Abstract
Phylogenetic reconstructions of the β-globin gene family in vertebrates have revealed that developmentally regulated systems of hemoglobin synthesis have been reinvented multiple times in independent lineages. For example, the functional differentiation of embryonic and adult β-like globin genes occurred independently in birds and mammals. In both taxa, the embryonic β-globin gene is exclusively expressed in primitive erythroid cells derived from the yolk sac. However, the “ε-globin” gene in birds is not orthologous to the ε-globin gene in mammals, because they are independently derived from lineage-specific duplications of a proto β-globin gene. Here, we report evidence that the early and late expressed β-like globin genes in monotremes and therian mammals (marsupials and placental mammals) are the products of independent duplications of a proto β-globin gene in each of these two lineages. Results of our analysis of genomic sequence data from a large number of vertebrate taxa, including sequence from the recently completed platypus genome, reveal that the ε- and β-globin genes of therian mammals arose via duplication of a proto β-globin gene after the therian/monotreme split. Our analysis of genomic sequence from the platypus also revealed the presence of a duplicate pair of β-like globin genes that originated via duplication of a proto β-globin gene in the monotreme lineage. This discovery provides evidence that, in different lineages of mammals, descendent copies of the same proto β-globin gene may have been independently neofunctionalized to perform physiological tasks associated with oxygen uptake and storage during embryonic development.
Keywords: β-globin gene family, platypus, hemoglobin, gene family evolution, gene duplication
In most vertebrates, developmentally regulated members of the α- and β-globin gene families direct the synthesis of functionally distinct hemoglobin isoforms in embryonic and adult (definitive) erythroid cells. The progenitors of the α- and β-globin gene families originated via tandem duplication of an ancestral, single-copy globin gene ≈450–500 Mya in the common ancestor of gnathostome vertebrates (1–3). The ancestral gene arrangement, consisting of a linked set of α- and β-like globin genes on the same chromosome, has been retained in teleosts such as the zebra fish (Danio rerio; ref. 4) and in amphibians such as Xenopus (5). Because the α- and β-globin gene clusters are located on different chromosomes in birds and mammals, the chromosomal translocation that broke up the ancestral linkage arrangement likely occurred in the common ancestor of these two vertebrate groups (6, 7).
Hemoglobin synthesis is also developmentally regulated in some invertebrates (8), which suggests that the capacity to express functionally distinct hemoglobins at different stages of development may have an ancient evolutionary origin (9–11). However, phylogenetic reconstructions of the β-globin gene family in vertebrates have revealed that developmentally regulated systems of blood oxygen transport have been reinvented multiple times in independent lineages. For example, the functional differentiation of embryonic and adult β-like globin genes occurred independently in birds and mammals. In both taxa, the embryonic β-globin gene is exclusively expressed in primitive erythroid cells derived from the yolk sac. However, the “ε-globin” gene in birds is not orthologous to the ε-globin gene in mammals (2, 12), because they are independently derived from lineage-specific duplications of a proto β-globin gene.
In placental mammals (subclass Eutheria), the β-globin gene cluster includes a linked set of three early expressed (prenatal) genes, ε-γ-η, at the 5′ end of the cluster, and a pair of late expressed (adult) genes, δ and β, at the 3′ end. There is extensive variation in the copy number of these different paralogs among species, and in a number of placental mammal lineages, the η- and δ-globin genes have been inactivated or deleted (13–15). In marsupials (subclass Metatheria), the β-globin gene cluster includes a single pair of genes, the early expressed ε-globin gene and the late expressed β-globin gene (16, 17). An additional β-like globin gene, ω-globin, was recently discovered at the 3′ end of the α-globin gene cluster in marsupials (18–20). The location of this “orphaned” ω-globin gene at the 3′ end of the α-globin gene cluster reflects the ancestral linkage arrangement of α- and β-globin genes before their translocation to different chromosomes. Because the ε- and β-globin genes are the only members of the gene family that are shared between marsupials and placental mammals, this single gene pair may have been inherited from the common ancestor of all mammals. Within the β-globin gene cluster of mammals, conservation of stage-specific expression is seen only for the embryonic ε-globin gene, which is always located at the 5′ end of the gene cluster in the position closest to the locus control region (LCR). The LCR is a cis-regulatory element that is typically located 30–50 kb upstream of the ε-globin gene (21–24). It appears that this proximity to the LCR accounts for the restriction of ε-globin gene expression to embryonic erythroid cells, followed by silencing in the fetal and adult lineage of erythroid cells (25).
In principle, comparative analysis of genomic sequence from the platypus (Ornithorhynchus anatinus) should provide much insight into the early evolution of the β-globin gene cluster in mammals. Because monotremes (subclass Prototheria) diverged from the ancestor of therian mammals (marsupials and placental mammals) between 231 and 217 Mya (26), comparison of the genomic structure of the platypus β-globin gene cluster with that of therian mammals should permit inferences regarding the ancestral state of the gene family at the stem of the mammalian radiation, and comparison with the avian β-globin gene cluster should reveal which genes and genomic features are uniquely mammalian innovations and which were inherited from the mammalian/sauropsid ancestor.
Here, we report evidence that the early and late expressed β-like globin genes in therians and monotremes are the products of independent duplications of a proto β-globin gene in each of these two lineages. Results of our analysis of genomic sequence data from a large number of vertebrate taxa, including sequence from the recently completed platypus genome, reveal that the ε- and β-globin genes of therian mammals arose via duplication of a proto β-globin gene after the therian/monotreme split. Our analysis of genomic sequence from the platypus also revealed the presence of a duplicate pair of β-like globin genes that originated via duplication of a proto β-globin gene in the monotreme lineage. Although the 3′ member of the duplicate gene pair encodes the β-chain subunit of adult hemoglobin (27, 28), the 5′ member of the duplicate gene pair has not been previously characterized. On the basis of positional homology with β-like globin genes of other vertebrates, expression of the 5′ gene is expected to be restricted to embryonic erythroid cells. This discovery provides evidence that, in different lineages of mammals, descendent copies of the same proto β-globin gene may have been independently neofunctionalized to perform physiological tasks associated with oxygen uptake and storage during embryonic development.
Results and Discussion
The goal of this study was to elucidate the early evolution of the β-globin gene cluster in mammals. To do this, we conducted a phylogenetic analysis of 168 β-like globin genes from a diverse array of vertebrate taxa, including cartilaginous and teleost fish, amphibians, reptiles, birds, and each of the three subclasses of mammals (monotremes, marsupials, and placental mammals). Importantly, the set of mammalian sequences in our dataset included β-like globin sequences from two monotremes, the platypus (O. anatinus) and the short-beaked echidna (Tachyglossus aculeatus).
Genomic Structure of the Platypus β-Globin Gene Cluster.
The β-globin gene cluster of the platypus contains a single pair of closely linked genes separated by 11,047 bp [supporting information (SI) Table 1]. We also found a putative β-like globin gene at the 3′ end of the α-globin gene cluster. Each of these three β-like globin genes was characterized by the canonical three exon/two intron structure typical of vertebrate globins.
Although the β-globin gene cluster of the platypus appears superficially similar to the marsupial gene cluster with its simple, two-gene structure (17, 19, 29), below, we report results indicating that the two monotreme genes have originated independently via duplication of a proto β-globin gene.
Phylogenetic Analysis of Coding Sequence.
It is generally thought that the duplication event that gave rise to the ε- and β-globin genes of mammals predated the radiation of therian mammals but occurred after divergence from the sauropsids ≈310 Mya (2, 29). Under this scenario, we would expect to recover a phylogenetic tree in which the monotreme 5′ β-like globin gene is placed sister to the early expressed genes of therian mammals and the monotreme 3′ gene is placed sister to the late expressed genes of therian mammals. Contrary to this expectation, our phylogeny reconstruction recovered a monophyletic clade containing all β-like globin genes of monotremes (the 5′ and 3′ genes of the platypus, Ornithorhyncus, and the 3′ gene of the echidna, Tachyglossus; Fig. 1). The surprising implication of this finding is that the pair of β-like globin genes in monotremes must have originated via duplication of a proto β-globin gene after the monotreme/therian split. Previous phylogenetic reconstructions have suggested the possibility of a monotreme-specific duplication, but taxon sampling was not sufficient to draw a definitive conclusion (3). The sister relationship between the 3′ β-like globin genes of platypus and echidna (Fig. 1) indicates that the monotreme-specific duplication occurred after divergence from the common ancestor of therian mammals. Consequently, the 5′ gene of monotremes is not a 1:1 ortholog of the early expressed therian ε-globin gene, and the 3′ gene of monotremes is not a 1:1 ortholog of the late expressed therian β-globin gene.
Fig. 1.
Bayesian phylogram describing relationships among the β-like globin genes of vertebrates. The β-globin sequences from spiny dogfish (S. acanthias) and arctic skate (A. hyperborea) were used as outgroups. Values on the nodes correspond to Bayesian posterior probabilities.
Topology Test.
To assess statistical support for our inferred scenario involving two independent duplication events in monotremes and therian mammals, we conducted a pair of topology tests in which we compared our Bayesian tree (Fig. 1) to the topology predicted under the single-duplication model and to the topology predicted under the two-duplication model. Under the single-duplication model, the platypus 5′ β-like globin gene is placed sister to the early expressed genes of therian mammals, and the monotreme (platypus and echidna) 3′ β-like globin genes are placed sister to the late expressed genes of therian mammals (SI Fig. 5A). Under the two-duplication model, the monophyletic clade containing all monotreme sequences is placed sister to all therian β-like globin genes (SI Fig. 5B). Results of the approximately unbiased topology test rejected the topology predicted by the single-duplication model (P = 0.022) but failed to reject the topology predicted by the two-duplication model (P = 0.310). This test result bolsters our initial conclusion that the 5′ and 3′ β-like globin genes of monotremes are the products of a lineage-specific duplication event that was distinct from the duplication event that gave rise to the ε- and β-globin genes of therian mammals.
Analysis of Flanking Sequences in Monotremes and Marsupials.
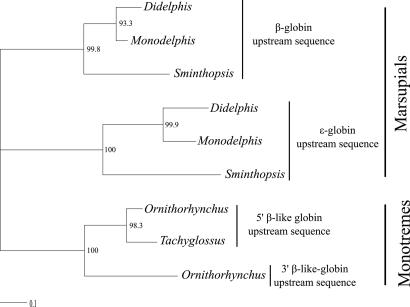
Phylogenetic analyses of multigene families often reveal cases in which paralogous genes from the same species are more similar to each other than they are to their orthologs in closely related species. This pattern is typically attributable to (i) concerted evolution between preexisting gene duplicates that were inherited from a common ancestor or (ii) recent ancestry between the daughter copies of a lineage-specific duplication event. Because mechanisms of concerted evolution such as interparalog gene conversion are often largely restricted to coding regions of globin genes (14, 30), the cause of sequence similarity between tandemly duplicated genes from the same species can often be determined by analyzing variation in flanking sequence (31–33). Accordingly, we conducted a phylogenetic analysis by using a 1-kb alignment of upstream flanking sequence for the 5′ and 3′ β-like globin genes of monotremes and upstream flanking sequence for the ε- and β-globin genes of marsupials. As shown in Fig. 2, our phylogeny reconstruction revealed three monophyletic clades with strong bootstrap support: (i) flanking regions of the marsupial ε-globin genes; (ii) flanking regions of the marsupial β-globin genes; and (iii) flanking regions of the monotreme 5′ and 3′ genes. In the case of the marsupials, phylogenetic analysis of ε- and β-globin flanking sequences recovered the same set of species relationships: [Sminthopsis (Didelphis, Monodelphis)]. In the case of the monotreme sequences, the flanking region of the platypus 3′ gene was placed sister to flanking regions of the platypus and echidna 5′ genes. The fact that each paralogous clade recovered the known species-level phylogeny indicates that the β-like globin genes have not been evolving in concert, and the fact that flanking sequences of the monotreme 5′ and 3′ genes are monophyletic relative to flanking sequences of the ε- and β-globin genes from therian mammals provides further evidence for an independent duplication of a proto β-globin gene in the monotreme lineage.
Fig. 2.
Maximum likelihood phylogram based on 1 kb of flanking sequence of β-like globin genes in two monotreme species, the platypus (O. anatinus) and the echidna (T. aculeatus), and three marsupial species, the North American opossum (Didelphis virginiana), the gray short-tailed opossum (Monodelphis domestica), and the stripe-faced dunnart (Sminthopsis macroura).
Interparalog Divergence.
To further investigate whether diversification of the β-globin gene cluster in monotremes and therian mammals was attributable to two separate lineage-specific duplication events, we compared the level of interparalog divergence between the ε- and β-globin genes of marsupials with the level of interparalog divergence between the 5′ and 3′ β-like globin genes of the platypus. For the platypus and each of the three marsupial species for which genomic sequence was available, we estimated interparalog divergence at third codon positions by using MEGA v3.1 (34). If monotremes and marsupials inherited the same pair of β-like globin genes from a common ancestor, then levels of interparalog divergence should be similar in each taxon. In contrast to this expectation, we found that levels of interparalog divergence in marsupials (range = 39.86% ± 3.95% to 43.24% ± 3.99%), were substantially higher than the level of divergence between the 5′ and 3′ genes of the platypus (21.62% ± 3.36%; SI Fig. 6). Given that we detected no evidence of interparalog gene conversion in either monotremes or marsupials (see above), the lower level of interparalog divergence in the platypus suggests that the 5′ and 3′ genes are the products of a more recent duplication event that was specific to the monotreme lineage. Similar results were obtained when estimates of interparalog divergence were based on first and second codon positions (data not shown).
Genomic Comparison of the β-Globin Gene Clusters in Monotremes and Marsupials.
The availability of genomic data allowed us to make comparisons involving sequence from flanking chromosomal regions in addition to coding sequence. In principle, comparison of the complete β-globin gene cluster of monotremes and marsupials should allow us to delineate boundaries of the duplication blocks in both groups. Dot-plot comparisons revealed different boundaries of the duplication blocks in monotremes and marsupials, suggesting that the tandem duplication that gave rise to the 5′ and 3′ genes of monotremes was distinct from the tandem duplication that gave rise to the ε- and β-globin genes of marsupials. In marsupials, the inferred boundaries of the duplication block are located immediately outside of the coding region, as indicated by locally alignable regions of high sequence identity that do not extend >300 bp into the upstream and downstream flanking regions (Fig. 3). In the platypus, by contrast, the duplication block appears to be substantially larger, because locally alignable regions of high sequence identity extend much further into the flanking regions (Fig. 3).
Fig. 3.
Dot plots of interparalog sequence identity between the 5′ and 3′ β-like globin genes of the platypus (O. anatinus) and between the ε- and β-globin genes of three marsupial species, the North American opossum (D. virginiana), the gray short-tailed opossum (M. domestica), and the stripe-faced dunnart (S. macroura). Light blue and light yellow vertical lines denote exons and introns, respectively, in the sequence aligned on the x axis. In each of the four interparalog comparisons, dot plots were based on the complete coding region in addition to 2 kb of upstream flanking sequence and 2 kb of downstream flanking sequence.
Presence of an ω-Globin Gene in Monotremes.
Consistent with previous studies of marsupials (20, 35), our analysis of genomic sequence from the platypus revealed a single-copy ω-globin gene that was located at the 3′ end of the α-globin gene cluster (SI Table 1). Our phylogeny reconstruction revealed a monophyletic clade of ω-globin sequences from monotremes and marsupials (Fig. 1). In this clade, the platypus ω-globin gene was sister to all remaining marsupial sequences, suggesting that this gene is a 1:1 ortholog of the marsupial ω-globin gene.
Evolution of the Mammalian β-Globin Gene Cluster.
We have presented several independent lines of evidence demonstrating that a developmentally regulated system of hemoglobin synthesis has been reinvented in monotremes and therian mammals. This surprising finding allows us to refine the current model for the evolution of the β-globin gene family in mammals.
In addition to the evidence for independent duplications of a β-globin pro-ortholog in the therian and monotreme lineages, our phylogeny reconstruction also indicates that the embryonic ε-globin genes of marsupials are orthologous to the early expressed genes in placental mammals, as suggested by Goodman et al. (2) and Koop and Goodman (29). This finding suggests that the duplication of a proto β-globin gene occurred after the monotreme/therian split but before the marsupial/eutherian split. Contrary to previous studies that suggested that a four-gene set of β-like globin genes (5′-ε-γ-δ-β-3′) was present in the common ancestor of all mammals (15), our phylogeny reconstruction indicates that the γ- and δ-globin genes are uniquely eutherian innovations: the early expressed γ-globin gene originated via duplication of the ε-globin gene, and the late expressed δ-globin gene originated via duplication of the β-globin gene. Our results indicate that both of these duplication events occurred after the marsupial/eutherian split.
Finally, consistent with the findings of Aguileta et al. (36), our results also indicate that avian β-like globin genes are not orthologous to the ω-globins of marsupials and monotremes, as was previously suggested by Wheeler et al. (19, 20, 36).
Our revised model for the early evolution of the β-globin gene cluster in vertebrates is graphically summarized in Fig. 4. According to this model, the ω-globin gene originated via duplication of an ancient β-globin gene that predated the divergence of birds and mammals but occurred after the amniote/amphibian split. The ω-globin gene has been retained in contemporary monotremes and marsupials, but it has been lost independently in birds and placental mammals. In the common ancestor of marsupials and placental mammals, a pair of ε- and β-globin genes originated via duplication of a proto β-globin gene after the therian/monotreme split. In the placental mammal lineage, subsequent duplications of the ε- and β-globin genes gave rise to the prenatally expressed γ-globin and the adult-expressed δ-globin, respectively. In the monotreme lineage, a pair of β-like globin genes (the 5′ and 3′ genes) originated via duplication of a proto β-globin gene sometime before the divergence of the platypus and echidna. The 3′ β-like globin gene of monotremes is expressed during adulthood, because conceptual translations of the platypus and echidna genes are 100% identical to the reported β-chain amino acid sequences of adult hemoglobin from each of the respective species (27, 28). On the basis of positional homology with other β-like globin genes, expression of the 5′ β-like globin gene is most likely restricted to embryonic erythroid cells. Despite this probable functional analogy in terms of timing of expression, the monotreme 5′ β-like globin gene is not a 1:1 ortholog of the therian ε-globin gene and the 3′ β-like globin gene of monotremes is not a 1:1 ortholog of the therian β-globin gene, because the two gene pairs derive from independent duplication events. In Figs. 1 and 4, we refer to the 5′ and 3′ β-like globin genes of monotremes as “εP-globin” and “βP-globin,” respectively, where the “P” superscript stands for subclass Prototheria. We use the “P” superscript to acknowledge that these genes are not 1:1 orthologs of the ε- and β-globin genes of therian mammals.
Fig. 4.
An evolutionary hypothesis regarding the evolution of the β-globin gene family. According to this model, the ω-globin gene originated via duplication of an ancient β-globin gene that occurred before the divergence of birds and mammals but after the amniote/amphibian split. The ω-globin gene has been retained in contemporary monotremes and marsupials, but it has been lost independently in birds and placental mammals. In the common ancestor of marsupials and placental mammals, a pair of ε- and β-globin genes originated via duplication of a proto β-globin gene after the therian/monotreme split. In the placental mammal lineage, subsequent duplications of the ε- and β-globin genes gave rise to the prenatally expressed γ-globin and the adult-expressed δ-globin, respectively. In the monotreme lineage, a pair of β-like globin genes (εP- and βP-globin) originated via duplication of a proto β-globin gene sometime before the divergence of the platypus and echidnas (the two monotreme lineages). The βP-globin gene is expressed during adulthood, and, on the basis of positional homology with other β-like globin genes, expression of the εP-globin gene is most likely restricted to embryonic erythroid cells.
In summary, we have demonstrated that the β-like globin genes of monotremes and therian mammals originated independently via lineage-specific duplication events. Additional functional experiments are required to test whether the εP- and βP-globin genes of monotremes are developmentally regulated in the same fashion as the embryonic and adult β-like globin genes of therian mammals. If this proves to be the case, then it will also be important to assess whether the reinvention of a developmentally regulated system of hemoglobin synthesis entailed parallel or convergent evolution of protein function and stage-specific transcriptional regulation.
Materials and Methods
DNA Sequence Data.
We obtained genomic DNA sequences for structural genes in the β-globin gene family from the High Throughput Genomic Sequences database (HTGS). All of the genomic sequences analyzed in this study were in phase 2, meaning that the order and orientation of the constituent sequence contigs had been firmly established. We characterized the genomic structure of the β-globin gene cluster in 36 mammalian species, 1 bird species, and 1 amphibian species. We also included sequences from shorter records based on genomic DNA or cDNA to attain a broad and balanced taxonomic coverage of β-like globin gene sequences. This approach allowed us to include sequences from fish (Danio rerio), amphibians (Xenopus laevis and Rana castebeina), reptiles (Geochelone chilensis, G. carbonaria, and Alligator mississipiensis), birds (Cairina and Taeniopygia), and some additional mammalian taxa (SI Table 2). The β-globin sequences from spiny dogfish (Squalus acanthias) and arctic skate (Amblyraja hyperborea) were used as outgroups. Our final dataset consisted of a 468-bp alignment of coding sequence from 168 β-like globin genes.
We identified globin genes in unannotated genomic sequences by using the program Genscan (37) and by comparing known exon sequences to genomic contigs by using the program BLAST 2, version 2.2 (38), available from the National Center for Biotechnology Information web site (www.ncbi.nlm.nih.gov/blast/bl2seq). Sequence alignment was carried out by using the program MUSCLE (39) as implemented in the Berkeley Phylogenomics group web server (http://phylogenomics.berkeley.edu), with manual adjustments performed to keep coding sequences in frame.
Phylogenetic Analyses.
We estimated phylogenetic relationships among the different β-like globin DNA sequences in our dataset by using a Bayesian approach as implemented in Mr. Bayes v3.1.2 (40). The program was used to simultaneously estimate the tree topology and parameter values for an independent GTR+Γ+I model of nucleotide substitution for each codon position. Two simultaneous independent runs were performed for 30 × 106 iterations of a Markov Chain Monte Carlo algorithm, with eight simultaneous chains, sampling every 1,000 generations. Support for the nodes and parameter estimates were derived from a majority rule consensus of the last 10,000 trees sampled after convergence. The average standard deviation of split frequencies remained <0.01 after the burn-in threshold. Topology tests were performed by using the approximately unbiased test (41), as implemented in the program TreeFinder, version June 2007 (42).
Supplementary Material
ACKNOWLEDGMENTS.
We thank two anonymous reviewers for helpful comments and suggestions. This work was funded by a Postdoctoral Fellowship in Population Biology (to F.G.H.) from the University of Nebraska, National Science Foundation Grant DEB-0614342 (to J.F.S.), a Layman Award (to J.F.S.), and an Interdisciplinary Research Grant (to J.F.S.) from the Nebraska Research Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710531105/DC1.
References
- 1.Czelusniak J, Goodman M, Hewett-Emmett D, Weiss ML, Venta PJ, Tashian RE. Nature. 1982;298:297–300. doi: 10.1038/298297a0. [DOI] [PubMed] [Google Scholar]
- 2.Goodman M, Czelusniak J, Koop BF, Tagle DA, Slightom JL. Cold Spring Harb Symp Quant Biol. 1987;52:875–890. doi: 10.1101/sqb.1987.052.01.096. [DOI] [PubMed] [Google Scholar]
- 3.Goodman M, Moore GW, Matsuda G. Nature. 1975;253:603–608. doi: 10.1038/253603a0. [DOI] [PubMed] [Google Scholar]
- 4.Chan FY, Robinson J, Brownlie A, Shivdasani RA, Donovan A, Brugnara C, Kim J, Lau BC, Witkowska HE, Zon LI. Blood. 1997;89:688–700. [PubMed] [Google Scholar]
- 5.Hosbach HA, Wyler T, Weber R. Cell. 1983;32:45–53. doi: 10.1016/0092-8674(83)90495-6. [DOI] [PubMed] [Google Scholar]
- 6.Deisseroth A, Velez R, Nienhuis AW. Science. 1976;191:1262–1264. doi: 10.1126/science.943846. [DOI] [PubMed] [Google Scholar]
- 7.Hughes SH, Stubblefield E, Payvar F, Engel JD, Dodgson JB, Spector D, Cordell B, Schimke RT, Varmus HE. Proc Natl Acad Sci USA. 1979;76:1348–1352. doi: 10.1073/pnas.76.3.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Terwilliger NB. J Exp Biol. 1998;201:1085–1098. doi: 10.1242/jeb.201.8.1085. [DOI] [PubMed] [Google Scholar]
- 9.Hardison R. J Exp Biol. 1998;201:1099–1117. doi: 10.1242/jeb.201.8.1099. [DOI] [PubMed] [Google Scholar]
- 10.Hardison R. In: Disorders of Hemoglobin: Genetics, Pathophysiology, and Clinical Management. Steinberg M, Forget B, Higgs D, Nagel R, editors. Cambridge, UK: Cambridge Univ Press; 2001. pp. 95–115. [Google Scholar]
- 11.Hardison RC. Proc Natl Acad Sci USA. 1996;93:5675–5679. doi: 10.1073/pnas.93.12.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reitman M, Grasso JA, Blumenthal R, Lewit P. Genomics. 1993;18:616–626. doi: 10.1016/s0888-7543(05)80364-7. [DOI] [PubMed] [Google Scholar]
- 13.Goodman M, Koop BF, Czelusniak J, Weiss ML. J Mol Biol. 1984;180:803–823. doi: 10.1016/0022-2836(84)90258-4. [DOI] [PubMed] [Google Scholar]
- 14.Hardies SC, Edgell MH, Hutchison CA., III J Biol Chem. 1984;259:3748–3756. [PubMed] [Google Scholar]
- 15.Hardison RC. Mol Biol Evol. 1984;1:390–410. doi: 10.1093/oxfordjournals.molbev.a040326. [DOI] [PubMed] [Google Scholar]
- 16.Cooper SJ, Hope RM. Proc Natl Acad Sci USA. 1993;90:11777–11781. doi: 10.1073/pnas.90.24.11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper SJ, Murphy R, Dolman G, Hussey D, Hope RM. Mol Biol Evol. 1996;13:1012–1022. doi: 10.1093/oxfordjournals.molbev.a025651. [DOI] [PubMed] [Google Scholar]
- 18.De Leo AA, Wheeler D, Lefevre C, Cheng JF, Hope R, Kuliwaba J, Nicholas KR, Westerman M, Graves JA. Cytogenet Genome Res. 2005;108:333–341. doi: 10.1159/000081528. [DOI] [PubMed] [Google Scholar]
- 19.Wheeler D, Hope R, Cooper SB, Dolman G, Webb GC, Bottema CD, Gooley AA, Goodman M, Holland RA. Proc Natl Acad Sci USA. 2001;98:1101–1106. doi: 10.1073/pnas.98.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wheeler D, Hope RM, Cooper SJ, Gooley AA, Holland RA. J Mol Evol. 2004;58:642–652. doi: 10.1007/s00239-004-2584-0. [DOI] [PubMed] [Google Scholar]
- 21.Forrester WC, Takegawa S, Papayannopoulou T, Stamatoyannopoulos G, Groudine M. Nucleic Acids Res. 1987;15:10159–10177. doi: 10.1093/nar/15.24.10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forrester WC, Thompson C, Elder JT, Groudine M. Proc Natl Acad Sci USA. 1986;83:1359–1363. doi: 10.1073/pnas.83.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardison R, Slightom JL, Gumucio DL, Goodman M, Stojanovic N, Miller W. Gene. 1997;205:73–94. doi: 10.1016/s0378-1119(97)00474-5. [DOI] [PubMed] [Google Scholar]
- 24.Tuan DY, Solomon WB, London IM, Lee DP. Proc Natl Acad Sci USA. 1989;86:2554–2558. doi: 10.1073/pnas.86.8.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson RM, Prychitko T, Gumucio D, Wildman DE, Uddin M, Goodman M. Proc Natl Acad Sci USA. 2006;103:3186–3191. doi: 10.1073/pnas.0511347103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Rheede T, Bastiaans T, Boone DN, Hedges SB, de Jong WW, Madsen O. Mol Biol Evol. 2006;23:587–597. doi: 10.1093/molbev/msj064. [DOI] [PubMed] [Google Scholar]
- 27.Thompson EO, Fisher WK, Whittaker RG. Aust J Biol Sci. 1973;26:1327–1335. doi: 10.1071/bi9731327. [DOI] [PubMed] [Google Scholar]
- 28.Whittaker RG, Thompson EO. Aust J Biol Sci. 1975;28:353–365. doi: 10.1071/bi9750353. [DOI] [PubMed] [Google Scholar]
- 29.Koop BF, Goodman M. Proc Natl Acad Sci USA. 1988;85:3893–3897. doi: 10.1073/pnas.85.11.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam KW, Jeffreys AJ. Proc Natl Acad Sci USA. 2006;103:8921–8927. doi: 10.1073/pnas.0602690103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardison R, Miller W. Mol Biol Evol. 1993;10:73–102. doi: 10.1093/oxfordjournals.molbev.a039991. [DOI] [PubMed] [Google Scholar]
- 32.Hardison RC, Gelinas RE. Mol Biol Evol. 1986;3:243–261. doi: 10.1093/oxfordjournals.molbev.a040392. [DOI] [PubMed] [Google Scholar]
- 33.Storz JF, Baze M, Waite JL, Hoffmann FG, Opazo JC, Hayes JP. Genetics. 2007;177:481–500. doi: 10.1534/genetics.107.078550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura K, Dudley J, Nei M, Kumar S. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 35.Cooper SJ, Wheeler D, De Leo A, Cheng JF, Holland RA, Marshall Graves JA, Hope RM. Mol Phylogenet Evol. 2006;38:439–448. doi: 10.1016/j.ympev.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 36.Aguileta G, Bielawski JP, Yang Z. Gene. 2006;380:21–29. doi: 10.1016/j.gene.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 37.Burge C, Karlin S. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 38.Tatusova TA, Madden TL. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 39.Edgar RC. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ronquist F, Huelsenbeck JP. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 41.Shimodaira H. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- 42.Jobb G, von Haeseler A, Strimmer K. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.