Abstract
The six-transmembrane helix (6 TM) tetrameric cation channels form the largest ion channel family, some members of which are voltage-gated and others are not. There are no reported channel structures to match the wealth of functional data on the non-voltage-gated members. We determined the structure of the transmembrane regions of the bacterial cyclic nucleotide-regulated channel MlotiK1, a non-voltage-gated 6 TM channel. The structure showed how the S1–S4 domain and its associated linker can serve as a clamp to constrain the gate of the pore and possibly function in concert with ligand-binding domains to regulate the opening of the pore. The structure also led us to hypothesize a new mechanism by which motions of the S6 inner helices can gate the ion conduction pathway at a position along the pore closer to the selectivity filter than the canonical helix bundle crossing.
Keywords: crystal structure, membrane protein, non-voltage-gated
In the superfamily of tetrameric cation channels, the ion pore domain is formed by four subunits or repeats with two transmembrane helices (TMs) (labeled S5 and S6 in Fig. 1A) and a pore loop (Ploop) or ion-selectivity region (1–3). Some of these channels, known as six-transmembrane helix (6 TM) channels, have an extra 4 TM domain in each repeat or subunit. The role of this domain (Fig. 1A), referred here as the S1–S4 domain, is well described in voltage-gated channels; it senses membrane electrical potential changes and undergoes a conformational change involving the translocation of approximately three charges across the electrical field (3–7), resulting in the opening/closing of the channel gate. Most of the voltage dependence is conferred by the fourth TM (S4), which has a conserved sequence pattern of a positively charged residue at every third position (4, 5). In 6 TM non-voltage-gated channels, the role of the S1–S4 domain is unclear; they are related to the S1–S4 domains of voltage-gated channels, but they do not act as voltage sensors. There are no reported structures of non-voltage-gated 6 TM channels.
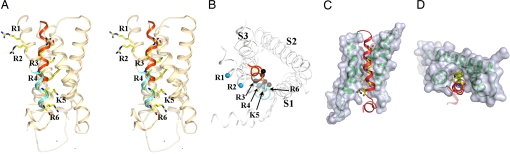
Fig. 1.
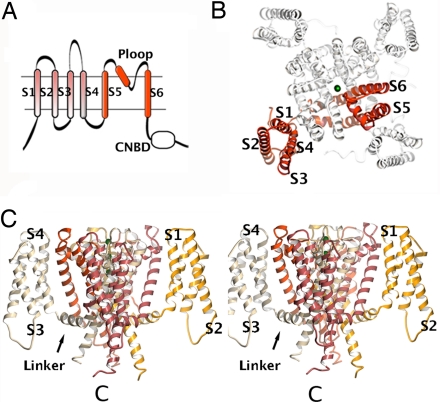
Architecture of 6 TM channel. (A) Illustration of a 6 TM channel. The ion pore regions (S5, Ploop, and S6) are shown in red. Also shown are the S1–S4 domain and a C-terminal cytoplasmic domain [CNB domain (CNBD)]. (B) MlotiK1 structure viewed from the extracellular side. One subunit is shown in red. TMs are labeled S1 to S6. Green spheres in the pore are K+. (C) Stereo side view of the MlotiK1 channel structure. Extracellular side at the top of figure. Subunits are shown in different colors. Some of the TMs and the S4–S5 linker are labeled. One of the C termini is indicated by C.
Cyclic nucleotide-regulated channels are 6 TM channels that have a C-terminal cytoplasmic cyclic nucleotide-binding (CNB) domain (Fig. 1A). A family of prokaryotic cyclic nucleotide-regulated channels has been characterized recently (8, 9). The functional characterization of the MlotiK1 channel from the bacterium Mesorhizobium loti, in a radioactive uptake assay, shows a 2- to 3-fold increase in channel activity in the presence of cAMP over basal flux. An unusual feature of this channel is that uptake is very slow and does not reach equilibrium even after an hour.
Despite a wealth of functional and structural information about tetrameric cation channels, there are crucial open questions about the molecular mechanisms of channel gating and, in particular, the role of the S1–S4 domain in non-voltage-gated channels. To address these fundamental questions, we determined the crystal structure of the bacterial cyclic nucleotide-regulated channel MlotiK1. This structure defines the transmembrane regions (pore and S1–S4 domains), but the CNB domains are not resolved. This study is focused on the transmembrane components of the first known structure of a non-voltage-gated 6 TM channel.
Results
MlotiK1 Architecture.
The best diffracting crystals of MlotiK1 were grown in the presence of substoichiometric amounts of ligand. We determined a 3.1Å (Rfree, 28.6%; Rwork 27.6%) crystal structure of the MlotiK1 potassium channel in the presence of 200 μM cAMP (Fig. 1 B and C). The high quality of the electron-density maps [supporting information (SI) Fig. 6] allowed us to define the transmembrane regions with good accuracy. The C-terminal CNB domains were disordered and not included in the model.
The MlotiK1 channel structure adopted a flower-like arrangement, with the pore domain at the center and the S1–S4 domains in the periphery (Fig. 1 B and C). The S1–S4 domains packed against the ion pore domain through an adjacent subunit. The S4 in MlotiK1 was partially exposed to the lipid environment and was connected to S5, in the same subunit, through a 3-turn helical linker (S4–S5 linker) that ran parallel to the plane of the membrane (Fig. 1C). The overall architecture of MlotiK1 was similar to that of the eukaryotic voltage-gated potassium channels (10, 11); however, there were important differences.
The Pore.
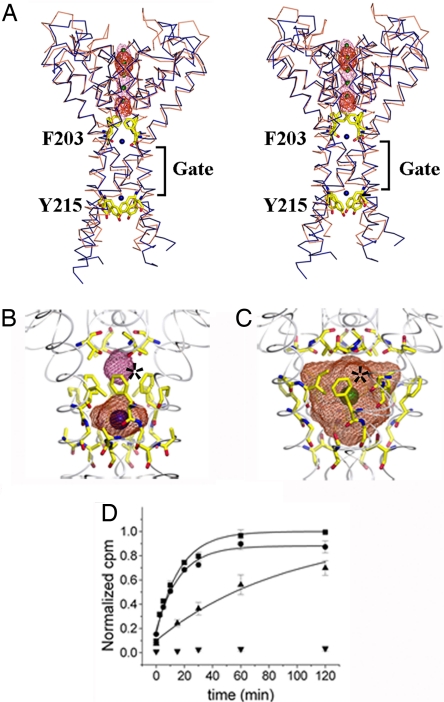
Fig. 2A shows the helices that lined the transmembrane pore of the MlotiK1 channel. The arrangement of these helices was very similar to that of other potassium channels in a closed state, such as KcsA (12, 13) (Fig. 2A). Although the helical positions were the same, there were two notable features of the MlotiK1 transmembrane pore that distinguished it from KcsA. Above and below the inner helix bundle crossing, labeled “Gate” in Fig. 2A, the pore was constricted by amino acids F203 (above the gate) and Y215 (below the gate). In addition to the constriction at the gate, F203 and Y215 should create high-energy barriers to the passage of ions. The constriction created by F203 was particularly interesting because the four side chains protrude into the central cavity (Fig. 2 A and B). The KcsA potassium channel also had phenylalanines in the same position, but their side chains were tucked back toward the wall of the cavity (Fig. 2C). In KcsA the central cavity (760 Å3) has been shown to contain a fully hydrated potassium ion (13). In contrast, in the MlotiK1 structure, there was no space for an ion in an equivalent position because of the conformation of the F203 side chains. The phenylalanines in MlotiK1 split the cavity into two (Fig. 2B). Above F203, the cavity was small (≈30 Å3); below F203, the cavity was ≈130 Å3 and contained electron density assigned to a water molecule. To confirm the absence of an ion, we grew crystals in which K+ was substituted by the permeant electron-dense Rb+ or Cs+. Difference Fourier maps (Fig. 2A) showed the expected positive density in the selectivity filter and not in any of the cavities.
Fig. 2.
The pore domain. (A) Stereoview of MlotiK1 (blue) and KcsA (brown) pores. TMs S1–S5 are not shown. Phe-203 and Tyr-215 shown as yellow sticks. The difference electron-density maps are shown in red (Cs+) and pink (Rb+) mesh. K+ is shown as green spheres. Water molecules are shown as blue spheres. The bundle crossing gate region is indicated. (B) MlotiK1 cavities. Water-accessible volumes are shown as mesh. Residues lining the cavity are shown as sticks. The asterisk marks the Phe-203 side chains. Water molecules are shown as blue spheres. (C) KcsA cavity as in B. The green sphere represents K+. The asterisk indicates Phe-103 side chains. (D) Time course of MlotiK1 activity as monitored by 86Rb+ flux assay. Wild types and mutants, F203A and Y215A, were assayed for activity in the presence of saturating cAMP. The points and error bars show mean and SD for three measurements. The data were fit to the single exponential, and the time constants were determined: F203A, 16.7 ± 1.8 min; Y215A, 24.6 ± 3.8 min; wild type, 94.4 ± 9.7 min. Triangles, wild type; squares, F203A; circles, Y215A; inverted triangle, control (liposomes without protein).
We wondered whether disruption of the central cavity in MlotiK1 by F203 may be relevant to conductivity or gating. We tested this by mutating this residue to alanine and evaluated its function in a rubidium-uptake assay in reconstituted proteoliposomes, the only available assay for MlotiK1 (8, 9). The mutation F203A showed a clear increase in the rate of uptake relative to the wild-type channel (Fig. 2D), with a time constant (17 min) that was ≈6-fold faster than the wild type (94 min). The mutant Y215A also had a faster uptake rate (25 min) relative to wild type. These structural and functional data raise the possibility of a gate in the central cavity of certain tetrameric cation channels. Such a gate may explain a set of electrophysiological data from eukaryotic cyclic nucleotide-gated channels that is addressed in Discussion.
The S4–S5 Linker in a Closed 6TM Channel.
The S4–S5 linker is one of the regions thought to be important during gating; in voltage-gated channels, interactions between the linker and the end of S6 have been demonstrated both functionally (14, 15) and structurally (10, 11). Because the Kv1.2 channel is in an open conformation, the MlotiK1 channel structure provides the first opportunity to inspect the S4–S5 linker in the context of a closed 6 TM channel.
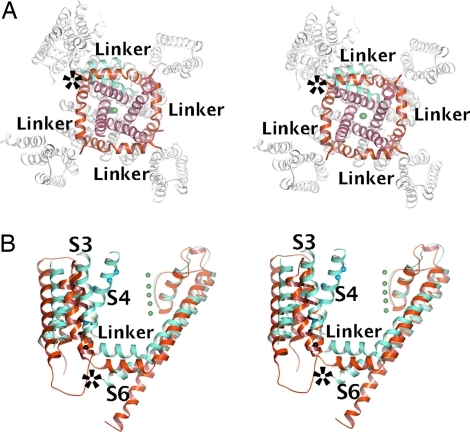
Fig. 3 A and B shows the superposition of the MlotiK1 and Kv1.2 structures via the pore regions. The S4–S5 linkers formed a cuff around the gate regions, restricting the movement of S6. The superpositions show the C-terminal end of the S6 helices from the open Kv1.2 (Fig. 3 A and B) clashing with the linkers of the closed MlotiK1. For the MlotiK1 channel gate to open to the same extent as in Kv1.2, it would be necessary for the S4–S5 linkers to be repositioned. Indeed, relative to MlotiK1, the Kv1.2 linker was shifted outward (≈5 Å), away from the pore (Fig. 3A), and upward (≈5 Å), toward the extracellular side (Fig. 3B). These differences were the result of the state of the gate and not of differences between the linkers. A superposition of the isolated S4–S5 linkers (SI Fig. 7) revealed that the linkers and their structural connections with S4, S5, and S6 were very similar in the two channels.
Fig. 3.
The S4–S5 linker and channel gate. (A) Stereoview of cytoplasmic side of MlotiK1 channel with the Kv1.2 subunit superposed via the pore region. In MlotiK1, the S4–S5 linkers and S5 helices are colored red, whereas the C-terminal end of S6 helices is purple. In Kv1.2, the S4–S5 linker, S5, and S6 are colored cyan. The rest of the molecules are colored white. The green spheres represent K+. The asterisk indicates steric clash between the S6 of Kv1.2 and the S4–S5 linker of MlotiK1. (B) Stereo side view of MlotiK1 (red) and Kv1.2 (cyan) subunits superposed via the pore region. The S4–S5 linker and some TMs are labeled. The asterisk indicates steric clash between the S4–S5 linker of MlotiK1 and the S6 of Kv1.2.
The S1–S4 Domain.
The first significant difference between the S1–S4 domain of MlotiK1 and that of the established voltage-gated channel was evident at an amino acid sequence level (Fig. 4A). The voltage-gated channels contain a positively charged amino acid at every third position of S4, referred to as R1, R2, R3, R4, K5, and R6 (3). The first four of these positions have been shown to account for most of the gating charge that enables a voltage sensor to respond to changes in membrane voltage (4, 5). All four of these residues in MlotiK1 are uncharged. In addition, two of the three important countercharges that are conserved in voltage-gated channels (E283 and E293 in the eukaryotic Shaker potassium channel) (16, 17) were also absent in MlotiK1 (SI Fig. 8). It is clear that MlotiK1 does not fall into the category of established voltage-gated potassium channels.
Fig. 4.
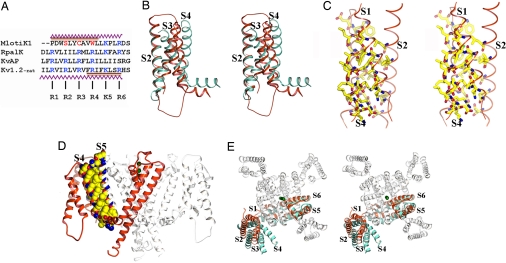
The S1–S4 domain. (A) Structure-based sequence alignment of S4 regions from the MlotiK1 (GI:14023393), RpalK (GI:39937293), KvAP (GI:30749950), and Kv1.2 (GI:73536154) channels. Conserved positively charged residues are shown in blue. MlotiK1 residues corresponding to R2, R3, and R4 are shown in red. The zigzag lines indicate residues in the S4 helices of MlotiK1 and Kv1.2. The brown bar indicates regions of S4 adopting 310 conformation in MlotiK1 and Kv1.2. (B) Stereoview of superposition of S1–S4 domains from MlotiK1 (red) and Kv1.2 (cyan) via S2 and S1. TMs are labeled. (C) Stereoview of residues within the core of S1–S4 domain. TMs are labeled. (D) Side view of MlotiK1. The extracellular side of the membrane is above the molecule. One subunit is shown in red. The residues involved in the interaction between S4 and S5 are shown in Corey–Pauling–Koltun representation. (E) Stereoview from the extracellular side. The MlotiK1 channel (in white and red) and a Kv1.2 subunit (in cyan) are superposed via the pore region. TMs are labeled. The green spheres represent K+.
The MlotiK1 S1–S4 domain was also different from the voltage sensors of the voltage-gated potassium channels in three important structural aspects. A superposition of the structures of the isolated S1–S4 domains from MotiK1 and Kv1.2 via the S1 and S2 helices showed the first of these differences (Fig. 4B). In MlotiK1, the S1–S4 domain formed a compact arrangement of the four transmembrane helices. This was in contrast to the voltage sensor of Kv1.2 in which the S3b–S4 paddle motif is tilted away from the S1 and S2 helices, creating a water-accessible crevice (10, 11, 18). The compact arrangement of helices in MlotiK1 was mediated by interdigitated side chains forming a solid protein core (Fig. 4C).
A second structural difference was the extent to which the S1–S4 domain interacts with the pore. This interaction buried a total surface area of ≈1,200 Å2 and involved residues along the entire length of S4, the extracellular end of S1, and, from the adjacent subunit, the entire length of S5 (Fig. 4D). This extensive interaction contrasted with the interface in Kv1.2 that is more limited to the intracellular half of S4 and the extracellular end of S1 (10, 11, 18). The S1–S4 domain in MlotiK1 appeared to be held more rigidly against the pore domain.
The third difference concerns the precise relation between the S1–S4 domain and the pore: In MlotiK1, the entire S1–S4 domain was rotated around the pore (Fig. 4E) relative to Kv1.2. The repositioning paralleled the position of the S4–S5 linker and the closed conformation of the gate discussed above.
A 310 Helix in S4.
A surprising finding from the comparison between S4 in MlotiK1 and Kv1.2 is that both adopted a 310 helical conformation over part of the helix but in different regions (Figs. 4A, 5 A and B, and SI Fig. 9). Unlike α-helices, where a main-chain hydrogen-bonding pattern is established between residues at positions i and i + 4, in a 310 helix, the pattern is i to i + 3, and the helix is more tightly wound. In MlotiK1, a consequence of the adoption of the 310 conformation, where every third residue was on the exact same helical face, was that residues equivalent to R2, R3, R4, K5, and R6 were buried within the core of the S1–S4 domain (Fig. 5 A and B).
Fig. 5.
The S4 helix. (A) Stereo side view of MlotiK1 and Kv1.2 S1–S4 domains superposed via S2 and S1. 310 regions in S4 of MlotiK1 and Kv1.2 are shown in red and cyan, respectively. Other TMs are represented by brown ribbons. S4 residues discussed in the text are labeled. (B) Extracellular view of S1–S4 domains from MlotiK1 and Kv1.2 superposed via S2 and S1. 310 regions in S4 as in A. Other helical regions are in white ribbons. Kv1.2 R1, R2, R3, R4, K5, and R6 and equivalent MlotiK1 Cα atoms are shown as blue and black spheres, respectively. (C) Surface representation of MlotiK1 S1–S4 domain. S4 is shown as a red ribbon, with residues equivalent to R2, R3, R4, K5, and R6 shown as sticks. Protein regions from S1 to S3 are shown as gray surface representations. S1, S2, and S3 helices are shown as green ribbons inside the surface. (D) Same as in C but viewed from the extracellular side.
Long 310 helices are rare (19). A recent survey of 689 nonredundant protein structures at 1.6 Å resolution or better has found 1,757 310 helices. Among these, the longest is 15 residues, the second longest is 11 residues, and there are only 53 with 8 residues. Remarkably, the 310 helix was 11 residues long in MlotiK1 and 8 residues in Kv1.2. Packing interactions between S4 and the rest of the S1–S4 domain probably stabilize these rare long 310 helices. In Kv1.2, the 310 was replaced by an α-helix in the region where S4 was splayed away from the S1–S4 domain (Fig. 5A). In MlotiK1, S4 was located within a narrow groove formed by the residues from S1, S2, and S3 (Fig. 5 C and D); this appeared to force the side chains in S4 into a ridge and favor the adoption of a 310 conformation. The presence of this rare secondary structure element in the S1–S4 domains from two different channels must have general functional implications.
Discussion
The structure of the S1–S4 domain in MlotiK1 displayed several fundamental differences relative to the voltage sensors of voltage-gated channels. It was a more compact domain (Fig. 4B), with tight interactions between all of the transmembrane helices (Fig. 4C). It had an extensive packing interface with the pore domain (Fig. 4D) and was positioned differently, rotated relative to the pore (Fig. 4E). Importantly, a structure-based alignment revealed that the S4 of MlotiK1 clearly lacked the positively charged residues, shown to be essential for voltage sensing, and some of their countercharges (Fig. 4A and SI Fig. 8). In light of these structural and sequence differences, we conclude that MlotiK1 is distinct from standard voltage-gated channels and that it is likely that the primary function of the S1–S4 domain is not as a voltage sensor.
What, then, is the role of the S1–S4 domain in the MlotiK1 channel? This question is also relevant to other 6 TM channels with S1–S4 domains that do not function as voltage sensors. Given that the S4–S5 linker encircled the pore at the level of the gate and that the domain was directly connected to the linker (Fig. 3 A and B), we assumed that the primary function of this domain is as a mediator of gating. One possibility is that the domain may restrict opening of the gate until a yet unknown stimulus, such as the binding of a small molecule or lipid, directly affects the domain conformation. This conformational change may not necessarily be attributable to the movement of S4 as in a voltage-gated channel but could result from a rigid body movement of the entire domain. A second possibility is that S1–S4 work against the ligand-binding domains to ensure that the pore remains tightly shut in the absence of cAMP binding.
The single most striking feature of the S1–S4 domain in MlotiK1 was the existence of a 310 helix over the length of 11 amino acids (Fig. 5 A and B). Long 310 helices are very rare because they are high-energy relative to α-helices; therefore, its presence implies an important functional role. This secondary structural element also has been detected in the S4 of eukaryotic voltage-gated channel structures, where it has been proposed to play a role in voltage sensing (18). Indeed, there were certain unexplained features of voltage sensing for which the properties of a long 310 helix offer an intriguing structural solution. For example, the three residues per turn of a canonical 310 helix (3.6 in an α-helix) would explain the distinctive S4 sequence motif. In MlotiK1, the adoption of a 310 conformation placed residues equivalent to R2, R3, R4, K5, and R6 all on the same helical face, shielded from the lipid environment, and in contact with the S1–S4 protein regions (Fig. 5A). Also, many voltage-sensing mechanisms (7, 20, 21) have proposed a rotation of the whole S4 to bring the charged residues into contact with the S1–S4 domain and away from the lipid environment. Reversible transitions between 310 and α-helices have been described in peptides (22–25) and in two enzymes (26, 27); this transition provides an interesting structural basis for the S4 rotational movements. Comparing residues R1–R4 in the region S4 of Kv1.2 with equivalents in MlotiK1 illustrated the effect of an α-helix/310 transition in S4 (Fig. 5B). Residues that were exposed to the lipid and away from the S1–S4 domain (Kv1.2), after the transition, would become buried within the domain (MlotiK1).
In a non-voltage-gated channel like MlotiK1, we can imagine that if a stimulus could alter the conformation of the S1–S4 domain, changing the packing interactions between the transmembrane helices, then the 310 would undergo a transition to an α-helix. The increase per residue along the helical axis is 1.5 Å in an α-helix and 1.8–2.0 Å in a 310 helix (19); conversion of 11 residues from a 310 helix to an α-helix would shorten the MlotiK1 S4 by 3–5 Å. When comparing the Kv1.2 (open gate) and the MlotiK1 (closed gate) channels, we noted that the linker was shifted by ≈5 Å toward the cytoplasm (Fig. 3B); therefore, shortening of S4 could affect the position of the S4–S5 linker and influence the state of the gate.
The structure of MlotiK1 led us to consider a new way in which motions of the S6 helices could gate conduction through the pore. A prominent feature of the KcsA potassium channel is a water-filled cavity at the center of the membrane (12). This cavity is known to be the receptor site where many potassium channel inhibitors bind and prevent ion conduction (28). In MlotiK1, the center of this cavity is filled with phenylalanine side chains (Fig. 2B). In KcsA, the equivalent residue is also a phenylalanine, but the conformation of its side chain did not occlude the cavity (Fig. 2C). This difference makes us wonder whether residue side chains that fill the cavity could function as a kind of gate. It is easy to imagine that the ligand-binding domain could control the position of these side chains by affecting the movement of the inner helices. This would represent a gate that is closer to the selectivity filter than the normal helix-bundle crossing gate (Fig. 2A). In MlotiK1, it was clear that this cavity gate and the bundle crossing gate are both closed. However, it is possible that some channels may be able to close the cavity gate without closing the bundle crossing gate. Such a gating mechanism could possibly explain the closed-state accessibility of deep regions of the pore in some channels. For example, in the CNG1 channel, residue Val-391 is positioned on the cytoplasmic end of the cavity and is accessible to small molecules applied to the cytoplasm even in a closed conformation (29). The cavity-gating mechanism that we are hypothesizing here would be compatible with these experimental observations.
Materials and Methods
Protein Purification, Functional Assay, and Crystal Conditions.
MlotiK1 protein was expressed and purified according to a variation of the protocol described previously (8, 9). The rubidium uptake assay was performed as described previously (8, 9). In functional assays, uptake was initially normalized by that mediated by valinomycin in the same sample. A further normalization was performed with all of the samples from a given day. In this second step, samples were normalized to the maximal sample-mediated uptake observed in the day. MlotiK1 crystals grew in sitting drop from a 1:1 mix of protein solution (10 mg/ml protein, 200 μM cAMP, 5 mM lauryl dimethylamide oxide, 150 mM KCl) with 10% Peg 2000, 100 mM trisodium citrate, pH 5.6. KCl was replaced by RbCl or CsCl by size exclusion as needed.
X-Ray Crystallography Methods.
Data were processed by using XDS and MOSFLM in space group R3. Cell dimensions were as follows: (native) 282.7 Å, 282.7 Å, 105.5 Å, 90°, 90°, 120°; (RbCl co-crystal) 282.4 Å, 282.4 Å, 105.3 Å, 90°, 90°, 120°; and (CsCl co-crystal) 284.3 Å, 284.3 Å, 105.8 Å, 90°, 90°, 120°. The initial maps were determined with phases calculated from a molecular replacement (PHASER) solution by using the KcsA structure and refined by averaging with RESOLVE. The final maps were calculated with phases determined by MIR by using RbCl and CsCl soaks; heavy-atom positions were determined from difference maps by using molecular replacement phases. The MIR maps were improved by 4-fold, multidomain averaging in DM and amplitude sharpening. Combination maps using averaged and multimodel phases were used to define regions that do not obey noncrystallographic symmetry. CNS, REFMAC, and TLS were used in the refinement of the final model. Building was performed in O. Figures were made with MOLSCRIPT, POVSCRIPT, and POVRAY. The surface plots were made in PYMOL. Cavity calculations were performed with VOIDOO. Details about the quality of the diffraction data and refined structure are presented in SI Tables 1 and 2.
Supplementary Material
ACKNOWLEDGMENTS.
We thank F. Sigworth, D. Engelman, J. Wang, Y. Xiong, and B. Wimberly for advice and comments and the staff at X12C, X25, and X29 beamlines at the National Synchrotron Light Source and at the Cornell High Energy Synchrotron Source. This work was supported, in part, by an American Heart Association grant-in-aid (to J.H.M.-C.), National Institutes of Health Grant GM068585 (to J.H.M.-C.), and United States Public Health Service Grants GM66145 and GM071590 (to V.M.U.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3BEH).
This article contains supporting information online at www.pnas.org/cgi/content/full/0711533105/DC1.
References
- 1.Hille B. Ion Channels of Excitable Membranes. Sunderland, MA: Sinauer; 2001. [Google Scholar]
- 2.Jan LY, Jan YN. Tracing the roots of ion channels. Cell. 1992;69:715–718. doi: 10.1016/0092-8674(92)90280-p. [DOI] [PubMed] [Google Scholar]
- 3.Sigworth FJ. Voltage gating of ion channels. Q Rev Biophys. 1994;27:1–40. doi: 10.1017/s0033583500002894. [DOI] [PubMed] [Google Scholar]
- 4.Aggarwal SK, MacKinnon R. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 1996;16:1169–1177. doi: 10.1016/s0896-6273(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 5.Seoh SA, Sigg D, Papazian DM, Bezanilla F. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron. 1996;16:1159–1167. doi: 10.1016/s0896-6273(00)80142-7. [DOI] [PubMed] [Google Scholar]
- 6.Tombola F, Pathak MM, Isacoff EY. How far will you go to sense voltage? Neuron. 2005;48:719–725. doi: 10.1016/j.neuron.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 7.Tombola F, Pathak MM, Isacoff EY. How does voltage open an ion channel? Annu Rev Cell Dev Biol. 2006;22:23–52. doi: 10.1146/annurev.cellbio.21.020404.145837. [DOI] [PubMed] [Google Scholar]
- 8.Nimigean CM, Shane T, Miller C. A cyclic nucleotide modulated prokaryotic K+ channel. J Gen Physiol. 2004;124:203–210. doi: 10.1085/jgp.200409133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clayton GM, Silverman WR, Heginbotham L, Morais-Cabral JH. Structural basis of ligand activation in a cyclic nucleotide regulated potassium channel. Cell. 2004;119:615–627. doi: 10.1016/j.cell.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 10.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 11.Long SB, Campbell EB, Mackinnon R. Voltage sensor of Kv1.2: Structural basis of electromechanical coupling. Science. 2005;309:903–908. doi: 10.1126/science.1116270. [DOI] [PubMed] [Google Scholar]
- 12.Doyle DA, et al. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 13.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 14.Lu Z, Klem AM, Ramu Y. Ion conduction pore is conserved among potassium channels. Nature. 2001;413:809–813. doi: 10.1038/35101535. [DOI] [PubMed] [Google Scholar]
- 15.Lu Z, Klem AM, Ramu Y. Coupling between voltage sensors and activation gate in voltage-gated K+ channels. J Gen Physiol. 2002;120:663–676. doi: 10.1085/jgp.20028696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papazian DM, et al. Electrostatic interactions of S4 voltage sensor in Shaker K+ channel. Neuron. 1995;14:1293–1301. doi: 10.1016/0896-6273(95)90276-7. [DOI] [PubMed] [Google Scholar]
- 17.Tiwari-Woodruff SK, Schulteis CT, Mock AF, Papazian DM. Electrostatic interactions between transmembrane segments mediate folding of Shaker K+ channel subunits. Biophys J. 1997;72:1489–1500. doi: 10.1016/S0006-3495(97)78797-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 19.Enkhbayar P, Hikichi K, Osaki M, Kretsinger RH, Matsushima N. 3(10)-helices in proteins are parahelices. Proteins. 2006;64:691–699. doi: 10.1002/prot.21026. [DOI] [PubMed] [Google Scholar]
- 20.Tombola F, Pathak MM, Gorostiza P, Isacoff EY. The twisted ion-permeation pathway of a resting voltage-sensing domain. Nature. 2007;445:546–549. doi: 10.1038/nature05396. [DOI] [PubMed] [Google Scholar]
- 21.Campos FV, Chanda B, Roux B, Bezanilla F. Two atomic constraints unambiguously position the S4 segment relative to S1 and S2 segments in the closed state of Shaker K channel. Proc Natl Acad Sci USA. 2007;104:7904–7909. doi: 10.1073/pnas.0702638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bellanda M, et al. Solvent polarity controls the helical conformation of short peptides rich in Calpha-tetrasubstituted amino acids. Chemistry. 2007;13:407–416. doi: 10.1002/chem.200600719. [DOI] [PubMed] [Google Scholar]
- 23.Dike A, Cowsik SM. Solution structure of amphibian tachykinin Uperolein bound to DPC micelles. J Struct Biol. 2006;156:442–452. doi: 10.1016/j.jsb.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 24.Kitagawa K, Morita T, Kimura S. A helical molecule that exhibits two lengths in response to an applied potential. Angew Chem Int Ed Engl. 2005;44:6330–6333. doi: 10.1002/anie.200502240. [DOI] [PubMed] [Google Scholar]
- 25.Mikhonin AV, Asher SA. Direct UV Raman monitoring of 3(10)-helix and pi-bulge premelting during alpha-helix unfolding. J Am Chem Soc. 2006;128:13789–13795. doi: 10.1021/ja062269+. [DOI] [PubMed] [Google Scholar]
- 26.Gerstein M, Chothia C. Analysis of protein loop closure. Two types of hinges produce one motion in lactate dehydrogenase. J Mol Biol. 1991;220:133–149. doi: 10.1016/0022-2836(91)90387-l. [DOI] [PubMed] [Google Scholar]
- 27.McPhalen CA, Vincent MG, Picot D, Jansonius JN, Lesk AM, Chothia C. Domain closure in mitochondrial aspartate aminotransferase. J Mol Biol. 1992;227:197–213. doi: 10.1016/0022-2836(92)90691-c. [DOI] [PubMed] [Google Scholar]
- 28.Zhou M, Morais-Cabral JH, Mann S, MacKinnon R. Potassium channel receptor site for the inactivation gate and quaternary amine inhibitors. Nature. 2001;411:657–661. doi: 10.1038/35079500. [DOI] [PubMed] [Google Scholar]
- 29.Flynn GE, Zagotta WN. Conformational changes in S6 coupled to the opening of cyclic nucleotide-gated channels. Neuron. 2001;30:689–698. doi: 10.1016/s0896-6273(01)00324-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.