Abstract
PTEN-induced putative kinase 1 (Pink1) is a recently identified gene linked to a recessive form of familial Parkinson's disease (PD). The kinase contains a mitochondrial localization sequence and is reported to reside, at least in part, in mitochondria. However, neither the manner by which the loss of Pink1 contributes to dopamine neuron loss nor its impact on mitochondrial function and relevance to death is clear. Here, we report that depletion of Pink1 by RNAi increased neuronal toxicity induced by MPP+. Moreover, wild-type Pink1, but not the G309D mutant linked to familial PD or an engineered kinase-dead mutant K219M, protects neurons against MPTP both in vitro and in vivo. Intriguingly, a mutant that contains a deletion of the putative mitochondrial-targeting motif was targeted to the cytoplasm but still provided protection against 1-methyl-4-phenylpyridine (MPP+)/1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced toxicity. In addition, we also show that endogenous Pink1 is localized to cytosolic as well as mitochondrial fractions. Thus, our findings indicate that Pink1 plays a functional role in the survival of neurons and that cytoplasmic targets, in addition to its other actions in the mitochondria, may be important for this protective effect.
Keywords: Parkinson's disease, neurodegeneration, neuroprotection
Parkinson's disease (PD) is a movement disorder with progressive loss of dopamine neurons in the substantia nigra pars compacta (SNc). The molecular events responsible for the loss of dopaminergic neurons in PD remain poorly understood. One common feature of PD is the dysfunction of mitochondria, which results in reduced complex I activity in the SNc (1, 2). Experimentally, inhibitors of complex I of the mitochondrial respiratory chain can recapitulate this selective dopaminergic neuronal loss and consequent behavioral deficits (1, 3–5). These observations support the hypothesis that nigral dopamine neurons are highly vulnerable to stress arising from mitochondrial dysfunction.
Recently, several genes have been identified that cause PD (6). These genes include α-synuclein, parkin, PTEN-induced putative kinase 1 (Pink1), DJ-1, and LRRK2. Although several of the genes have been partially localized to the mitochondria, Pink1 is the only gene with a putative mitochondrial targeting motif. Several studies have shown that the mitochondrial targeting motif at the N-terminal region of Pink1 is sufficient to direct proteins to the mitochondria (7). Pink1 was initially identified as a PTEN-inducible transcript and contains a serine/threonine kinase domain (8). Interestingly, the G309D mutation of the kinase domain leads to a mild decrease in mitochondrial complex I activity, elevation of superoxide radicals, and increased lipid peroxidation (9). Studies with Drosophila lacking Pink1 showed mitochondrial pathology with the similar phenotype as seen in Parkin knockout flies (10, 11). The above observations suggest that mitochondrial dysfunction may be linked to the Pink1 PD phenotype.
The mechanisms by which Pink1 affects the neuronal death process are not completely clear. Expression of Pink1 has been shown to reduce the death of cell lines induced by staurosporine (12). Moreover, siRNA-induced Pink1 suppression sensitizes SH-SY5Y and/or HeLa cell lines to multiple apoptotic stressors (13, 14). However, the specific ways in which Pink1 affects cell survival and how this is related to mitochondrial function is unknown.
Therefore, in the present study, we examined the functional role of Pink1 and mutants in MPTP/MPP+ models of neuron loss in vivo and in vitro. Our present results indicate that Pink1 kinase activity is critical for neuronal survival in response to mitochondrial stressors. Surprisingly, we found evidence that mitochondrial targeting of Pink1 activity is not absolutely required for its protective effects. This finding suggests a model by which cytosolic targets/transduction pathways may be modified by Pink1 to affect neuronal survival.
Results
Knockdown of Mouse Pink1 Gene Sensitizes Neurons to MPP+ Insult.
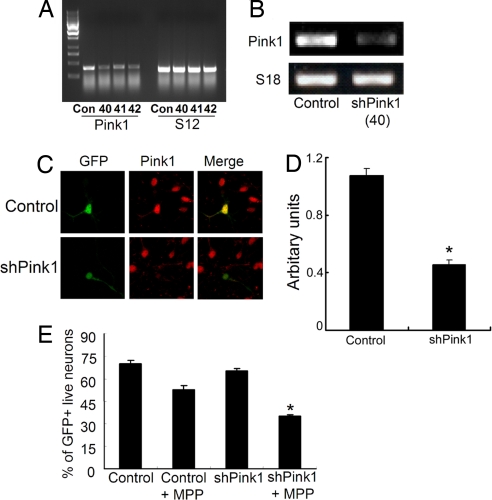
To assess whether endogenous Pink1 is important for the survival of primary neurons, we first examined the effects of Pink1 down-regulation on neuronal survival after exposure to the mitochondrial toxin MPP+. We first screened three potential siRNA sequences directed against mouse Pink1 to down-regulate endogenous Pink1 in NIH 3T3 cells. As shown in Fig. 1A, transfection of all three oligonucleotides Pink1 siRNA complexes, but not a control sequence, significantly reduced Pink1 message, but had no effect on control S12 message. Because the #40 sequence appeared to have the most prominent effect, we used this sequence in subsequent experiments. Transfection of NIH 3T3 cells with a hairpin vector construct (shRNA) based on this targeting sequence down-regulated the Pink1 message (data not shown) and protein [supporting information (SI) Fig. 6B]. We then constructed a recombinant adenovirus expressing this shRNA Pink1 sequence and showed the down-regulation in cultured cortical neurons in two ways. First, neurons were infected with the Pink1 shRNA virus or control. Cultures were then harvested and evaluated for Pink1 message by RT-PCR. As shown in Fig. 1B, a decrease in Pink1 message was detected. To further confirm, we evaluated levels of Pink1 protein in individual neurons infected with the shRNA Pink1 virus, followed by immunofluorescence analyses. The virus also coexpresses GFP, which was used to identify infected neurons. To detect endogenous Pink1, we used a rabbit polyclonal antibody raised against residues 125–149 of Pink1 (characterization of this antibody is shown in SI Fig. 6A). The antibody recognizes endogenous bands of expected size to Pink1 in control HEK293 cells extracts. These bands are similar in mobility to those observed with transfected V5-tagged wild-type (WT) Pink1 as recognized by either the V5 or our Pink1 antibodies. In addition, we also performed shRNA down-regulation experiments on mouse 3T3 cells to confirm the specificity of our antibody. Our results indicate that there is a major band observed corresponding to a putative Pink1, which is substantially decreased after Pink1 shRNA expression. We also detected bands of lower molecular weight that appeared variably depending on the particular experiment and cells used. Using this antibody, we also observed that Pink1 protein was detectably reduced in neurons infected with the Pink1 shRNA virus (Fig. 1 C and D). Interestingly, Pink1 expression also appeared very broadly within the cell (see below).
Fig. 1.
Knockdown of Pink1 in NIH 3T3 cell lines or cortical neurons. (A) NIH 3T3 cells were transfected with the siRNA targeting the mouse Pink1 gene, and 24 h after the transfection cells were collected to isolate the RNA; RT-PCR was carried out by using Pink1 primers and S12 primer as an internal control. (B) Alternatively, cortical neurons were infected with adenovirus harboring shRNA Pink1 at the time of plating, and cells were processed for RT-PCR. (C) Cortical neurons after infection with shRNA Pink1 virus or control were processed for immunostaining with Pink1 antibody. (D) Quantification of fluorescent intensity of Pink1-immunostained neurons infected with control and shRNA Pink1 virus. The asterisk denotes significance (P < 0.05). Values are mean ± SEM (n = 30–34). (E) Pink1 knockdown by shRNA Pink1 in cortical neurons sensitizes the cells to MPP+. Cortical neurons are transfected with shRNA plasmid 3 days after plating. Cells are treated with 20 μM MPP+ for 24 h, and GFP-positive cells are counted based on nuclear integrity. The asterisk denotes significance (P < 0.05) when compared with other conditions.
We next determined the effects of this shRNA Pink1 construct on the survival of cortical neurons exposed to MPP+, an agent that was previously shown to induce apoptotic neuronal death (15). Neuron cultures were cotransfected with GFP and control or Pink1 shRNA sequences and then untreated or exposed for 24 h to MPP+. As shown in Fig. 1E, neurons transfected with Pink1 shRNA showed significant sensitization to the effects of MPP+ when compared with control sequences (≈53% survival in control vs. ≈35% survival of shRNA for Pink1). This result implicates endogenous Pink1 as important in the regulation of primary neurons to exogenous stressors such as mitochondrial toxins.
Effects of the Expression of Pink1 Against MPP+ in Neurons.
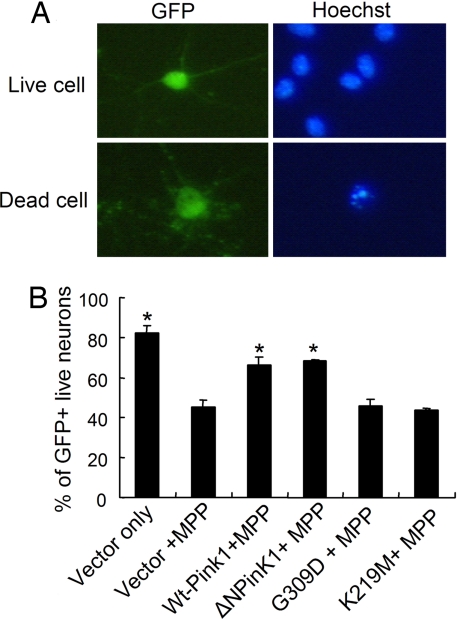
We next performed the converse experiment and determined how increased expression of Pink1 might affect the survival of cultured neurons. In this case, we were particularly interested in the mechanism by which Pink1 may be protective and the domains required for its protective effect. We first determined the effects of WT and kinase-deficient Pink1 expression. Twenty-four hours after transfection, cultured cortical neurons were treated with 20 μM MPP+ and incubated for a further 48 h. As shown in Fig. 2B, expression of WT Pink1 significantly protected neurons from the toxic effects of MPP+ treatment (≈45% survival in control vs. ≈67% survival with WT or ΔNPink1-transfected cells). However, expression of a mutant of Pink1 associated with PD in humans (G309D) failed to provide any protection when compared with the vector control. Importantly, we show that the kinase activity is required for the protective effects of Pink1. To demonstrate this idea conclusively, we constructed a mutant of Pink1 with the kinase activity abolished (K219M). Lys-219 is critical for the ATP binding, and a mutation at this site destabilizes the ATP-binding property of the gene and results in the loss of Pink1 kinase activity (12). Expression of the K219M mutant, similar to the G309D mutant, failed to protect cultured neurons against MPP+ toxicity.
Fig. 2.
Primary cortical neurons were cultured for 3 days and then transfected with different constructs by calcium phosphate. (A) Cells were treated with 20 μM MPP+ for 48 h, and neuronal survival was evaluated by assessing the nuclear integrity of GFP-positive neurons. (B) Neuroprotective effect of WT Pink1 or ΔNPink1 (N-terminal truncated Pink1) in cortical neurons against MPP+. The mutant G309D or K219M did not show any protection. The data are the mean ± SEM (n = 3). The asterisk denotes significance (P < 0.05) when compared with MPP+ control.
Mitochondrial Localization of Pink1 and Its Survival-Promoting Effect in Vitro.
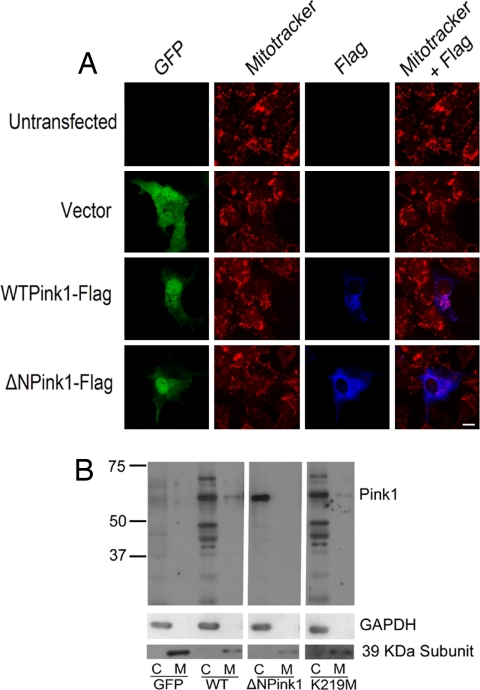
Pink1 contains a putative mitochondrial localization signal (7). To test whether this domain is essential for the protective effects of Pink1, we generated a Pink1 mutant (ΔNPink1) lacking the first 111 amino acids from the N terminus. This Pink1 mutant (ΔNPink1) impaired targeting of Pink1 to the mitochondria. This impaired targeting was demonstrated by two different means. First, COS-7 cells were transfected with either GFP alone or with WT Pink1 or the ΔNPink1 mutant. GFP-positive cells were then evaluated for exogenous Pink1 localization by Flag staining, as well as for mitochondria by mitotracker staining. As shown in Fig. 3A, the expression of WT Pink1 showed both cytoplasmic and mitochondrial localization. In contrast, there was little if any overlap costaining with the ΔNPink1 flag and mitotracker signal. Similar results were obtained with cultured cortical neurons (data not shown). The ΔNPink1 localization appeared diffuse and cytoplasmic indicating that deletion of the first 111 amino acids of Pink1 inhibited mitochondrial targeting. In support of this observation, we also performed cell fractionation studies after transfection of our WT and ΔNPink1 constructs in NIH 3T3 cells. As shown in Fig. 3B, WT Pink1 was localized to both mitochondrial and cytoplasmic fractions. Interestingly, Pink1 is predominately cytoplasmic at least when expressed in this cell line. Similar localization was detected for the kinase-dead K219M mutant, indicating that the kinase activity of Pink1 does not affect its localization to mitochondrial fractions. However, predominately cytoplasmic localization of the ΔNPink1 mutant was detected, indicating again that the ΔNPink1 mutant displays impaired mitochondrial targeting. Importantly, this ΔNPink1 mutant protected neurons from MPP+ toxicity identically to that of wild-type Pink1 (Fig. 2B). Taken together our results suggest that, although kinase activity is essential for the protective effects of Pink1, the mitochondrial targeting sequence and mitochondrial localization are not.
Fig. 3.
Immunostaining of COS-7 cell lines with WT Pink1 or ΔNPink1. (A) Cells are transfected with WT or ΔNPink1, and, 24 h after transfection, cells are incubated with mitotracker and fixed for immunostaining. The cells stained with anti-Flag antibody to visualize Pink1 and images are taken by confocal microscopy. (B) NIH 3T3 cells are transfected with GFP, WT, ΔNPink1, and K219M Pink1 construct. Then, 24 h after transfection, cells are fractionated into cytosolic and mitochondrial fraction with an equal volume of buffer. Electrophoresis was carried out with an equal volume from each fraction.
Overexpression of Pink1, but Not the Mutant Forms, Can Provide Protection Against MPTP in Vivo.
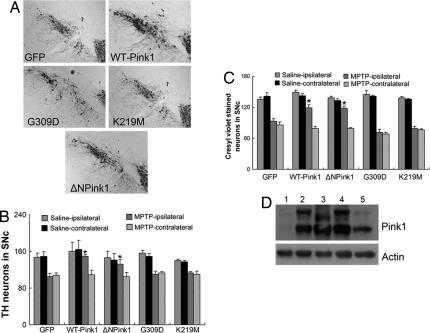
To validate our in vitro evidence indicating the potential relevance of cytoplasmic Pink1 kinase activity in mediating neuronal survival, we also evaluated whether similar conclusions would be obtained in vivo. Accordingly, we targeted WT, G309D, K219M, and ΔNPink1 mutants of Pink1 or GFP control vector to the substantia nigra of adult mice by using recombinant adenoviruses. One week after unilateral adenoviral injections, mice were challenged with a chronic paradigm of MPTP treatment (25 mg/kg for 5 consecutive days) or saline as described in Methods and Materials. The midbrain was sectioned, and the number of tyrosine hydroxylase (TH) (a marker for dopamine neurons)-immunopositive neurons in both the virus-uninjected (contralateral) and virus-injected (ipsilateral) SNc was assessed. Similar to our in vitro results, we found that the expression of WT Pink1 provided significant protection against MPTP insult (Fig. 4 A and B). Protection required kinase activity because both the G309D mutant associated with PD and the kinase-dead K219M mutant failed to provide any protection. Intriguingly, the ΔNPink1 mutant protected to a similar degree to that of WT Pink1 (Fig. 4 A and B). To ensure that the observed protective/death effects generated with evaluation of TH was not simply due to the loss of a phenotypic dopamine marker, we also evaluated the number of healthy neurons in the SNc region by cresyl violet staining (Fig. 4C), which produced similar results. Finally, we confirmed the expression of the Pink1 constructs by Western blot and immunohistochemical analyses as shown in Fig. 4D and SI Fig. 7, respectively. Taken together both in vitro and in vivo results suggest the potential importance of cytoplasmic kinase activity in mediating Pink1 protection.
Fig. 4.
WT or ΔNPink1 provides protection against MPTP administration. The adenoviruses (2 μl, 1 × 107 particles per μl) expressing WT, ΔNPink1, G309D, and K219M Pink1 were injected directly into the striatum of animals 7 days before the initiation of MPTP treatment. A GFP-expressing virus was used as a control. Brains were sectioned into 14-μm slices for TH staining. (A) Representative images of TH-immunopositive neurons of the ipsilateral side of the animals 2 weeks after treatment with MPTP. (B and C) TH-immunopositive (B) or cresyl violet-stained (C) neurons from the ipsilateral or the contralateral region of SNc were quantified. The data are the mean ± SEM (n = 3–4). (D) The expression of Pink1/mutants 1 week after viral delivery of GFP vector (lane 1), WT Pink1 (lane 2), ΔNPink1 (lane 3), G309D (lane 4), and K219M (lane 5) to mouse brain and protein are processed for immunoblotting. The asterisk denotes significance (P < 0.05) when compared with GFP control.
Endogenous Localization of Pink1.
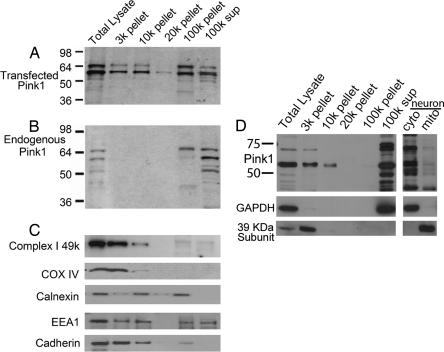
Our fractionation studies with expressed Pink1 indicate that Pink1 is both cytoplasmic and mitochondrial in localization despite the mitochondrial localization signal in Pink1. Indeed, the presence of Pink1 in the cytoplasm is consistent with the functional data, suggesting the critical nature of cytoplasmic kinase activity in mediating survival. However, overexpression of Pink1 may lead to improper localization. To verify that endogenous Pink1 also was present in the cytosol, we used the antibody that we generated against Pink1. We transfected HEK293 cells with WT Pink1 as a positive control and determined Pink1 distribution by using our Pink1 antibody. As previously described, we detected multiple immunoreactive species that associated with both membrane and cytosolic fractions similar to that observed previously with the detection of Flag-tagged Pink1 by using FLAG antibody. Differential centrifugation revealed that overexpressed Pink1 is widely distributed and sedimented with markers of multiple organelles, including mitochondria (Fig. 5 A and C). We also examined for distribution of endogenous Pink1 in multiple cell types, including HEK293 cells (Fig. 5B), 3T3 cells (Fig. 5D), and cultured cortical neurons (Fig. 5D). We found that the majority of endogenous Pink1 corresponding to full-length Pink1 (shown to be down-regulated by shRNA treatment) (see SI Fig. 6B) was found in cytoplasmic fractions, with significantly lower amounts associated with the mitochondria (Fig. 5B). As previously mentioned, there were other lower-molecular-weight bands that also appeared variably depending on the experiment and cell type. It is not clear whether these bands correspond to processed fragments of Pink1 or nonspecific bands. However, it is clear that a significant amount of full-length Pink1 is present in the cytosol.
Fig. 5.
Subcellular distribution of Pink1 in HEK293, NIH 3T3 cells, and cortical neurons. (A, B, D) Supernatants collected after centrifugation at 1,000 × g of cell lysate from WT Pink1 transfected (A), untransfected (B), HEK293, or NIH 3T3 (D) cells were subjected to sequential centrifugation, and pellets were analyzed by SDS/PAGE and probed by antibodies to either Pink1 (A, B, D) or to various organelle markers as indicated (C, D). 3k, nuclei, heavy mitochondria, and plasma membrane; 10k, mitochondria, lysosomes, and Golgi; 20k, lysosomes, large vesicles, and rER; 100k, ER vesicles, plasma membrane, Golgi, and endosomes; 100k sup, soluble cytoplasmic proteins. (D) Alternatively, neurons were fractionated into cytoplasmic and mitochondrial fraction in a one-step differential centrifugation as noted in Materials and Methods. This procedure was performed because of the limited neuronal extract available relative to that obtained from cultured fibroblasts.
Finally, we examined endogenous Pink1 localization by immunofluorescence in mouse embryonic fibroblasts (MEFs) (SI Fig. 8). We observed that Pink1 was localized clearly in the mitochondria, but also broadly in the cytoplasm. Similar results were obtained from skin fibroblasts obtained from a control individual and from a 74-year-old male with PD carrying compound heterozygous (E240K/L489P) Pink1 mutations (data not shown). No differences in Pink1 localization were observed between these cells. Overall, our results indicate that substantial endogenous Pink1 is localized to the cytosol despite the presence of a mitochondrial localization signal.
Discussion
The identification of several genes linked to familial PD has provided potential insight into the mechanisms of neurodegeneration (16). Important biological themes such as protein aggregation, protein turnover, and mitochondrial dysfunction have been highlighted. The latter is particularly interesting given that several of the genes, including DJ-1, Parkin, and Pink1, have at least been partially localized to the mitochondria (7, 17–19). The importance of mitochondria also is supported by observations of mitochondrial defects with loss of function of several of these genes, including Parkin and Pink1. Mitochondrial dysfunction in PD is further supported by reports of impaired mitochondria in PD patients, as well as sensitivity of dopamine neurons to mitochondrial toxins (2–5). The mitochondrial hypothesis in PD is consistent with the essential role of this organelle in ATP production, organization of a large number of signaling networks, and as a major source of ROS that also has been implicated in PD (20, 21). However, the involvement of mitochondria as a principal source/reason of degeneration in PD is controversial and not completely clear.
The importance of Pink1 in mitochondria is supported mainly by two observations. First, the protein contains putative mitochondrial localization sequence and has been reported to be localized to mitochondria (7). Second, the down-regulation of Pink1 in the fly results in abnormal mitochondrial morphology. However, the mechanism by which Pink1 may affect mitochondria, whether the abnormal effect is directly due to its absence in the mitochondria, and how any such activity might impact cell survival are unknown.
In the present study, we evaluated the functional role of Pink1 in mitochondrial toxin-induced neuronal death both in vitro and in vivo. We also determined the role of the kinase domain and the mitochondrial localization sequence in this neuroprotective role. We demonstrated that Pink1 plays an important functional role because siRNA down-regulation of Pink1 sensitizes neurons to MPP+. In addition, expression of WT Pink1 protects both cultured and dopamine neuronal loss induced by MPP+/MPTP. Importantly, mutations that impair kinase activity (the PD-associated mutation G309D and the artificial kinase-dead construct K219M) failed to protect both in vitro and in vivo, indicating that kinase activity is required for the neuroprotective function of Pink1. This result is supported by evidence that, although both WT Pink1 and G309D mutant can be localized to the mitochondria, only WT Pink1 prevents mitochondrial membrane potential change induced by MG-132 (7).
Interestingly, our present results also suggest that cytoplasmic kinase function of Pink1 is essential for Pink1-mediated protection. Deletion of the mitochondrial localization sequence impairs targeting of Pink1 to the mitochondria. Surprisingly, the expression of this Pink1 mutant is still protective both in vitro and in vivo. It must be made clear that there are several caveats to our interpretation. First, it is possible that a small amount of N-terminally truncated Pink1 might still localize to the mitochondria. Indeed, a recent report has suggested that Pink1 might be shuttled into the mitochondria by interacting with HtrA2 (omi), a serine protease with both survival/apoptotic functions (22). Clearly, however, the N-terminal mitochondrial localization signal is not required for the protective capacity of Pink1. Second, our protection studies were performed with overexpressed Pink1. Whether endogenous mitochondrial Pink1 is essential for any protective effects of Pink1 in neurons remains unclear and difficult to determine. However, a significant portion of endogenous Pink1 is localized to the cytoplasm, which is similar to previous reports with transfected Pink1 (23).
Although the nature of a potentially critical cytoplasmic function of Pink1 in survival is unclear, there are some tantalizing possibilities. An important hypothesis involves the relationship between Pink1 and Parkin. Pink1 mutant phenotype, at least in the fly, can be rescued by parkin gene overexpression (10, 11). However, the converse does not occur, suggesting that parkin acts downstream of Pink1 (10, 11). Interestingly, Drosophila Parkin mutant flies show increased oxidative stress and decreased antioxidant activity with a similar phenotype observed in the Pink1 mutant fly (24, 25). Pink1 also is reported to directly interact with Parkin (26), although the significance of the Pink–Parkin interaction in survival has not been verified. The vast majority of Parkin is localized to the cytoplasm, suggesting that if their interaction were functionally relevant, Parkin would likely be a critical cytoplasmic target of Pink1. Recently, it also has been reported that Pink1 physically interacts with TRAP1, a mitochondrial chaperone also known as HSP-75. Pink1-mediated phosphorylation of TRAP1 is important for survival in response to oxidative stress (27). However, TRAP1 also can reside in the cytoplasm (28), suggesting that Pink1 could modulate TRAP1 outside of the mitochondria.
Finally, although our results imply that cytoplasmic Pink1 activity is critical for cell survival, it does not contradict the model that Pink1 has central functions in the mitochondria. It is important to clarify that it is unclear whether Pink1 is integral to mitochondrial function and whether loss of Pink1 necessarily leads to mitochondrial dysfunction in all cell types. This issue is highlighted by the observation that the dPink1 inactivation phenotype could be rescued by antioxidants like SOD1 and Vitamin E (29). SOD1 is localized to the cytosol. How cytoplasmic actions of an antioxidant enzyme reverse some of the observed Pink1 deficiency phenotypes is unclear (29). Importantly, whether the increased level of ROS can lead to changes in mitochondrial morphology similar to that reported with Pink1 deficiency is unknown. It is therefore unclear whether Pink1 deficiency acts primarily in the mitochondria, which eventually leads to increased ROS, or whether a more generalized ROS increase occurs after Pink1 deficiency, which secondarily leads to mitochondrial disruption. If the latter were true, it might suggest that the cytoplasmic actions of Pink1 might regulate the antioxidant environment similar to SOD1. This notion is not without precedence with PD genes. The most notable example is with DJ-1, which has been shown to regulate the stability of NRF2, a factor essential for the coordinated expression of a number of antioxidant enzymes (30).
Taken together our results strongly imply that the cytoplasmic actions of Pink1 are critical for the survival properties of Pink1. How Pink1 may perform at least a part of its function in the cytoplasm will need to be examined in further and careful detail.
Materials and Methods
Mice.
All procedures involving animals were approved by the University of Ottawa Animal Care Committee and were maintained in strict accordance with the Guidelines for the Use and Treatment of Animals put forth by the Animal Care Council of Canada and endorsed by the Canadian Institutes of Health Research.
Plasmid Construction.
Multiple Pink1 constructs [WT Pink1, G309D mutant, K219M mutant, and deletion mutant of ΔNPink1 (deletion of first 111 amino acids from N-terminal end)] were generated with or without C-terminal Flag tags. In brief, Pink1 (8) was excised from pcDNA3.1(+) by using NotI and BamHI. A C-terminal 3XFlag tag was introduced by the removal of a stop codon from the gene and ligated into the pAdtrack-CMV vector for amplification into recombinant adenovirus as previously described (31). The resulting construct/virus expressed Pink1 variants and GFP from separate CMV promoters.
The Pink1 siRNA oligos (Ambion ID nos. 180640, 180641, and 180642) directed to silence mouse Pink1 as well as negative control siRNA (nontargeting siRNAs) were purchased from Ambion and cloned into the pSilencer 3.0-H1 siRNA vector. The shRNA fragment, including the H1 promoter, was subcloned to pAdtract vector to generate adenovirus as described previously (31).
Cell Line Culture and Transfection.
COS7 cells were grown in Opti-MEM supplemented with 10% (vol/vol) FBS (Invitrogen) and were transiently transfected with WT or mutant Pink1 for immunohistochemical analysis by using FuGENE (Roche Applied Science, Indianapolis). Similarly, mouse NIH 3T3 cells were maintained in DMEM supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells were seeded into 60-mm dishes for siRNA transfection according to the manufacturer's instructions. The required amount of Pink1 siRNAs and lipofectamine 2000 (Invitrogen) was diluted into Opti-MEM I (Gibco) and performed the transfection. After transfection, cells were incubated for 24–30 h, cells were lysed by using TRIzol reagent, and total RNA was isolated by chloroform/isopropanol precipitation.
Adenoviral Gene Delivery in Vivo.
Male 8- to 10-week-old C57BL/6 mice (Charles River Laboratories) were administered recombinant adenoviruses that expressed either WT Pink1, mutant Pink1, or the reporter gene GFP alone as control. Adenoviruses (2 μl; 1 × 10 7 particles μl−1 per construct) were stereotaxically injected into the right dorsolateral striatum (0.5 mm rostral and 2.2 mm to the right of bregma and 3.4 mm below the skull surface) at an infusion rate of 0.5 μl/min by using a syringe pump (PHD2000; Harvard Apparatus). The expression of recombinant proteins was confirmed by Western blot analyses as previously described (32).
MPTP Administration in Vivo.
Mice were challenged with MPTP once a day for five consecutive days (25 mg/kg, i.p., measured as free base; MPTP-HCl; Sigma–Aldrich) 1 week after adenovirus injection to permit sufficient time for retrograde transport and expression of the adenovirus-derived proteins (32, 33). Mice used as controls received an equivalent volume of saline (0.9%) once daily. Assessment of dopamine neuron survival was performed 2 weeks after the start of the MPTP dosing regimen.
Immunohistochemical Analysis of TH-Positive Neurons.
Brain tissues from mice injected with MPTP or saline were collected for immunohistochemical analyses as described previously (32, 33). Antibodies used were TH (1:10,000; Immunostar), and immunoreactivity was visualized by using an avidin–biotin complex peroxidase reaction. To examine the viral expression of the gene in TH neurons, a double-labeling immunofluorescence approach was used by using specific primary antibody to GFP (Abcam) or TH (Immunostar) and secondary antibodies Alexa 488-conjugated anti-rabbit IgG and Alexa 594-conjugated anti-mouse IgG (Molecular Probes).
Assessment of Dopamine Neuronal Loss in Vivo.
The loss of neurons in the SNc was determined by serial section analysis of the total number of TH+ neurons in the medial terminal nucleus region because intrastriatal administration of adenovirus results in the retrograde expression of the injected gene only those subpopulation of SNc neurons (32). The estimation of total TH+ nigral neuron populations in both ipsilateral and contralateral regions of control or MPTP-treated animals was counted in at least three sections for each animal and expressed in Fig. 4C. Cresyl violet staining and counting were similarly performed to validate the result of TH immunostaining as previously reported (32).
Neuronal Culture and Assessment of Death.
Primary culture of mouse cortical neurons was carried out as described previously (34–36). Three days after plating, cortical neurons were transiently transfected by using a modified calcium phosphate precipitation protocol (34, 37, 38). Cultures were treated with 20 μM MPP+ for 48 h after transfection. For the survival of transfected neurons, cultures were fixed and stained with the nuclear marker Hoechst. GFP-expressing neurons were counted as either alive or dead according to the appearance of Hoechst. Survival was expressed as the percentage of total cells that were classified as alive. Alternatively, neurons were infected with adenovirus as previously described (37).
Immunofluorescence.
COS-7 cells were transfected with WT Pink1/ΔNPink1 and stained with MitoTracker (Molecular Probes). Pink1 was stained with anti-Flag antibody (1:200 dilution; Sigma–Aldrich). For measurement of endogenous Pink1 in MEFs, cells from CD-1 mice were fixed with 4% paraformaldehyde and probed with rabbit polyclonal anti-Pink1 and mouse monoclonal cytochrome C antibodies (BD PharMingen). Staining was revealed with secondary anti-mouse and anti-rabbit antibodies coupled to Alexa Fluor 488 and 594 (Molecular Probes), respectively. To analyze the reduction of endogenous Pink1 by shRNA, cortical neurons after infection with shRNA Pink1 virus or control processed for immunostaining with Pink1 antibody. Quantification of the fluorescent intensity of Pink1-immunostained neurons infected with control and shRNA Pink1 virus (which express GFP) was performed in a randomly selected area.
Subcellular Fractionation.
Subcellular fractionation was performed by multiple methods. NIH 3T3 cells transfected with Pink1 (Fig. 3) or cortical neurons (Fig. 5D) were grossly fractionated into cytosolic and mitochondria containing fractions by single-step centrifugation. Cells were homogenized with buffer containing 250 mM sucrose, 20 mM HEPES, 3 mM EDTA, and protease inhibitor (pH 7.5) and spun at 3,000 rpm for 10 min. The supernatant was removed by using Eppendorf Centrifuge 5417R and centrifuged at 13,000 rpm for 10 min. The supernatant was retained, and the pellet was washed again with the same buffer and centrifuged as previously described. Both fractions of the supernatant were combined and designated as the cytosolic fraction. The pellet containing the crude mitochondrial fraction was suspended in the same buffer. Equal volumes of samples from mitochondrial and cytosolic fraction were processed for immunoblotting. Mouse monoclonal antibody for GAPDH (Chemicon) and a 39-kDa subunit of complex I (Molecular Probes) was used as a marker for cytosolic and mitochondrial fraction, respectively. For subcellular fractionation studies using larger amounts of HEK293 or NIH 3T3 extract (Fig. 5), cells were scraped into buffer [0.25 M sucrose, 5 mM Hepes (pH 7.2), 1 mM EDTA, and protease inhibitor mixture] and disrupted by 20 passages through a 26-gauge needle. The homogenate was cleared by centrifugation at 1,000 × g, and the resulting supernatant was sequentially centrifuged at 3,000, 10,000, 20,000, and 100,000 × g. Pellets were resuspended in equal volumes of extraction buffer. Equal volumes of samples were analyzed by polyacrylamide gel electrophoresis and Western blotting with a polyclonal rabbit anti-Pink1 antibody (raised to synthetic peptides corresponding to residues 125–149 of human Pink1). To detect mitochondria, antibodies such as complex I 49-kDa, 39-kDa subunit of complex I (Molecular Probe) and COX IV (Abcam) were used. Calnexin (BD Biosciences), EEA1 (BD Bioscience), and Cadherin (Abcam) antibodies were used as markers for ER, early endosomes, and plasma membrane, respectively. GAPDH was used as a marker of cytosol.
RT-PCR.
Semiquantitative RT-PCR was used to evaluate Pink1 mRNA. In brief, cDNA synthesis reaction was amplified in a total volume of 25 μl containing 10 pmol of each primer by using one-step RT-PCR kits (Qiagen). The sequences of the primers are: Pink1-F, 5′-GTTTTCCGCGCCTTCACCTCATCT-3′; and Pink1-R, 5′-GCCATTGCCACCACGCTCTACACT-3′. We used ribosomal protein S12 (37) or S18 mRNA as a loading control. S12 cDNA was amplified as described previously (37). Alternatively, primers for S18 are forward, 5′-ATAACAGGTCTGTGATGCCCTTAG-3′; and reverse, 5′-ATAGTCAAGTTCGACCGTCTTCTC-3′.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Dr. Motoko Unoki (University of Tokyo, Tokyo) for providing human PINK1 cDNA, and Dominque Vaillant (University of Ottawa/Ottawa Health Research Institute Virus Core Facility, Ottawa, ON, Canada) for making adenovirus for the experiment. This work was supported by the Parkinson's Society of Canada (D.S.P.), the Canadian Institutes of Health Research (A.T., P.F., and D.S.P.), the Parkinson's Disease Foundation (D.S.P.), the U.S. Army (D.S.P.), the Parkinson's Research Consortium (D.S.P.), the Heart and Stroke Foundation Ontario (D.S.P.), Neuroscience Canada, Brain Repair Program (D.S.P.), the Michael J. Fox Foundation (P.F.), a Parkinson's Society of Canada Fellowship (to M.E.H.), and a Canadian Institutes of Health Research Fellowship (to C.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705363105/DC1.
References
- 1.Schapira AH, Cooper JM, Dexter D, Jenner P, Clark JB, Marsden CD. Lancet. 1989;333:1269. doi: 10.1016/s0140-6736(89)92366-0. [DOI] [PubMed] [Google Scholar]
- 2.Greenamyre JT, Sherer TB, Betarbet R, Panov AV. IUBMB Life. 2001;52:135–141. doi: 10.1080/15216540152845939. [DOI] [PubMed] [Google Scholar]
- 3.Beal MF. Nat Rev Neurosci. 2001;2:325–334. doi: 10.1038/35072550. [DOI] [PubMed] [Google Scholar]
- 4.Langston JW. Life Sci. 1985;36:201–216. doi: 10.1016/0024-3205(85)90059-1. [DOI] [PubMed] [Google Scholar]
- 5.Langston JW, Irwin I, Langston EB, Forno LS. Neurosci Lett. 1984;48:87–92. doi: 10.1016/0304-3940(84)90293-3. [DOI] [PubMed] [Google Scholar]
- 6.Hardy J, Cai H, Cookson MR, Gwinn-Hardy K, Singleton A. Ann Neurol. 2006;60:389–398. doi: 10.1002/ana.21022. [DOI] [PubMed] [Google Scholar]
- 7.Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, et al. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 8.Unoki M, Nakamura Y. Oncogene. 2001;20:4457–4465. doi: 10.1038/sj.onc.1204608. [DOI] [PubMed] [Google Scholar]
- 9.Hoepken HH, Gispert S, Morales B, Wingerter O, Del Turco D, Mulsch A, Nussbaum RL, Muller K, Drose S, Brandt U, et al. Neurobiol Dis. 2007;25:401–411. doi: 10.1016/j.nbd.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 11.Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, Chung J. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 12.Petit A, Kawarai T, Paitel E, Sanjo N, Maj M, Scheid M, Chen F, Gu Y, Hasegawa H, Salehi-Rad S, et al. J Biol Chem. 2005;280:34025–34032. doi: 10.1074/jbc.M505143200. [DOI] [PubMed] [Google Scholar]
- 13.Deng H, Jankovic J, Guo Y, Xie W, Le W. Biochem Biophys Res Commun. 2005;337:1133–1138. doi: 10.1016/j.bbrc.2005.09.178. [DOI] [PubMed] [Google Scholar]
- 14.MacKeigan JP, Murphy LO, Blenis J. Nat Cell Biol. 2005;7:591–600. doi: 10.1038/ncb1258. [DOI] [PubMed] [Google Scholar]
- 15.Offen D, Beart PM, Cheung NS, Pascoe CJ, Hochman A, Gorodin S, Melamed E, Bernard R, Bernard O. Proc Natl Acad Sci USA. 1998;95:5789–5794. doi: 10.1073/pnas.95.10.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wood-Kaczmar A, Gandhi S, Wood NW. Trends Mol Med. 2006;12:521–528. doi: 10.1016/j.molmed.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L, Shimoji M, Thomas B, Moore DJ, Yu SW, Marupudi NI, Torp R, Torgner IA, Ottersen OP, Dawson TM, et al. Hum Mol Genet. 2005;14:2063–2073. doi: 10.1093/hmg/ddi211. [DOI] [PubMed] [Google Scholar]
- 18.Kuroda Y, Mitsui T, Kunishige M, Shono M, Akaike M, Azuma H, Matsumoto T. Hum Mol Genet. 2006;15:883–895. doi: 10.1093/hmg/ddl006. [DOI] [PubMed] [Google Scholar]
- 19.Dodson MW, Guo M. Curr Opin Neurobiol. 2007;17:1–7. doi: 10.1016/j.conb.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 20.Jenner P. Mov Disord. 1998;13(Suppl 1):24–34. [PubMed] [Google Scholar]
- 21.Przedborski S. Parkinsonism Relat Disord. 2005;11(Suppl 1):S3–S7. doi: 10.1016/j.parkreldis.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 22.Plun-Favreau H, Klupsch K, Moisoi N, Gandhi S, Kjaer S, Frith D, Harvey K, Deas E, Harvey RJ, McDonald N, et al. Nat Cell Biol. 2007;9:1243–1252. doi: 10.1038/ncb1644. [DOI] [PubMed] [Google Scholar]
- 23.Beilina A, Van Der Brug M, Ahmad R, Kesavapany S, Miller DW, Petsko GA, Cookson MR. Proc Natl Acad Sci USA. 2005;102:5703–5708. doi: 10.1073/pnas.0500617102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greene JC, Whitworth AJ, Andrews LA, Parker TJ, Pallanck LJ. Hum Mol Genet. 2005;14:799–811. doi: 10.1093/hmg/ddi074. [DOI] [PubMed] [Google Scholar]
- 25.Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Proc Natl Acad Sci USA. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore DJ. Biochem Soc Trans. 2006;34:749–753. doi: 10.1042/BST0340749. [DOI] [PubMed] [Google Scholar]
- 27.Pridgeon JW, Olzmann JA, Chin LS, Li L. PLoS Biol. 2007;5:e172. doi: 10.1371/journal.pbio.0050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cechetto JD, Gupta RS. Exp Cell Res. 2000;260(1):30–39. doi: 10.1006/excr.2000.4983. [DOI] [PubMed] [Google Scholar]
- 29.Wang D, Qian L, Xiong H, Liu J, Neckameyer WS, Oldham S, Xia K, Wang J, Bodmer R, Zhang Z. Proc Natl Acad Sci USA. 2006;103:13520–13525. doi: 10.1073/pnas.0604661103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clements CM, McNally RS, Conti BJ, Mak TW, Ting JP. Proc Natl Acad Sci USA. 2006;103:15091–15096. doi: 10.1073/pnas.0607260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith PD, Crocker SJ, Jackson-Lewis V, Jordan-Sciutto KL, Hayley S, Mount MP, O'Hare MJ, Callaghan S, Slack RS, Przedborski S, et al. Proc Natl Acad Sci USA. 2003;100:13650–13655. doi: 10.1073/pnas.2232515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crocker SJ, Lamba WR, Smith PD, Callaghan SM, Slack RS, Anisman H, Park DS. Proc Natl Acad Sci USA. 2001;98:13385–13390. doi: 10.1073/pnas.231177098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Qu D, Morris EJ, O'Hare MJ, Callaghan SM, Slack RS, Geller HM, Park DS. J Neurosci. 2006;26:8819–8828. doi: 10.1523/JNEUROSCI.2593-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fortin A, Cregan SP, MacLaurin JG, Kushwaha N, Hickman ES, Thompson CS, Hakim A, Albert PR, Cecconi F, Helin K, et al. J Cell Biol. 2001;155:207–216. doi: 10.1083/jcb.200105137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiang H, Hochman DW, Saya H, Fujiwara T, Schwartzkroin PA, Morrison RS. J Neurosci. 1996;16:6753–6765. doi: 10.1523/JNEUROSCI.16-21-06753.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aleyasin H, Cregan SP, Iyirhiaro G, O'Hare MJ, Callaghan SM, Slack RS, Park DS. J Neurosci. 2004;24:2963–2973. doi: 10.1523/JNEUROSCI.0155-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia Z, Dudek H, Miranti CK, Greenberg ME. J Neurosci. 1996;16:5425–5436. doi: 10.1523/JNEUROSCI.16-17-05425.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.