Abstract
In all eukaryotic cells, origins of DNA replication are characterized by the binding of the origin recognition complex (ORC). How ORC is positioned to sites where replication initiates is unknown, because metazoan ORC binds DNA without apparent sequence specificity. Thus, additional factors might be involved in ORC positioning. Our experiments indicate that a family member of the high-mobility group proteins, HMGA1a, can specifically target ORC to DNA. Coimmunoprecipitations and imaging studies demonstrate that HMGA1a interacts with different ORC subunits in vitro and in vivo. This interaction occurs mainly in AT-rich heterochromatic regions to which HMGA1a localizes. Fusion proteins of HMGA1a and the DNA-binding domain of the viral factor EBNA1 or the prokaryotic tetracycline repressor, TetR, can recruit ORC to cognate operator sites forming functional origins of DNA replication. When HMGA1a is targeted to plasmid DNA, the prereplicative complex is assembled during G1 and the amount of ORC correlates with the local concentration of HMGA1a. Nascent-strand abundance assays demonstrate that DNA replication initiates at or near HMGA1a-rich sites. Our experiments indicate that chromatin proteins can target ORC to DNA, suggesting they might specify origins of DNA replication in metazoan cells.
Keywords: chromatin, DNA replication, EBNA1, Epstein–Barr virus, Orc6
Eukaryotic cells duplicate their genomes with remarkable precision and in a timely coordinated fashion. The process of DNA replication initiates at multiple origins of replication, which are recognized by the heterohexameric origin recognition complex (ORC), consisting of Orc1–6 (1). Human ORC is a dynamic cell cycle-regulated complex and can be regarded as an interactive platform for the assembly of the prereplicative complex (preRC), consisting of Orc1–6, Cdt1, Cdc6, and the MCM2–7 complex (1). The assembly of preRCs licenses origins for replication initiation. The human ORC subunits Orc2–5 form a stable core complex, whereas the association of the largest subunit Orc1 is cell-cycle-regulated (1). Biochemical studies indicate that the smallest subunit Orc6 is only loosely attached, and the existence of a hexameric holocomplex has only recently been postulated in human cells (2). ORC and most other proteins involved in initiation of DNA replication are conserved among eukaryotes, but specification of origins in mammalian cells remains elusive and is controversially discussed (3, 4).
How metazoan ORC recognizes origins is unknown, because ORC does not bind to DNA sequence specifically (3–5). The positioning of ORC at origins might be determined by veiled DNA sequence motifs, local chromatin structures, or accessory targeting factors such as AIF-C, Trf2, Ku80, or EBNA1, which can specify sites of ORC binding (6–10), and recently a direct role of Myc in replication initiation has been suggested (11). In Schizosaccharomyces pombe (Sp), origins contain large stretches of AT-rich sequences, and SpORC is recruited to these origins via the SpOrc4 subunit. It contains a unique N-terminal extension with nine AT-hook motifs, which meditate origin binding (12). In metazoan cells, AT-hook motifs are a hallmark of the HMGA family of high-mobility group (HMG) proteins. One member, HMGA1a, binds with high specificity to the minor groove of AT tracks and induces conformational changes (13). It is known as a structural nonhistone chromatin constituent that competes and antagonizes histone H1-mediated repression of genes (14), thus contributing to cell proliferation (15–17). However, no direct role in origin definition or DNA replication has been ascribed to HMGA proteins.
In this study, we describe a functional interaction between HMGA1a and ORC. Targeting HMGA1a to specific sites on plasmid DNA recruits ORC and generates artificial origins of DNA replication. Replication initiates at or in close vicinity of HMGA1a-rich sites, and preRCs form at these loci during the G1 phase of the cell cycle. We demonstrate that HMGA1a and ORC directly interact in vivo and in vitro. An HMGA1a variant with mutated AT-hook motifs and competition experiments with Hoechst 33342 both indicate that this interaction occurs mainly in AT-rich chromatin domains. Our data suggest that genuine chromatin proteins might contribute to the specification of chromosomal origins of DNA replication and their recognition by ORC at the molecular level.
Results
HMGA1a Supports Replication When Targeted to Plasmid DNA.
Studying mammalian origins of DNA replication in the context of chromosomal DNA is difficult because of the scarcity of tractable model systems. Therefore, we concentrated on plasmid models, which can be addressed genetically and biochemically. We took advantage of the latent origin of Epstein–Barr virus (EBV), oriP. It mediates extrachromosomal replication of EBV genomes and has a modular bipartite structure. The family of repeats (FR) binds the EBV-encoded protein EBNA1 (Fig. 1A) and tethers oriP to chromatin mediating replication-independent nuclear retention and long-term plasmid stability. The dyad symmetry element (DS) acts as a bona fide eukaryotic replication origin (18). Deleting DS results in a replication-defective oriP mutant, indicating that EBNA1, when bound to FR, does not contribute to oriP replication (19). The transactivation domain of EBNA1 can be functionally substituted by HMGA1a (Fig. 1A) and HMGA1a:EBNA1-DBD confers replication competence of oriP plasmids (20, 21) and EBV genomes (22). Thus, we asked whether HMGA1a could recruit ORC to DNA and thereby contribute to the molecular definition of a replication origin. Transient replication assays with an oriP reporter were performed in two HEK293 cell lines, which express EBNA1 or HMGA1a:EBNA1-DBD (Fig. 1A). Two to three weeks after transfection and selection, low-molecular-weight DNA was isolated and digested with DpnI to frequently cleave nonreplicated plasmid DNA, which had retained the dam methylation pattern acquired in the prokaryotic host. Escherichia coli cells were transformed with 500 ng of DNA, and ampicillin-resistant colonies were determined. Similar colony numbers were obtained with DNA from cells expressing EBNA1 or HMGA1a:EBNA1-DBD (Fig. 1A). Parental HEK293 cells or HEK293 cells expressing the DNA-binding domain of EBNA1, only, did not give rise to colonies (data not shown), confirming the function of HMGA1a in this system (20). ChIP experiments in synchronized HEK293 derivatives [supporting information (SI) Fig. 5] clearly indicated that HMGA1a:EBNA1-DBD mediates DNA replication of oriP in an ORC-dependent manner in synchrony with the cell cycle as expected (18).
Fig. 1.
HMGA1a supports plasmid replication and interacts with ORC. (A) Design of the transacting factors HMGA1a:EBNA1-DBD and EBNA1. HMGA1a (green) was fused to the DNA binding and dimerization domain (EBNA1-DBD, gray) and nuclear localization signal (black) of EBNA1. AT hooks are depicted in blue, HMGA1a's acidic domain in orange. The N-terminus of EBNA1 contains two linking domains [light red and green (LR1 and LR2)]. Each LR comprises a Gly-Arg-repeat. HEK293 cells (293) and derivatives, expressing either EBNA1 (EBNA1) or HMGA1a fused to EBNA1-DBD (HMGA1a:EBNA1-DBD), were transfected with the oriP plasmid shown. After selection, low-molecular-weight DNA was isolated and digested with DpnI. E. coli DH10B cells were electroporated with 500 ng of DNA, and ampicillin-resistant colonies of three independent experiments were counted. Mean and standard deviations are provided. (B) HMGA1a interacts with ORC subunits. For affinity purification experiments, HMGA1a:EBNA1-DBD with a C-terminal Strep-tagII was stably introduced in HEK293 cells. Pull-down experiments with nuclear protein of 2 × 107 cells indicate an interaction of HMGA1a:EBNA1-DBD and different ORC subunits (lane 6). Western blots of nuclear extracts of different cell numbers are shown in lanes 1–3. An Orc2-specific antibody was used to coprecipitate HMGA1a:EBNA1-DBD from 2 × 107 cells in lanes 8–10, which was detected with an EBNA1-specific antibody. Protein G Sepharose beads were eluted with 5% N-Lauroylsarcosine, which preferentially released HMGA1:EBNA1-DBD but not Orc2 as shown in lane 8. Remaining Orc2 was eluted with Laemmli buffer (IP; lane 9). The unbound fraction of the protein lysate is shown in lane 10 (unbd). An isotype antibody did not precipitate HMGA1a:EBNA1 (lane 7). (C) Endogenous Orc6 and HMGA1a coprecipitate (Left) and interact directly in the absence of DNA (Right). Nuclear extracts of 2 × 107 HeLa cells were precipitated with an HMGA1a-specific rabbit antibody or an isotype control. The former coprecipitated Orc6 (Left). Nuclear extracts of 2 × 104 and 2 × 105 cells are shown for comparison. In pull-down experiments, recombinant His-tagged Orc6 (purified from Baculovirus-infected insect cells) was coprecipitated with bacterially expressed and highly purified Strep-tagged HMGA1a protein (10 μg each) immobilized to Strep-Tactin (IBA) (Right). Ten and one nanograms of recombinant Orc6 protein samples are shown for comparison.
HMGA1a and ORC Associate.
The replication functions of EBNA1 depend on the interaction between this viral protein and ORC (7, 9). To assess whether HMGA1a:EBNA1-DBD interacted with ORC, we generated a HEK293 cell line with a Strep-tagged version of HMGA1a:EBNA1-DBD. Pull-down experiments indicated that it coprecipitated the four ORC subunits Orc1, 2, 4, and 6 (Fig. 1B, lane 6), suggesting that HMGA1a interacts with the entire ORC holocomplex. Albeit the interaction between HMGA1a:DBD-Strep and different ORC subunits was weak, it depended on HMGA1a, because the DNA-binding domain of EBNA1 alone, EBNA1-DBD, did not precipitate ORC subunits (data not shown). Conversely, an Orc2-specific antibody coprecipitated HMGA1a:EBNA1 (Fig. 1B, lane 8). Endogenous HMGA1a interacts with the Orc6 component of ORC (Fig. 1C Left and SI Fig. 6), as shown by coimmunoprecipitation with an HMGA1a-specific antibody but not an isotype control. Pull-down experiments with recombinant HMGA1a and Orc6 proteins indicate this interaction is direct and independent of DNA (Fig. 1C Right). Despite the relatively weak coprecipitation of ORC and HMGA1a:EBNA1-DBD, transient replication (Fig. 1A) and ChIP experiments (SI Fig. 5) provided clear evidence that the HMGA1a fusion could mediate specific binding of ORC, generating a functional origin of DNA replication.
HMGA1a and ORC Interact in AT-Rich Heterochromatin in Living Cells.
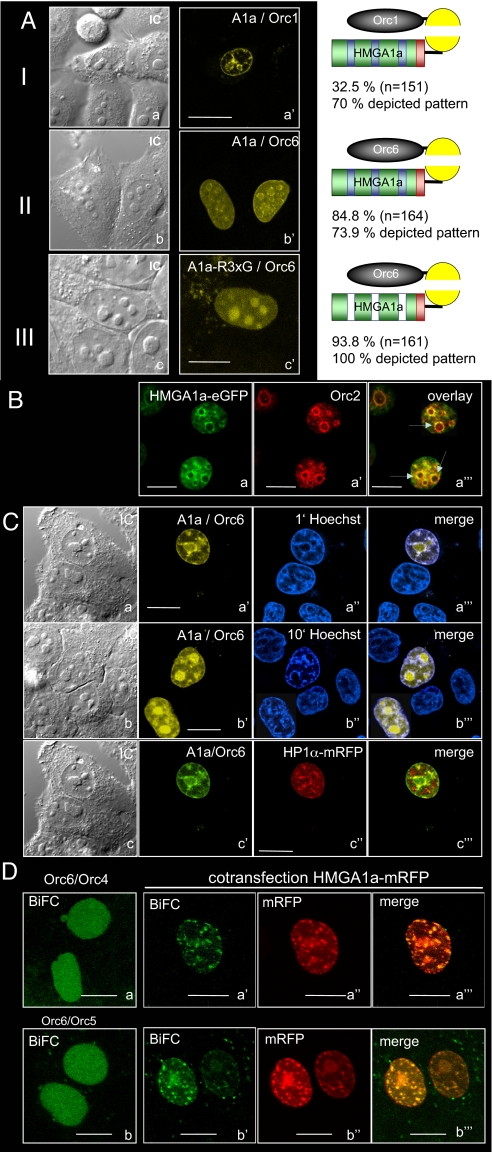
To explore the character of the HMGA1a-ORC interaction in living cells, we used bimolecular fluorescent complementation (BiFC), which visualizes protein–protein interactions (23). HMGA1a was fused to the C-terminal fragment of YFP and YFP's N-terminal part to different ORC subunits. We observed fluorescence complementation in living and fixed cells in the combination of HMGA1a and Orc1 and HMGA1 and Orc6 (Fig. 2A and SI Fig. 7) but not with other ORC subunits (SI Table 1). Seventy percent of Orc1:HMGA1a BiFC-positive cells and 73.9% of Orc6:HMGA1a BiFC-positive cells displayed interactions at the nuclear periphery and perinucleolar regions (Fig. 2A a and b; for detailed numbers and intensity profiles, see SI Fig. 8). In living cells, the BiFC efficiencies between HMGA1a and Orc1 or Orc6 were 32.5% and 84.8%, respectively (Fig. 2A). Orc1 is cell cycle-regulated and becomes unstable after entry into S phase, which might explain the lower efficiency of the Orc1:HMGA1a BiFC complex.
Fig. 2.
BiFC analysis and colocalization of HMGA1a and Orc2. Fluorescence images of HepG2 cells expressing the indicated proteins were acquired 24 h after DNA transfection. (A) Analysis of interaction of HMGA1a with Orc1 and Orc6. HMGA1a fused to the N-terminal domain of YFP was cotransfected with Orc1 (I) and Orc6 (II) linked to the C-terminal fragment of YFP. (III) An HMGA1a variant with mutated AT-hooks (R3xG) was cotransfected with the Orc6-BiFC construct. To calculate BiFC efficiencies, cells were cotransfected with BiFC vectors and an mRFP-expression vector. For example, for Orc1, 32.5% of all cotransfected cells (n = 151) showed BiFC and 70% of BiFC-positive cells showed the depicted pattern. For intensity profiles, see SI Figs. 8 and 9. (B) Colocalization of Orc2 and HMGA1a-eGFP was visualized in fixed HepG2 cells by immunofluorescence using an Orc2-specific antibody. HMGA1a-eGFP and Orc2 colocalize especially in perinucleolar regions (arrows). (C) HMGA1a and Orc6 BiFC experiments in the presence of 5 μg/ml Hoechst 33342 incubated for 1 (a) and 10 min (b). (Scale bars: 10 μm.) (c) Signal complexes of HMGA1a/Orc6 BiFC- and HP1α-mRFP partially overlap. Orc6 and HMGA1a-BiFC plasmids were cotransfected with an HP1α-mRFP expression plasmid (see also SI Fig. 9D). To increase contrast, the yellow BiFC signal was changed to green. (D) Orc4/Orc6 and Orc5/Orc6 BIFC-signals altered after overexpression of HMGA1a-mRFP. Orc4/Orc6 (a) and Orc5/Orc6 were cotransfected into HepG2-cells. Cotransfection of the indicated BiFC-plasmids (a′ and b′) and HMGA1a-mRFP (a″ and b″) resulted in a significant relocalization of ORC (a‴ and b‴). In A–C, Left shows interference contrast figures of the transfected cells. (Scale bars: 10 μm.)
Our coimmunoprecipitation experiments indicated that HMGA1a interacts with ORC. To validate whether fluorescence complementation between Orc components and HMGA1a (Figs. 1 B and C and 2A) is representative for this interaction, we investigated the localization of the core component Orc2 in relation to an HMGA1a-eGFP fusion in HepG2 cells. Consistent with our BiFC assays (Fig. 2A), a significant colocalization of HMGA1a-eGFP with endogenous Orc2 was observed in perinucleolar regions (Fig. 2B; SI Fig. 9A). We also asked whether HMGA1a might direct the localization of ORC via HMGA1a's AT-hook domains, which mediate its preference for AT-rich regions. We used an HMGA1a mutant, HMGA1a(R3xG), in which glycines replaced the arginines (R28G, R69G, and R86G) of the AT-hook consensus motifs (GRP). This HMGA1a mutant shows a diffuse nuclear localization pattern (16). HeG2 cells expressing Orc6 and HMGA1a(R3xG)-BiFC proteins (93.6%) showed fluorescence complementation, and most Orc6/HMGA1a complexes were now found in nucleoli (Fig. 2 Ac; SI Fig. 8), indicating that HMGA1a can alter the localization of Orc6. All BiFC cells analyzed displayed a healthy morphology, and the diameter of the nucleoli of transfected cells was similar to those of untransfected cells (SI Fig. 8c).
HMGA1a preferentially binds through its AT-hooks to the minor groove of AT-rich sequences and can be displaced by several dyes binding to DNA (13). In living cells, Hoechst 33342 competed with the prevalent binding of HMGA1a/Orc6 complexes to AT-rich domains in a time-dependent manner (Fig. 2C a and b). The intensity profiles of the BiFC complexes were unaltered when the cells were incubated with Hoechst 33342 for 1 min, but HMGA1a/Orc6 relocated to the nucleoli after 10 min (SI Fig. 9 B and C). The prominent nucleolar localization of the HMGA1a/Orc6 complex after competition with this dye is most likely mediated by interactions of Orc6 with components of the ribosome biogenesis pathway (24) (M. Rohrmoser and A.W.T., unpublished data). Cotransfecting the heterochromatin protein HP1α-mRFP revealed that HMGA1a/Orc6 partially colocalizes with HP1α (Fig. 2C and SI Fig. 9D for intensity profiles). This observation further indicates that the interactions of HMGA1a and ORC-subunits occur mainly in AT-rich heterochromatic domains but not in the entire heterochromatin. In summary, our data suggested that HMGA1a's AT-hook domains mediate the preferred heterochromatic positioning of the ORC/HMGA1a complex, but the integrity of this complex is independent of HMGA1a's subnuclear localization.
It is likely that nuclear factors other than HMGA1a can also determine ORC localization and specify replication origins, because immunofluorescence experiments with Orc2 and HMGA1a-eGFP clearly showed Orc2 localization beyond HMGA1a rich domains (25, 26). These results suggested that a certain molecular ratio between HMGA1a and ORC proteins might determine ORC localization to AT-rich heterochromatic regions. To analyze the targeting functions of HMGA1a, we first monitored the localization of ORC via BiFC between Orc6/Orc4 and Orc6/Orc5. The diffuse nuclear distribution is characteristic for interphase nuclei in Orc6/Orc4 and Orc6/Orc5 BiFC-experiments (Fig. 2D a and b) (26). Altering the molecular ratio between ORC after overexpression of HMGA1a-mRFP caused relocation of the diffuse BiFC patterns of Orc4/Orc6 and Orc5/Orc6 resulting in colocalization with HMGA1a-mRFP (Fig. 2D a‴ and b‴). It thus appeared that HMGA1a could target ORC to AT-rich chromatin regions. Moreover, and consistent with our observations in Fig. 1C, the BiFC experiments confirmed Orc6 as an integral part of the human origin recognition complex in living cells.
HMGA1a-Dependent Replication of Extrachromosomal Plasmids.
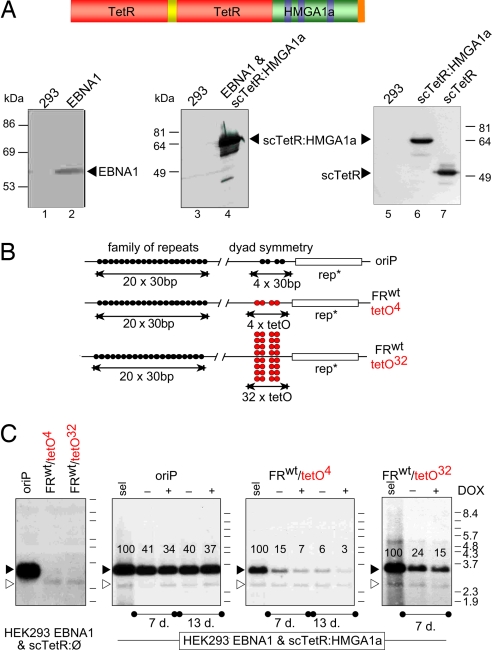
The HMGA1a/ORC interaction could also point to nonreplicative functions of ORC, such as heterochromatin formation (27), but the potential role of HMGA1a in the definition of replication origins is of immediate interest. HMGA1a-specific and ChIP grade antibodies are not available for analysis of endogenous HMGA1a at chromosomal origins. Therefore, we asked whether HMGA1a might be sufficient to target ORC to DNA creating a functional origin of DNA replication in a plasmid model system. HMGA1a was fused to the single-chain tetracycline repressor scTetR to form the chimeric scTetR:HMGA1a gene. scTetR:HMGA1a and, as a control, the scTetR gene alone were stably introduced in HEK293/EBNA1+ cells (Fig. 3A) (28). Both HEK293 cell lines, EBNA1+/scTetR+ and EBNA1+/scTetR:HMGA1a+, were transfected with three reporter plasmids: a wild-type oriP control plasmid (oriP) or two test plasmids with four tetO sites (FRwttetO4) or 32 tetO sites (FRwttetO32) to create clusters of binding sites with different density for scTetR:HMGA1a (Fig. 3B; SI Table 2). In all tetO reporter plasmids, EBNA1-binding sites within FR confer nuclear retention of plasmid DNA molecules only, but no DNA replication. Both FRwttetO plasmids did not replicate in EBNA1+/scTetR+-cells as expected, indicating that the tetracycline-repressor scTetR does not confer a functional replicator/initiator interaction (Fig. 3C). Similarly, an oriP-mutant with a deleted DS element (FRwttetOΔ) was replication-deficient in 293/EBNA1+ cells (SI Fig. 10B). Under selection the FRwttetO4 and FRwttetO32-plasmids stably replicated in EBNA1+/scTetR:HMGA1a+ HEK293 cells similar to oriP (Fig. 3C) according to the once-per-cell cycle rule as assessed in Meselson–Stahl experiments (data not shown). Under nonselective conditions, both tetO plasmids were relatively unstable. Doxycyclin, which prevents binding of scTetR:HMGA1a to tetO motifs (data not shown), further diminished their copy numbers in contrast to oriP (Fig. 3C).
Fig. 3.
HMGA1a mediates conditional DNA replication. (A) scTetR::HMGA1a consists of a single-chain (sc) dimer of the DNA-binding domain of the tetracycline repressor (TetR; red) fused via an artificial linker [(SG4)5, yellow] to the entire HMGA1a coding sequence. HEK293 cells (lanes 1 and 3) and a derivative expressing both EBNA1 (lane 2) and scTetR:HMGA1a (lane 4) were analyzed with EBNA1- and TetR-specific antibodies in Western blots. HEK293 cells expressing scTetR:HMGA1a (lane 6) and scTetR (lane 7) were analyzed with a TetR-specific antibody. (B) OriP has a bipartite structure: FR is an array of 20 high-affinity-EBNA1-binding sites (black circles); DS encompasses two pairs of EBNA1-binding sites (black circles). Four tet-operator sites (tetO4, red circles) replace DS in FRwttetO4. FRwttetO32 contains eight tetO-clusters. (C) Reporter plasmids were transfected into EBNA1+/scTetR+- (Left) or EBNA1+/scTetR:HMGA1a+-HEK293 cells (Right) and selected for 2–3 weeks (sel.) Low-molecular-weight DNA was prepared and digested with DpnI and HindIII. The radioactive probe used in the Southern blot hybridizations recognized a DNA fragment of 3.5 kbp (black arrowhead). A background signal appears at 2.5 kbp (open arrowhead). Addition of doxycyclin (+; 2 μg/ml) without hygromycin selection caused a modest reduction in copy number of the FRwttetO plasmids at the indicated time points (7 and 13 days). The drug did not affect the copy number of the oriP control plasmid, as indicated by the relative signal intensities in percent.
Pull-down and imaging experiments (Figs. 1C and 2) suggested an interaction between HMGA1a and ORC subunits. To assess this observation for scTetR:HMGA1a, we performed coimmunoprecipitations with nuclear extracts of scTetR:HMGA1a+ and control scTetR+-HEK293 cell lines. Orc2- and Orc6-specific antibodies coprecipitated scTetR:HMGA1a but not scTetR alone (SI Fig. 6). Previous experiments with recombinant human ORC largely consisted of Orc subunits 1–5 but lacked the weakly associated Orc6 (29). Orc2-specific antibody coprecipitated reproducibly the Orc1 and Orc6 subunits, verifying the existence of an ORC holocomplex (2).
Replication Efficacy and ORC Binding Correlate with the Local Density of HMGA1a.
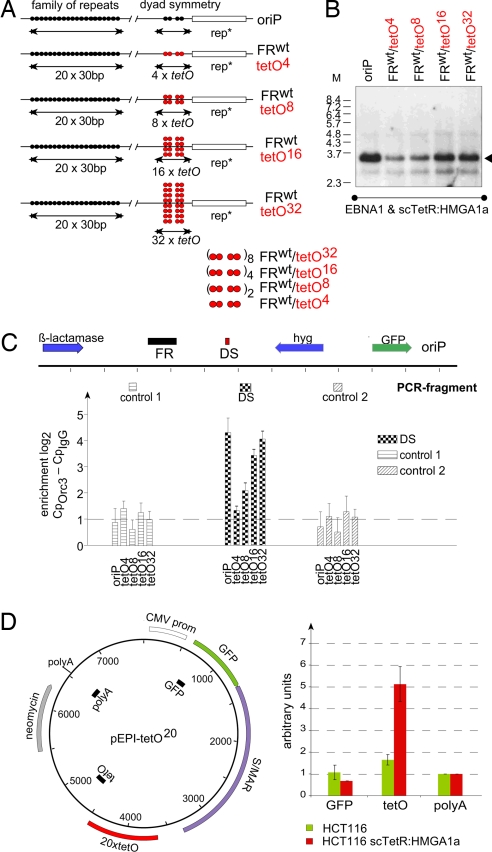
In comparison to FRwttetO32, the FRwttetO4 plasmid showed a relatively low copy number (Fig. 3C), indicating that additional tetO sites might increase the replication efficacy in a dose-dependent manner. Two, four, and eight DS-like tetO elements were integrated into reporter plasmids (Fig. 4A; SI Table 2). Each member of this plasmid family was separately transfected into EBNA1+/scTetR:HMGA1a+-HEK293 cells, which were selected for 2–3 weeks. All plasmids were maintained episomally and the different signal intensities correlated with the number of tetO motifs (Fig. 4B).
Fig. 4.
Multiple tetO sites increase the copy number of scTetR:HMGA1a-dependent plasmids and recruit ORC to the origin of DNA replication in a dose-dependent manner. (A) Different reporter plasmids with one (FRwttetO4), two (FRwttetO8), four (FRwttetO16), and eight (FRwttetO32) arrays of tetO4-motifs. (B) EBNA1+/scTetR:HMGA1a+-HEK293 cells were transfected with the indicated plasmid and replication was assessed as described in Fig. 3. (C) ChIP analysis indicates site-specific ORC binding. For each experiment, 500-μg chromatin of HEK293 cells stably transfected with the indicated plasmids was used. The location of the primer pairs is designated: DS is in close proximity to the tetO array, control primer pairs 1 and 2 are located 1.4 kbp downstream and 2.0 kbp upstream of the tetO sites, respectively. The heights of the columns indicate the relative enrichment (mean values and standard deviation of three experiments) on a logarithmic scale expressed as the difference between PCR values (Cp) obtained with the Orc3-specific antibody vs. controls obtained with preimmune serum (IgG). (D) High local concentration of HMGA1a promote site-specific initiation. A pEPI-based plasmid (pEPI-tetO20; Left) encompassing an array of 20 tetO sites. Nascent-strand analysis was performed in HCT116 cells and a derivative expressing scTetR:HMGA1a. Nascent DNA was purified from parental HCT116 cells (green bars) and scTetR:HMGA1a+ HCT116 (red bars) stably transfected with pEPI-tetO20. DNA was quantified by real-time PCR 2 weeks after transfection. The histogram shows the relative ratio of three different primer sets. The primer pair polyA was arbitrarily set to one.
To assess whether scTetR:HMGA1a could target ORC specifically to tetO motifs, ChIP was performed. Drug-selected EBNA1+/scTetR:HMGA1a+-HEK293 cells carrying the four different tetO plasmids or the oriP control plasmid (Fig. 4A) were cross-linked, and the fragmented chromatin was subjected to immunoprecipitations using a human Orc3-specific antibody and an appropriately matched Ig control. The association of Orc3 at or near the tetO motifs was compared by quantitative PCR to that at distal reference sites (see map in Fig. 4C). As expected, oriP showed reproducible enrichment of Orc3 at the tetO proximal PCR fragment in relation to two distal control sites (Fig. 4C). Similar results were obtained with an Orc2-specific antibody (data not shown). The abundance of ORC components correlated directly with the number of tetO motifs and led to a higher replication efficacy of the test plasmids.
HMGA1a Specifies Replication Origins.
Efficient targeting of HMGA1a to DNA correlated with efficient corecruitment of ORC (Fig. 4C). This result also suggested that a certain numbers of DNA-bound HMGA1a molecules could generate a strong and dominant replication origin. To test this hypothesis, we made use of the pEPI plasmid (30, 31). pEPI consists of several genetic elements including a promoter element and a scaffold/matrix attachment region (S/MAR) as part of the transcribed region (Fig. 4D). These elements ensure extrachromosomal maintenance and once-per-cell-cycle replication in an ORC-dependent manner (30, 31). pEPI does not contain a dedicated origin, but DNA replication initiates in a sequence-independent manner from multiple sites (31). We introduced an array of 20 tetO sites into a nontranscribed region of pEPI (Fig. 4D). The resulting plasmid pEPI-tetO20 was introduced in parental HCT116 cells and a derivative expressing scTetR:HMGA1a. Replication start sites were determined by nascent strand analysis after G418 selection. In parental HCT116 cells, pEPI-tetO20 did not show any locus-specific preference of nascent strand plasmid DNA (green bars, Fig. 4D), as expected (31). pEPI-tetO20 exhibited a 5-fold abundance of nascent-strand DNAs near the integrated tetO sites relative to reference sites in cells expressing scTetR:HMGA1a, only (compare red and green bars, Fig. 4D). These data indicated that high local concentrations of HMGA1a not only recruit ORC in a site-specific manner but also form a dominant origin in a replication system with otherwise multiple, apparently sequence-independent initiation sites.
Discussion
HMGA1a fused to the DNA-binding and dimerization domains of EBNA1 or TetR targets ORC to the cognate operator sites generating functional origins of DNA replication. ORC binding and replication competence did not rely on the plasmid background or an oriP-like configuration of tetO sites, because an array of tetO sites integrated into the pEPI-vector generated a dominant and site-specific origin (Fig. 4D). This study is not the first example that origins can be specified on plasmid DNAs (32, 33). However, a direct interaction between ORC and a chromatin constituent like HMGA1a in vivo, and a possible contribution to origin formation has not been described before.
Our in vivo and in vitro data indicate a direct interaction between HMGA1a and ORC subunits, corroborating HMGA1a's interaction with the ORC holocomplex. Coimmunoprecipitation experiments with tagged and endogenous proteins point to a transient interaction between HMGA1a and Orc6 (Figs. 1 and 2). Orc6 is biochemically less tightly associated with other ORC subunits but is essential for ORC DNA binding in Drosophila (34). A higher local concentration of HMGA1a at a given site might stabilize this interaction leading to a more efficient recruitment of ORC and thus increased replication efficiency (Fig. 4). Assuming that HMGA1a is also involved in cellular DNA replication, only a subset of chromosomal AT-sites might become competent for HMGA1a-mediated ORC binding and DNA replication when HMGA1a is present at high local density. Our BiFC experiments indicate that these sites are preferentially located in AT-rich heterochromatin domains.
HMGA1a is a multifunctional chromatin protein. Our data suggest now that it plays a role in recruiting ORC to AT-rich heterochromatin domains. All members of the HMGA protein family have been described as architectural transcription factors, and several studies link HMGA proteins to cell proliferation. Overexpression of HMGA1a can transcriptionally up-regulate cell cycle and growth regulators, i.e., cyclin A, p38 MAPK or N-myc (17). In addition, steady-state protein levels of HMGA1a are elevated in cancer cells with high proliferative potential (16) but decreased in differentiated cells. In contrast, deletion of one Hmga allele results in a pygmy phenotype in mice (15), and the expression of antisense RNA or a dominant-negative HMGA1a variant diminishes proliferation of tumor cells (17). Our own results suggest an additional nontranscriptional function of HMGA1a in proliferation control, on the basis of chromosomal DNA replication.
A connection between ORC and heterochromatin is well documented and seems to be conserved throughout evolution. For example, ScORC is essential for silencing the HMR-loci (35, 36), and mutations in the Orc2 subunit of Drosophila reduce the ability to spread heterochromatin (37). Therefore, Leatherwood and Vas hypothesized that heterochromatin might require additional ORC to replicate these tightly condensed regions (38). In analogy to the S. pombe Orc4 subunit, our data suggest a cofactor model in which HMGA1a recruits ORC to certain sites in heterochromatin to form functional origins of DNA replication in metazoan cell DNA. Further experiments shall reveal whether HMGA1a is instrumental in replicating chromosomal origins.
Materials and Methods
DNA Transfection, Plasmid Rescue, and Southern Blotting.
Plasmid DNAs were transfected with Polyfect into HEK293 cells and derivatives, which were selected with 100 μg/ml hygromycin, 250 μg/ml puromycin, or 200 μg/ml neomycin. Five hundred nanograms of low-molecular-weight DNA was digested with DpnI and electroporated into E. coli DH10B, which were selected with ampicillin. For Southern blotting, 6 μg of DNA was separated, transferred onto nylon membranes (Amersham), and probed with a radiolabeled prokaryotic probe. Signals were quantified with the Imager FLA-5100 (Fuji).
ChIP and Real-Time PCR Analysis.
ChIP experiments were performed as described (9). Nuclei (1 × 107) were isolated, cross-linked with formaldehyde for 10 min at 37°C, washed, and lysed by adding N-laurylsarcosine (2% final concentration). After washing, the chromatin was resuspended in 2 ml of TE buffer (10 mM Tris, pH 8.0; 1 mM EDTA) and sonicated. Five hundred micrograms of nucleoprotein was immunoprecipitated with 10 μg of polyclonal (Orc2, Orc3, and Mcm7) or monoclonal (EBNA1) antibodies in 50 mM Tris, 150 mM NaCl, 0.5 mM EDTA, and 0.5% Nonidet P-40 (NET). Coprecipitated DNA was isolated and purified, and quantitative real-time PCR was performed as described (9). Primer pairs are listed in SI Table 3.
Immunofluorescence and Live-Cell Microscopy.
Fluorescence complementation (23) was analyzed with a Leica TCS-SP2/AOBS instrument 22-26 h after transfection of 1 μg of the indicated expression plasmids. For immunocolocalizations, cells were fixed in 2% formaldehyde/PBS, washed, and permeabilized. Antibodies to detect HA- and Flag-tags were used as described in SI Text; DNA was visualized with 5 μg/ml Hoechst 33342.
Coimmunoprecipitation Assays and Western Blot.
Chromatin-bound proteins were isolated by high salt extraction from 2.5 × 107 cells and incubated with 5–10 μg of antibodies coupled to protein A or G Sepharose. Bound proteins were eluted, separated on SDS/PAGE, blotted, and detected with the indicated antibodies.
Nascent-Strand Analysis.
Nascent DNA of HCT116 cells transfected with pEPI-tetO20 was analyzed as described (39).
Supplementary Material
ACKNOWLEDGMENTS.
We thank the members of our laboratory for discussion, M. J. Deutsch for help with confocal microscopy, and P. B. Becker and P. Varga-Weisz for critical reading of the manuscript. We are grateful to B. Stillman (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) for providing mouse monoclonal antibody #920 and to H. J. Lipps (University of Witten/Herdecke, Witten, Germany) for the pEPI plasmid. This work was supported by the following institutional grants: Deutsche Forschungsgemeinschaft Grants Sche470/4, SFB646 and SPP1230 (to A.S. and W.H.), GK639 and Ho1804/5 (to R.H.), and SFB455 (to W.H.) and National Institutes of Health Grant CA70723 (to W.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.M.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707260105/DC1.
References
- 1.Sivaprasad U, Dutta A, Bell SP. Assembly of Pre-replication Complexes in DNA Replication and Human Disease. In: DePamphilis ML, editor. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2006. pp. 63–88. [Google Scholar]
- 2.Siddiqui K, Stillman B. ATP-dependent assembly of the human origin recognition complex. J Biol Chem. 2007;282:32370–32383. doi: 10.1074/jbc.M705905200. [DOI] [PubMed] [Google Scholar]
- 3.Cvetic C, Walter JC. Eukaryotic origins of DNA replication: could you please be more specific? Sem Cell Dev Biol. 2005;16:343–353. doi: 10.1016/j.semcdb.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert DM. In search of the holy replicator. Nat Rev Mol Cell Biol. 2004;5:848–855. doi: 10.1038/nrm1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilbert DM. Making sense of eukaryotic DNA replication origins. Science. 2001;294:96–100. doi: 10.1126/science.1061724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atanasiu C, Deng Z, Wiedmer A, Norseen J, Lieberman PM. ORC binding to TRF2 stimulates OriP replication. EMBO Rep. 2006;7:716–721. doi: 10.1038/sj.embor.7400730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhar SK, et al. Replication from oriP of Epstein–Barr virus requires human ORC, is inhibited by geminin. Cell. 2001;106:287–296. doi: 10.1016/s0092-8674(01)00458-5. [DOI] [PubMed] [Google Scholar]
- 8.Minami H, Takahashi J, Suto A, Saitoh Y, Tsutsumi K. Binding of AlF-C, an Orc1-binding transcriptional regulator, enhances replicator activity of the rat aldolase B origin. Mol Cell Biol. 2006;26:8770–8780. doi: 10.1128/MCB.00949-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schepers A, et al. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein–Barr virus. EMBO J. 2001;20:4588–4602. doi: 10.1093/emboj/20.16.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sibani S, Price GB, Zannis-Hadjopoulos M. Ku80 binds to human replication origins prior to the assembly of the ORC complex. Biochemistry. 2005;44:7885–7896. doi: 10.1021/bi047327n. [DOI] [PubMed] [Google Scholar]
- 11.Dominguez-Sola D, et al. Nontranscriptional control of DNA replication by c-Myc. Nature. 2007;448:445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 12.Chuang RY, Kelly TJ. The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc Natl Acad Sci USA. 1999;96:2656–2661. doi: 10.1073/pnas.96.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reeves R, Nissen MS. The AT-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J Biol Chem. 1990;265:8573–8582. [PubMed] [Google Scholar]
- 14.Zhao K, Kas E, Gonzalez E, Laemmli UK. SAR-dependent mobilization of histone H1 by HMG-I/Y in vitro: HMG-I/Y is enriched in H1-depleted chromatin. EMBO J. 1993;12:3237–3247. doi: 10.1002/j.1460-2075.1993.tb05993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benson KF, Chada K. Mini-mouse: phenotypic characterization of a transgenic insertional mutant allelic to pygmy. Gen Res. 1994;64:27–33. doi: 10.1017/s0016672300032511. [DOI] [PubMed] [Google Scholar]
- 16.Hock R, Furusawa T, Ueda T, Bustin M. HMG chromosomal proteins in development and disease. Trends Cell Biol. 2007;17:72–79. doi: 10.1016/j.tcb.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reeves R, Edberg DD, Li Y. Architectural transcription factor HMGI(Y) promotes tumor progression and mesenchymal transition of human epithelial cells. Mol Cell Biol. 2001;21:575–594. doi: 10.1128/MCB.21.2.575-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammerschmidt W, Sugden B. In: DNA replication and human disease. DePamphilis ML, editor. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 687–706. [Google Scholar]
- 19.Leight ER, Sugden B. The cis-acting family of repeats can inhibit and stimulate establishment of an oriP replicon. J Virol. 2001;75:10709–10720. doi: 10.1128/JVI.75.22.10709-10720.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung SC, Kang MS, Kieff E. Maintenance of Epstein–Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc Natl Acad Sci USA. 2001;98:1865–1870. doi: 10.1073/pnas.031584698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sears J, Kolman J, Wahl GM, Aiyar A. Metaphase chromosome tethering is necessary for the DNA synthesis and maintenance of oriP plasmids but is insufficient for transcription activation by Epstein–Barr nuclear antigen 1. J Virol. 2003;77:11767–11780. doi: 10.1128/JVI.77.21.11767-11780.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altmann M, et al. Transcriptional activation by EBV nuclear antigen 1 is essential for the expression of EBV's transforming genes. Proc Natl Acad Sci USA. 2006;103:14188–14193. doi: 10.1073/pnas.0605985103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 24.Killian A, et al. Inactivation of the RRB1-Pescadillo pathway involved in ribosome biogenesis induces chromosomal instability. Oncogene. 2004;23:8597–8602. doi: 10.1038/sj.onc.1207845. [DOI] [PubMed] [Google Scholar]
- 25.McNairn AJ, Gilbert DM. Epigenomic replication: linking epigenetics to DNA replication. BioEssays. 2003;25:647–656. doi: 10.1002/bies.10305. [DOI] [PubMed] [Google Scholar]
- 26.Prasanth SG, Prasanth KV, Siddiqui K, Spector DL, Stillman B. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J. 2004;23:2651–2663. doi: 10.1038/sj.emboj.7600255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chesnokov IN. Multiple functions of the origin recognition complex. Int Rev Cyt. 2007;256:69–109. doi: 10.1016/S0074-7696(07)56003-1. [DOI] [PubMed] [Google Scholar]
- 28.Krueger C, Berens C, Schmidt A, Schnappinger D, Hillen W. Single-chain Tet transregulators. Nucleic Acids Res. 2003;31:3050–3056. doi: 10.1093/nar/gkg421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ranjan A, Gossen M. A structural role for ATP in the formation and stability of the human origin recognition complex. Proc Natl Acad Sci USA. 2006;103:4864–4869. doi: 10.1073/pnas.0510305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baiker A, et al. Mitotic stability of an episomal vector containing a human scaffold/matrix-attached region is provided by association with nuclear matrix. Nat Cell Biol. 2000;2:182–184. doi: 10.1038/35004061. [DOI] [PubMed] [Google Scholar]
- 31.Schaarschmidt D, Baltin J, Stehle IM, Lipps HJ, Knippers R. An episomal mammalian replicon: sequence-independent binding of the origin recognition complex. EMBO J. 2004;23:191–201. doi: 10.1038/sj.emboj.7600029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Danis E, et al. Specification of a DNA replication origin by a transcription complex. Nat Cell Biol. 2004;6:721–730. doi: 10.1038/ncb1149. [DOI] [PubMed] [Google Scholar]
- 33.Takeda DY, Shibata Y, Parvin JD, Dutta A. Recruitment of ORC or CDC6 to DNA is sufficient to create an artificial origin of replication in mammalian cells. Genes Dev. 2005;19:2827–2836. doi: 10.1101/gad.1369805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balasov M, Huijbregts RP, Chesnokov I. Role of the Orc6 protein in origin recognition complex-dependent DNA binding and replication in Drosophila melanogaster. Mol Cell Biol. 2007;27:3143–3153. doi: 10.1128/MCB.02382-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Micklem G, Rowley A, Harwood J, Nasmyth K, Diffley JFX. Yeast origin recognition complex is involved in DNA replication and transcriptional silencing. Nature. 1993;366:87–89. doi: 10.1038/366087a0. [DOI] [PubMed] [Google Scholar]
- 36.Foss M, McNally FJ, Laurenson P, Rine J. Origin recognition complex (ORC) in transcriptional silencing and DNA replication in S. cerevisiae. Science. 1993;262:1838–1844. doi: 10.1126/science.8266071. [DOI] [PubMed] [Google Scholar]
- 37.Pak DT, et al. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell. 1997;91:311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- 38.Leatherwood J, Vas A. Connecting ORC, heterochromatin: Why? Cell Cycle. 2003;2:573–575. [PubMed] [Google Scholar]
- 39.Ghosh M, Liu G, Randall G, Bevington J, Leffak M. Transcription factor binding and induced transcription alter chromosomal c-myc replicator activity. Mol Cell Biol. 2004;24:10193–10207. doi: 10.1128/MCB.24.23.10193-10207.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.