTs in complexes with TMC278. The final models for the three structures were obtained after cycles of model building in COOT (25) and restrained refinement with REFMAC (26) and CNS 1.1 (27). The x-ray data and refinement statistics are listed in Table 2. The high-resolution structure of the RT/TMC278 complex revealed no metal binding at the polymerase active site. Also, no metal ion with clear coordination geometry could be located at the RNase H active site. An electron-density peak that is nearly the positional equivalent of a metal cation at the RNase H active site, however, was assigned as a water because its lacks the proper metal coordination. Coordinates and structure factors for the structures of wild-type RT/TMC278, K103N/Y181C/TMC278, and L100I/K103N/TMC278 complexes are available from the PDB with ID codes 2ZD1, 3BGR, and 2ZE2, respectively.
Supplementary Material
Acknowledgments
We acknowledge personnel at the Cornell High Energy Synchrotron Source (CHESS) and Brookhaven National Laboratory (BNL) for support of data collection, the members of our laboratories for valuable conversations and assistance, and Robin Hochstrasser for discussions. E.A. is grateful to the National Institutes of Health (Grants AI 27690 MERIT Award and P01 GM 066671) for support of RT structural studies. S.H.H. was supported by the Intramural Research Program of National Institutes of Health, National Cancer Institute, Center for Cancer Research, and National Institute of General Medical Sciences.
Note Added in Proof.
TMC125 (etravirine/Intelence), a member of the DAPY chemical class that includes TMC278, was recently approved by the U.S. Federal Drug Administration for treatment of patients infected with HIV-1 variants resistant to other antiretroviral drugs.
Footnotes
The authors declare no conflict of interest.
Data deposition: Coordinates and structure factors for the structures of wild-type RT/TMC278, K103N/Y181C/TMC278, and L100I/K103N/TMC278 complexes are available from the Protein Data Bank, www.pdb.org (PDB ID codes 2ZD1, 3BGR, and 2ZE2, respectively).
This article contains supporting information online at www.pnas.org/cgi/content/full/0711209105/DC1.
References
- 1.Ludovici DW, et al. Evolution of anti-HIV drug candidates. Part 3: Diarylpyrimidine (DAPY) analogues. Bioorg Med Chem Lett. 2001;11:2235–2239. doi: 10.1016/s0960-894x(01)00412-7. [DOI] [PubMed] [Google Scholar]
- 2.Guillemont J, et al. Synthesis of novel diarylpyrimidine analogues and their antiviral activity against human immunodeficiency virus type 1. J Med Chem. 2005;48:2072–2079. doi: 10.1021/jm040838n. [DOI] [PubMed] [Google Scholar]
- 3.Janssen PA, et al. In search of a novel anti-HIV drug: Multidisciplinary coordination in the discovery of 4-[[4-[[4-[(1E)-2-cyanoethenyl]-2,6-dimethylphenyl]amino]-2- pyrimidinyl]amino]benzonitrile (R278474, rilpivirine). J Med Chem. 2005;48:1901–1909. doi: 10.1021/jm040840e. [DOI] [PubMed] [Google Scholar]
- 4.Das K, Lewi PJ, Hughes SH, Arnold E. Crystallography and the design of anti-AIDS drugs: Conformational flexibility and positional adaptability are important in the design of non-nucleoside HIV-1 reverse transcriptase inhibitors. Prog Biophys Mol Biol. 2005;88:209–231. doi: 10.1016/j.pbiomolbio.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Goebel F, et al. Short-term antiviral activity of TMC278–A novel NNRTI–in treatment-naive HIV-1-infected subjects. AIDS. 2006;20:1721–1726. doi: 10.1097/01.aids.0000242818.65215.bd. [DOI] [PubMed] [Google Scholar]
- 6.Pozniak A, et al. 48-week primary analysis of trial TMC278–C204: TMC278 demonstrates potent and sustained efficacy in ARV-naïve patients. 14th Conference on Retroviruses and Opportunistic Infections (CROI); February 25–28, 2007; Los Angeles, CA. 2007. paper 144LB. [Google Scholar]
- 7.Das K, et al. Roles of conformational and positional adaptability in structure-based design of TMC125–R165335 (etravirine) and related non-nucleoside reverse transcriptase inhibitors that are highly potent and effective against wild-type and drug-resistant HIV-1 variants. J Med Chem. 2004;47:2550–2560. doi: 10.1021/jm030558s. [DOI] [PubMed] [Google Scholar]
- 8.Pauwels R. Aspects of successful drug discovery and development. Antiviral Res. 2006;71:77–89. doi: 10.1016/j.antiviral.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 9.Frank KB, Noll GJ, Connell EV, Sim IS. Potent and selective inhibition of HIV-1 replication in vitro by a novel series of TIBO derivatives. Nature. 1990;343:470–474. doi: 10.1038/343470a0. [DOI] [PubMed] [Google Scholar]
- 10.Lewi PJ, et al. On the detection of multiple-binding modes of ligands to proteins, from biological, structural, and modeling data. J Comput Aided Mol Des. 2003;17:129–134. doi: 10.1023/a:1025313705564. [DOI] [PubMed] [Google Scholar]
- 11.Clark ADJ, Jacobo-Molina A, Clark P, Hughes SH, Arnold E. Crystallization of human immunodeficiency virus type 1 reverse transcriptase with and without nucleic acid substrates, inhibitors and an antibody Fab fragment. Methods Enzymol. 1995;262:171–185. doi: 10.1016/0076-6879(95)62017-6. [DOI] [PubMed] [Google Scholar]
- 12.Rodgers DW, et al. The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1995;92:1222–1226. doi: 10.1073/pnas.92.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsiou Y, et al. Structure of unliganded HIV-1 reverse transcriptase at 2.7 Å resolution: Implications of conformational changes for polymerization and inhibition mechanisms. Structure (London) 1996;4:853–860. doi: 10.1016/s0969-2126(96)00091-3. [DOI] [PubMed] [Google Scholar]
- 14.Ren J, et al. High-resolution structures of HIV-1 RT from four RT-inhibitor complexes. Nat Struct Biol. 1995;2:293–302. doi: 10.1038/nsb0495-293. [DOI] [PubMed] [Google Scholar]
- 15.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 16.Lindberg J, et al. Structural basis for the inhibitory efficacy of efavirenz (DMP-266), MSC194 and PNU142721 towards the HIV-1 RT K103N mutant. Eur J Biochem. 2002;269:1670–1677. doi: 10.1046/j.1432-1327.2002.02811.x. [DOI] [PubMed] [Google Scholar]
- 17.Himmel DM, et al. Crystal structures for HIV-1 reverse transcriptase in complexes with three pyridinone derivatives: A new class of non-nucleoside inhibitors effective against a broad range of drug-resistant strains. J Med Chem. 2005;48:7582–7591. doi: 10.1021/jm0500323. [DOI] [PubMed] [Google Scholar]
- 18.Ren J, et al. Relationship of potency and resilience to drug resistant mutations for GW420867X revealed by crystal structures of inhibitor complexes for wild-type, Leu100Ile, Lys101Glu, and Tyr188Cys mutant HIV-1 reverse transcriptases. J Med Chem. 2007;50:2301–2309. doi: 10.1021/jm061117m. [DOI] [PubMed] [Google Scholar]
- 19.Das K, et al. Crystal structures of clinically relevant Lys103Asn/Tyr181Cys double mutant HIV-1 reverse transcriptase in complexes with ATP and non-nucleoside inhibitor HBY 097. J Mol Biol. 2007;365:77–89. doi: 10.1016/j.jmb.2006.08.097. [DOI] [PubMed] [Google Scholar]
- 20.Ren J, et al. Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors. J Mol Biol. 2004;336:569–578. doi: 10.1016/j.jmb.2003.12.055. [DOI] [PubMed] [Google Scholar]
- 21.Fang C, et al. Two dimensional infrared spectra reveal relaxation of the nonnucleoside inhibitor TMC278 complexed with HIV-1 reverse transcriptase. Proc Natl Acad Sci USA. 2008;105:1472–1477. doi: 10.1073/pnas.0709320104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nanni RG, Ding J, Jacobo-Molina A, Hughes SH, Arnold E. Review of HIV-1 reverse transcriptase three-dimensional structure: Implications for drug design. Perspect Drug Disc Design. 1993;1:129–150. [Google Scholar]
- 23.Spence RA, Kati WM, Anderson KS, Johnson KA. Mechanism of inhibition of HIV-1 reverse transcriptase by nonnucleoside inhibitors. Science. 1995;267:988–993. doi: 10.1126/science.7532321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otwinowski Z, Minor W. DENZO and SCALEPACK. Boston: Kluwer; 2001. [Google Scholar]
- 25.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 27.Brunger AT, Adams PD, Rice LM. Recent developments for the efficient crystallographic refinement of macromolecular structures. Curr Opin Struct Biol. 1998;8:606–611. doi: 10.1016/s0959-440x(98)80152-8. [DOI] [PubMed] [Google Scholar]
- 28.Heeres J, Lewi PJ. The medicinal chemistry of the DATA and DAPY series of HIV-1 non-nucleoside reverse transcriptase inhibitors (NNRTIs). In: De Clercq E, editor. Advances in Antiviral Drug Design. Vol. 5. Amsterdam: Elsevier; 2007. pp. 213–242. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.