Abstract
Hsp90 is a highly conserved molecular chaperone that is involved in modulating a multitude of cellular processes. In this study, we identify a function for the chaperone in RNA processing and maintenance. This functionality of Hsp90 involves two recently identified interactors of the chaperone: Tah1 and Pih1/Nop17. Tah1 is a small protein containing tetratricopeptide repeats, whereas Pih1 is found to be an unstable protein. Tah1 and Pih1 bind to the essential helicases Rvb1 and Rvb2 to form the R2TP complex, which we demonstrate is required for the correct accumulation of box C/D small nucleolar ribonucleoproteins. Together with the Tah1 cofactor, Hsp90 functions to stabilize Pih1. As a consequence, the chaperone is shown to affect box C/D accumulation and maintenance, especially under stress conditions. Hsp90 and R2TP proteins are also involved in the proper accumulation of box H/ACA small nucleolar RNAs.
Introduction
Hsp90 is a highly conserved and abundant molecular chaperone that aids in the folding of a large number of proteins, including protein kinases and transcription factors involved in cellular signaling pathways. Hsp90 function requires ATP binding and hydrolysis as well as a cohort of cochaperones and cofactors such as Hsp70, Hsp40, Hop/Sti1, p23, Aha1, Cdc37, cyclophilins, and immunophilins (for review see Pearl and Prodromou, 2006). The physiological function of Hsp90 is typically dictated by the associated cochaperones and client proteins. In addition to protein-folding activities, Hsp90 has been implicated in several other activities such as ribosomal subunit nuclear export (Schlatter et al., 2002), transcription regulatory complex disassembly (Freeman and Yamamoto, 2002), and inhibition of β-amyloid aggregation (Evans et al., 2006).
In the Saccharomyces cerevisiae (yeast) cytoplasm, there are two virtually identical isoforms of Hsp90: one termed Hsp82, which is heat shock induced, and the other is termed Hsc82, which is constitutively expressed. Structural studies, sequence conservation, and proteolysis analyses of Hsp90s have indicated the presence of at least three domains (Fig. 1 A). The N-terminal domain is the site of ATP binding and hydrolysis and is connected to a middle domain. The middle domain has been proposed to be the site of cochaperone binding. Finally, the C-terminal domain provides a strong dimerization interface (for review seePearl and Prodromou, 2006). In this paper, the term Hsp90 is used when we do not explicitly distinguish between Hsp82 and Hsc82.
Figure 1.
Mapping the interactions between Hsp90, Tah1, and Pih1. (A) Schematic representation of the domains in Tah1, Pih1, Sti1, and Hsp82. (B) His6-tagged proteins were incubated with untagged proteins, and complexes were then isolated using Ni–nitrilotriacetic acid resin, resolved on SDS-PAGE gels, and stained with silver stain (left) or Coomassie Brilliant blue (right). (C) The interaction between Hsp82-C and Pih1 was characterized using yeast 2H. The 2H analysis was performed in WT diploid cells (top) or cells deleted of TAH1 (middle). The bottom panel shows control cells containing pOBD2 empty vector and pOAD2-Pih1. Cells were grown in selection medium without histidine. (D) Interactions between Hsp82, Tah1, and Pih1 were analyzed by SE chromatography on a Superdex 200 10/30 column. The positions of the molecular mass standards (in kilodaltons) are given along the top x axis. Tah1–Pih1 complexes are boxed for clarity. Proteins were detected on SDS-PAGE gels by silver staining. v.v., void volume. (E) A protein interaction model for Hsp82, Tah1, and Pih1. N, N-terminal domain of Hsp82; M, middle domain of Hsp82; C, C-terminal domain of Hsp82.
Previously, in an effort to globally analyze Hsp90 function and to identify novel Hsp90 substrates and cochaperones, we conducted a set of high-throughput physical and genetic interaction screens of Hsp90 in yeast (Zhao et al., 2005). In that work, we identified a new complex interacting with Hsp90 consisting of four proteins: Rvb1, Rvb2, Tah1, and Pih1 (also called Nop17). We termed the complex R2TP. Tah1 contains two tetratricopeptide repeats (TPRs), whereas Pih1 has no known motifs (Fig. 1 A). Both Tah1 and Pih1 were shown to be novel Hsp90 interactors. Tah1 and Pih1 were found to interact with Hsp82 from a large-scale two-hybrid (2H) screen and were shown to affect the yeast Hsp90 chaperone activity toward the classical model substrates v-Src and glucocorticoid receptor (Zhao et al., 2005). Rvb1 and Rvb2 belong to the AAA+ (ATPase associated with diverse cellular activities) superfamily of ATPases and show weak homology to the bacterial DNA helicase RuvB. Rvb1 and Rvb2 are essential for yeast growth (Qiu et al., 1998) and are found to be critical components of the chromatin remodeling complexes Ino80 (Jonsson et al., 2001) and SWR-C (Krogan et al., 2003; Kobor et al., 2004; Mizuguchi et al., 2004) as well as of the TIP60 histone acetylase complex (Ikura et al., 2000). Furthermore, it was recently suggested that Rvb1 and Rvb2 play an important role in DNA damage repair and transcription (Radovic et al., 2007).
Rvb1 and Rvb2 homologues in the mouse were found to associate with small nucleolar RNP (snoRNP) complexes and were shown to be important for snoRNP assembly and function (Newman et al., 2000; Watkins et al., 2002, 2004). It was also shown that depletion of yeast Rvb2 affects the localization of snoRNP core proteins (King et al., 2001). snoRNPs are RNP complexes made up typically of either box H/ACA or box C/D small nucleolar RNAs (snoRNAs) associated with a set of core proteins. snoRNPs are trans-acting regulators that are responsible for cleavage and modifications of small nuclear RNA (snRNA), ribosomal RNA (rRNA), and tRNAs (Matera et al., 2007). Association of the core proteins with box C/D or box H/ACA snoRNAs is required for the stability and proper localization of the snoRNPs (Filipowicz and Pogacic, 2002). Other protein factors have also been found to promote the stability of snoRNAs, such as Naf1 for box H/ACA (Dez et al., 2002; Fatica et al., 2002) and Bcd1 for Box C/D snoRNAs (Peng et al., 2003). These factors are not part of the final complex but are present during the biogenesis and assembly of the snoRNPs.
Interestingly, analysis of pih1Δ yeast cells suggests that Pih1/Nop17 is also involved in pre-rRNA processing (Gonzales et al., 2005; Granato et al., 2005). Pih1 has been shown to directly interact with the box C/D core protein, Nop58 (Gonzales et al., 2005; Granato et al., 2005). Also, it has recently been suggested that Hsp90 is involved in the maintenance of ribosomes in mammalian cells through its interaction with the ribosomal proteins RpS3 and RpS6 (Kim et al., 2006).
In this study, we initially characterize the Hsp90–Tah1–Pih1 and R2TP complexes. We then show that Pih1 is an unstable protein interacting with Hsp90. We demonstrate in vivo and in vitro that Hsp90 together with Tah1 stabilize Pih1. Subsequently, we show that Hsp90 and R2TP play a role in box C/D snoRNP assembly/maintenance, especially under stress conditions. Such an effect of Hsp90 on snoRNP seems to be partially mediated by its effect on Pih1 stability. Furthermore, Hsp90 and R2TP proteins are found to affect the accumulation of other noncoding RNAs. Thus, we provide evidence that Hsp90 is required for proper RNA processing. This represents a novel activity for this essential chaperone.
Results
The Hsp90–Tah1–Pih1 complex
Our identification of Tah1 and Pih1 as novel interactors of Hsp90 and the finding that Tah1 and Pih1 form a tight complex with the essential helicases Rvb1 and Rvb2 to form the R2TP complex (Zhao et al., 2005) prompted us to investigate the consequences of the interaction of Hsp90 with the R2TP complex. We had previously suggested that Hsp90, Tah1, and Pih1 might form a ternary complex (Zhao et al., 2005). Therefore, experiments were initially performed to further characterize the interactions between the three proteins.
Pull-down experiments were performed using full-length proteins as well as protein domains (Fig. 1 A) that were expressed and purified in Escherichia coli. Tah1 can be divided into two folded, stable, and soluble domains (unpublished data): an N-terminal domain containing two TPR motifs (residues 1–75; Tah1-TPR) and a C-terminal region (residues 76–111; Tah1-C). Two fragments of Hsp82 were also used: Hsp82-NM (residues 1–560) and Hsp82-MC (residues 260–709). As shown in Fig. 1 B (left), full-length Tah1 interacted with Pih1 and Hsp82, which is consistent with our previous observations (Zhao et al., 2005). Furthermore, full-length Tah1 interacted with Hsp82-MC but not with the Hsp82-NM fragment (Fig. 1 B, right), which lacks the extreme C-terminal fragment containing the MEEVD motif. This suggests that the Tah1–Hsp90 interaction is a typical TPR domain–MEEVD interaction (Scheufler et al., 2000). As a positive control, the TPR2a domain from Sti1 (Fig. 1 A) interacted with Hsp82-MC but not with Hsp82-NM (Fig. 1 B, right). Interestingly, neither Tah1-TPR nor Tah1-C interacted with Hsp82-MC. Tah1-TPR also did not interact with Hsp82-NM. This suggests that full-length Tah1 is required for interaction with Hsp82. Surprisingly, Tah1-C is sufficient to bind Pih1 (Fig. 1 B, right). Control pull downs of individual proteins are also shown in Fig. 1 B.
The interaction between purified Hsp82 and Pih1 was also tested using the pull-down assays, but no significant binding between the two proteins was observed (Fig. 1 B, left), suggesting that the interaction is weak. The binding between Hsp82 C-terminal fragment (residues 599–709) and Pih1 was previously observed by yeast 2H (Zhao et al., 2005). To determine whether or not the interaction between Hsp82 and Pih1 is mediated by Tah1, yeast 2H interaction analysis was performed in a diploid TAH1 knockout strain. As shown in Fig. 1 C, Pih1 still interacted with the Hsp82 C-terminal fragment in diploid cells when both copies of TAH1 genes were deleted. Thus, the interaction between Hsp82 and Pih1 is not dependent on Tah1.
The interaction between Hsp82, Tah1, and Pih1 was also analyzed using size exclusion (SE) chromatography (Fig. 1 D). The results were similar in the absence or presence of ADP. Hsp82 eluted at the position of a dimer, whereas Tah1 and Pih1 eluted as monomers. It should be pointed out that Pih1 forms soluble aggregates (Pih1agg; see section Hsp90 and Tah1 disaggregate Pih1 aggregates) in addition to the monomeric species (referred to here as Pih1). Under the conditions of the SE experiment, it was difficult to detect a significant interaction between Hsp82 and Tah1 or Pih1; however, a complex of Tah1 and Pih1 was evident (Fig. 1 D, boxes).
Based on the in vitro experiments and in vivo 2H assays, a putative model for the interactions between Hsp82, Tah1, and Pih1 is proposed (Fig. 1 E). The interaction of Pih1 with Hsp82 is weak, whereas the interaction of Tah1 with Hsp82 and Pih1 is more robust. This seems to suggest that Pih1 acts a substrate of Hsp82, whereas Tah1, with its TPR domain, acts as a true cofactor of the chaperone. This is further explored in the next three sections.
The Rvb1–Rvb2–Tah1–Pih1 complex
In the next set of experiments, the protein–protein interactions in the R2TP complex were characterized. Purified Rvb1 (50.5 kD) and Rvb2 (51.6 kD) proteins were analyzed by SE chromatography. Each protein individually eluted at fractions corresponding to an apparent molecular mass of ∼50 kD (Fig. 2 A), suggesting that each component alone exists predominantly as a monomer in the absence or presence of ADP. When Rvb1 and Rvb2 were incubated together for 4 h before being separated on the SE column, the two helicases formed a complex of an apparent molecular mass of ∼300–600 kD in the presence or absence of ADP (Fig. 2 A). The complex formed was found to be a heterohexamer of Rvb1 and Rvb2 (Gribun et al., 2008).
Figure 2.
Mapping the interactions between proteins of the R2TP complex. (A and B) Interactions between Hsp82, Tah1, Pih1, Rvb1, and Rvb2 were examined by SE chromatography on a Superdex 200 10/30 column. The positions of the molecular mass standards (in kilodaltons) are given along the top x axis. Tah1–Pih1 complexes are boxed for clarity. Proteins were detected on SDS-PAGE gels by silver staining. v.v., void volume. (C) A 3xFLAG tag was added to the chromosomal TAH1 or PIH1 genes in WT, tah1Δ, or pih1Δ cells. Tah1–3FLAG or Pih1–3FLAG complexes were isolated using αFLAG antibody resin from cells grown to late log phase at 30°C. The same amount of complexes was loaded on the gels. Proteins were visualized by Coomassie staining and were identified by mass spectrometry. (D) Western blot analysis of equal amounts of total cell lysates from different strains. Note that the Hsp90 antibody does not distinguish between Hsp82 and Hsc82, which are 98% identical. (E) A protein interaction model for the R2TP complex.
When Tah1 or Pih1 was added individually to the Rvb1/2 complex, Pih1 comigrated with Rvb1/2 (Fig. 2 B), whereas Tah1 eluted at the position of the monomer (compare Fig. 2 B with Fig. 1 D). When both Tah1 and Pih1 were mixed with the Rvb1/2 complex, coelution of Rvb1, Rvb2, Tah1, and Pih1 was observed over several fractions, indicating that the complex might be dynamic (Fig. 2 B). In addition, the Tah1–Pih1 complex was also observed at ∼60 kD (compare Fig. 2 B with Fig. 1 D). These results show that the R2TP complex can be reconstituted in vitro using purified proteins in the absence (not depicted) or presence (Fig. 2 B) of ADP. Importantly, Pih1 directly interacts with Rvb1/2, whereas the presence of Tah1 in the R2TP complex is through its interaction with Pih1 and not with Rvb1/2.
The aforementioned results using purified proteins were further confirmed by immunoprecipitation experiments from different yeast strains containing a chromosomal copy of TAH1-3FLAG or PIH1-3FLAG replacing the wild-type (WT) genes. Immunoprecipitations with anti-FLAG antibody indicated that Rvb1 and Rvb2 are associated with Tah1 and Pih1 when isolated from WT cells (Fig. 2 C, left). In pih1Δ cells, neither Rvb1 nor Rvb2 coimmunoprecipitated with Tah1-3FLAG (Fig. 2 C, left); however, in tah1Δ cells, the helicases coimmunoprecipitated with Pih1-3FLAG (Fig. 2 C, right). It should be noted that the levels of Rvb1, Rvb2, Hsp90, Tah1 (if not deleted), and Pih1 (if not deleted) in these different strains are the same (Fig. 2 D). The pull-down results are in agreement with the SE chromatography in vitro results. Thus, Pih1 mediates the interaction between Tah1 and the Rvb1/2 complex, whereas Pih1 interaction with Rvb1/2 does not require Tah1.
Based on these results, an interaction model for components of the R2TP complex is proposed (Fig. 2 E). In this model, Rvb1 and Rvb2 form a tight complex, and this complex interacts with Pih1 directly, forming an Rvb1–Rvb2–Pih1 (R2P) complex. Tah1 is associated with Rvb1/2 via Pih1. Thus, any effect on Rvb1/2 by Tah1 and Hsp90 is likely mediated by Pih1.
Hsp90 and Tah1 stabilize Pih1 in vivo
To determine any effect of Hsp90 on the R2TP complex, Tah1 and Pih1 were added to Rvb1/2 in the presence of Hsp82 (Fig. 2 B). The chaperone did not affect the assembly of the preformed R2TP complex in the absence or presence of ADP (Fig. 2 B) or ATP (not depicted). Direct binding of Hsp90 to the preformed R2TP complex was also not observed by SE chromatography (Fig. 2 B), indicating that the binding is probably weak or transient and that the chaperone does not affect/modulate the complex once the purified complex is formed. Consequently, in vivo experiments were performed to assess the effect of Hsp90 on the R2TP complex.
The R2TP complex was isolated from yeast cells in which the constitutive PIH1 gene was C-terminally 3FLAG tagged by immunoprecipitation with anti-FLAG antibody as described for Fig. 2 C. The complex was then immunoblotted with anti-Hsp90 antibody. As shown in Fig. 3 A, Hsp90 was not significantly associated with the R2TP complex purified from log-phase cells; however, more of the chaperone was associated with the complex isolated from stationary-phase cells, which indicates that Hsp90, Tah1, and Pih1 are in the same complex under these conditions. Surprisingly, higher levels of Hsp90 were found to be associated with the R2P complex purified from tah1Δ cells at both log- and stationary-phase cells. Considering that Tah1 might act as a cochaperone of Hsp90 (Zhao et al., 2005), it is likely that Hsp90 compensates for the depletion of Tah1 by increasing its association with R2P to preserve the function of the complex. Furthermore, the increased association of the chaperone with the R2P and R2TP complexes in stationary-phase cells compared with log-phase cells seems to indicate that one or more proteins in the complexes require added chaperoning under stress conditions.
Figure 3.
Hsp90 and Tah1 stabilize Pih1 in vivo. (A) The R2TP complex was isolated from log- and stationary-phase WT and tah1Δ cells expressing a chromosomal copy of PIH1–3FLAG using αFLAG resin. The interaction of Hsp90 with the complex was assessed by Western blot analysis. (B) The change in the levels of Pih1, Rvb2, Rvb1, Hsp90, and Tah1 in different strain backgrounds was monitored by Western blot analysis at three growth stages as indicated by OD600. Cells had a chromosomal copy of PIH1–3FLAG in a WT background (sample 1; PIH1–3FLAG), in a background deleted of TAH1 containing an empty vector (sample 2; PIH1–3FLAG, tah1Δ, and p414), or in a background deleted of TAH1 but expressing Tah1 from a p414 plasmid (sample 3; PIH1-3FLAG, tah1Δ, and p414Tah1). 6.5 μg of total proteins was typically loaded and blotted with specific antibodies. (C) WT or deg-TAH1 yeast cells were exposed to heat shock for 2 h in the presence of 40 μg/ml cycloheximide. For each sample, proteins from cells with equivalent OD600 were loaded onto SDS-PAGE gels and blotted with specific antibodies.
To examine the exact effect of Tah1 and, consequently, of Hsp90 on the R2TP complex, we determined the steady-state levels of R2TP proteins at different growth stages in WT cells and in cells deleted of the TAH1 gene. It was surprising to find that Pih1 levels decreased in tah1Δ cells compared with WT cells as cell densities increased (Fig. 3 B). The decrease of Pih1 levels in tah1Δ cells was dramatic when cells reached late stationary phase, at which the OD600 of the culture approached 4.5. The decrease of Pih1 levels in tah1Δ cells is specifically caused by the TAH1 deletion because endogenous Pih1 levels are recovered if tah1Δ cells express exogenous Tah1 from a plasmid (Fig. 3 B, sample 3). The decrease in Pih1 levels was not caused by a decrease in PIH1 mRNA levels as measured by semiquantitative RT-PCR (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200709061/DC1). This implies that Pih1 is probably unstable and that when Tah1 is deleted, Pih1 is degraded, especially at stationary phase. It should be noted that the deletion or overexpression of Tah1 does not significantly alter the steady-state expression levels of Rvb1, Rvb2, or Hsp90 in any growth stage tested (Fig. 3 B).
Therefore, together with Hsp90, Tah1 seems to play an important role in stabilizing Pih1. To further confirm that Pih1 is degraded upon the deletion of Tah1, a deg-TAH1 strain, in which the chromosomal copy of TAH1 is fused to the Ubi-Arg-DHFRts degron cassette (Dohmen et al., 1994), was constructed. Upon heat shock, deg-Tah1 is specifically and rapidly degraded, and, after 2 h, no Tah1 could be detected by Western blot analysis (Fig. 3 C). Pih1 levels were found to be reduced upon depletion of Tah1 in the deg-TAH1 strain in both log- and stationary-phase cells as compared with WT cells (Fig. 3 C). No effect of Tah1 depletion was observed on Rvb2 levels (Fig. 3 C) or Rvb1 levels (not depicted). This strongly suggested that Pih1 is an unstable protein and that Tah1 and Hsp90 are required to maintain Pih1 stability in vivo.
Hsp90 and Tah1 disaggregate Pih1 aggregates
Our in vivo results on the role of Hsp90–Tah1 in stabilizing Pih1 were further complemented with in vitro experiments. We had noticed that during the expression and purification of Pih1 from E. coli, the main peak of Pih1 eluted at the position of the monomer; in addition, a significant fraction of Pih1 formed a soluble aggregate that eluted in the void volume of the SE column and was also partially bound to the precolumn 0.2-μM filter. Thus, to determine whether Hsp90 and its cochaperone Tah1 influence Pih1 stability in vitro, we investigated the possible effect of Hsp82 and Tah1 on Pih1agg (Pih1 soluble aggregates collected during purification). Pih1agg used in these assays was >90% pure as judged by SDS-PAGE analysis and had no measurable ATPase activity.
When 100 μg of Pih1agg was injected onto the SE column in the absence or presence of ATP, most Pih1agg did not enter the column because it was bound to the precolumn filter (Fig. 4 A). Addition of Tah1 ± ATP, Hsp82 2 ATP, or Hsp82 + Tah1 2 ATP had no effect on the behavior of Pih1agg. However, incubation of Pih1agg with an equimolar amount of Hsp82 and ATP for 1 h at 30°C and subsequent loading of the mixture onto the SE column resulted in the appearance of smaller Pih1 oligomers that entered the column. The formation of smaller Pih1 oligomers seemed to be more pronounced when Pih1agg was incubated with Hsp82, Tah1, and ATP. These results indicate that Hsp82 and Tah1 can break down the soluble Pih1 aggregates to smaller oligomers in an ATP-dependent manner.
Figure 4.
Nucleotide-dependent disaggregation of Pih1agg by Hsp82. (A) SE chromatography of 100 μg (10 μM) Pih1agg preincubated with equal molar amounts of Hsp82 (210 μg) and Tah1 (30 μg) for 1 h at 30°C with or without ATP. The positions of the molecular mass standards (in kilodaltons) are given along the top x axis. Proteins were detected on SDS-PAGE gels by silver staining. v.v., void volume. (B) Disaggregation of Pih1agg by Hsp82 and Tah1 was monitored by light scattering at 350 nm. 120 μg Pih1agg was incubated with 84 μg Hsp82 and 12 μg Tah1 in the presence or absence of ATP. (C) AFM images of 60 μg Pih1agg incubated with 42 μg Hsp82 and 4 mM ATP monitored as a function of time. Control samples of Pih1agg + 4 mM ATP and of Hsp82 + 4 mM ATP after 60 min of incubation are also shown. Bar, 0.5 μm. (D) Change in particle volume distribution of Pih1agg in the presence of Hsp82 and ATP monitored as a function of time by AFM. Particle shapes were assumed to be partially filled spheres. Errors are derived from three measurements. Error bars represent SD.
This effect of Hsp82 and Tah1 on Pih1agg was also observed upon examining the change in Pih1agg particle size by light scattering. When Pih1agg was incubated with Hsp82 and ATP, the intensity of the scattered light gradually decreased as a function of time (Fig. 4 B), indicating the disaggregation of Pih1agg. This ATP-dependent disaggregation of Pih1agg by Hsp82 was further accelerated in the presence of Tah1 (Fig. 4 B), although Pih1agg was not completely disaggregated.
We were able to monitor the effect of Hsp82 on Pih1agg particles by atomic force microscopy (AFM). As shown in Fig. 4 C, the addition of Hsp82 and ATP to Pih1agg resulted in a dramatic reduction in the particle size of Pih1agg as a function of time (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200709061/DC1). No significant change in particle size was observed when Pih1agg alone was incubated with ATP (Fig. 4 C). Quantitative analysis of the AFM data (Fig. 4 D) indicated that the addition of Hsp82 caused a 30-fold decrease in the number of large Pih1agg particles whose size was >300 nm3 and a 24-fold increase in the number of Pih1agg particles ranging in size from 0 to 300 nm3 in 60 min. Similar results were obtained when the disaggregation of Pih1agg was monitored by electron microscopy (Fig. S2 B).
This disaggregase activity of Hsp90 toward Pih1agg is rather surprising. Hsp90 is commonly described as a molecular chaperone that is required for the proper folding of a set of client proteins at a late stage of their folding process (for reviews seePratt and Toft, 2003; Pearl and Prodromou, 2006). Recent studies have also indicated that Hsp90 may play a role in preventing protein aggregation and amyloid formation in several types of neurodegenerative disorders (Sittler et al., 2001; Auluck and Bonini, 2002; Mitsui et al., 2002; Ansar et al., 2007). In our study, we observed that yeast Hsp90 can also disaggregate a yeast protein aggregate formed in vitro. The mechanism of this novel activity of Hsp90 is currently under investigation.
These in vitro (Fig. 4) and in vivo (Fig. 3) results specifically indicate that Tah1 is a cofactor of Hsp90 that directs the chaperone to stabilize Pih1. Furthermore, based on the models for the Hsp90–Tah1–Pih1 complex (Fig. 1 E) and for the R2TP complex (Fig. 2 E), our data indicate that the effect of Hsp90 on the R2TP complex is through modulating Pih1 stability and that this activity of Hsp90 is assisted by the Tah1 cofactor. Subsequently, experiments were performed to determine the consequence of this activity of Hsp90–Tah1 toward Pih1.
R2P proteins are genetically linked to pre-rRNA processing
As mentioned in the Introduction, there are published reports of a role of Pih1 and Rvb1/Rvb2 in pre-rRNA processing. Because of this, we speculated that the R2TP complex might play such a role and that Hsp90 would also have an effect on pre-rRNA processing, partially (at least) through its effect on Pih1. Thus, experiments were performed to investigate this possibility by first examining the cellular function of the R2TP complex.
Synthetic genetic analysis (SGA) screens (Tong et al., 2004) were performed to determine common genetic interactors of RVB1, RVB2, TAH1, and PIH1 (Fig. 5 A) in an effort to elucidate the cellular function of the R2TP complex.tah1Δ and pih1Δ cells were used for this analysis. Rvb1 and Rvb2 are essential for cell viability and cannot be knocked out; thus, hypomorphic DAmP (decreased abundance by mRNA perturbation) alleles of RVB1 and RVB2 (referred to as rvb1-DAmP and rvb2-DAmP, respectively) were constructed (Schuldiner et al., 2005). In these strains, the steady-state levels of Rvb1 and Rvb2 proteins are reduced to ∼30–50% of WT levels by insertion of the nourseothricin (NAT) resistance gene at the 3′ untranslated region of the respective genes (Fig. S5 A, available at http://www.jcb.org/cgi/content/full/jcb.200709061/DC1; and Table I).
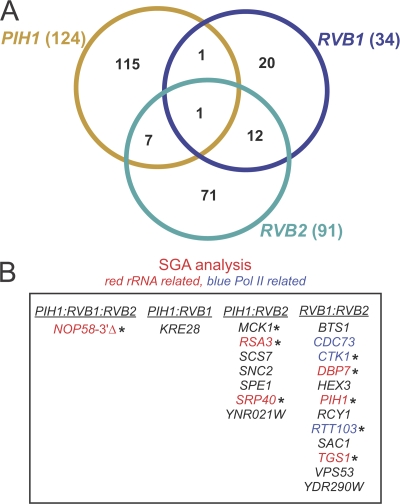
Figure 5.
SGA analysis of pih1Δ, rvb1-DAmP, and rvb2-DAmP strains. (A) Venn diagram showing the overlap in the hits obtained from the SGA analysis of pih1Δ, rvb1-DAmP, and rvb2-DAmP strains. (B) Common genetic interactors of PIH1, RVB1, and RVB2 are listed. Hits that were verified by random spore analysis are indicated by asterisks. Note that PIH1:RVB2, for example, means the hits that overlap between PIH1 and RVB2 only and not with RVB1.
Table I. Strains used in this study.
| Name | Background | Genotypes | References |
|---|---|---|---|
| W303 | W303 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 | http://www.yeastgenome.org/ community/W303.html |
| iG170D | W303 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 hsc82Δ∷LEU2 hsp82Δ∷LEU2 hsp82G170D∷HIS3 | Nathan et al., 1997 |
| R0023 | W303 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 pih1Δ∷KAN | Zhao et al., 2005 |
| R0024 | W303 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 tah1Δ∷NAT | Zhao et al., 2005 |
| R0066 | W303 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 hsc82Δ∷LEU2 hsp82Δ∷LEU2 hsp82G170D∷HIS3 TAH1-3FLAG∷KAN | This study |
| R0067 | W303 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 hsc82Δ∷LEU2 hsp82Δ∷LEU2 hsp82G170D∷HIS3 PIH1-3FLAG∷KAN | This study |
| R0068 | W303 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 hsc82Δ∷LEU2 hsp82Δ∷LEU2 hsp82G170D∷HIS3 PIH1-3FLAG∷KAN tah1Δ∷URA3 | This study |
| R0069 | W303 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 hsc82Δ∷LEU2 hsp82Δ∷LEU2 hsp82G170D∷HIS3 TAH1-3FLAG∷KAN pih1Δ∷URA3 | This study |
| R0074 | W303 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 hsc82Δ∷LEU2 hsp82Δ∷LEU2 hsp82G170D∷HIS3 PIH1-3FLAG∷KAN tah1Δ∷URA3 [p414GPD] | This study |
| R0075 | W303 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 hsc82Δ∷LEU2 hsp82Δ∷LEU2 hsp82G170D∷HIS3 PIH1-3FLAG∷KAN tah1Δ∷URA3 [p414GPDTah1] | This study |
| Y7092 | S288C | MATα can1Δ∷STE2pr-Sp_his5 lyp1Δ his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 | This study |
| Y3068 | S288C | MATα can1Δ∷MFA1pr-HIS3 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 lys2Δ0 | Zhao et al., 2005 |
| R0055 | S288C | MATα can1Δ∷MFA1pr-HIS3 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 lys2Δ0 pih1Δ∷NAT | Zhao et al., 2005 |
| R0061 | S288C | MATa can1Δ∷STE2pr-Sp_his5 lyp1Δ his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 tah1Δ∷URA3 | This study |
| R0013 | S288C | MATα can1Δ∷MFA1pr-HIS3 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 lys2Δ0 hsc82Δ∷LEU2 hsp82ts-URA3 | Zhao et al., 2005 |
| nop58-3′Δ | S288C | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 yor309cΔ∷KAN | Winzeler et al., 1999 |
| rvb1-DAmP | S288C | MATα can1Δ∷STE2pr-Sp_his5 lyp1Δ∷STE3pr-LEU2 rvb1∷NAT | Schuldiner et al., 2005 |
| rvb2-DAmP | S288C | MATα can1Δ∷STE2pr-Sp_his5 lyp1Δ∷STE3pr-LEU2 rvb2∷NAT | Schuldiner et al., 2005 |
| deg-TAH1 | W303 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 KAN-URA3pr-Ubi-Arg-DHFRts-TAH1 | This study |
| PJ69-4a-tah1Δ | PJ69-4a | MATα trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2∷GAL1-HIS3 GAL2-ADE2 met2∷GAL7-lacZ tah1Δ∷KAN | This study |
| PJ69-4α-tah1Δ | PJ69-4α | MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2∷GAL1-HIS3 GAL2-ADE2 met2∷GAL7-lacZ tah1Δ∷KAN | This study |
| YK3 | S288C | MATa leu2Δ0 his3Δ1 ura3Δ0 lys2Δ0 NOP56-3FLAG∷KAN | This study |
| YK5 | S288C | MATa leu2Δ0 his3Δ1 lys2Δ0 can1Δ∷STE2pr-Sp_his5 NOP56-3FLAG∷KAN pih1Δ∷NAT | This study |
| YK7 | S288C | MATa leu2Δ0 his3Δ1 lys2Δ0 can1Δ∷STE2pr-Sp_his5 NOP56-3FLAG∷KAN tah1Δ∷NAT | This study |
| YK9 | S288C | MATa leu2Δ0 his3Δ1 lys2Δ0 can1Δ∷STE2pr-Sp_his5 NOP56-3FLAG∷KAN rvb1∷NAT | This study |
| YK11 | S288C | MATa leu2Δ0 his3Δ1 lys2Δ0 can1Δ∷STE2pr-Sp_his5 NOP56-3FLAG∷KAN rvb2∷NAT | This study |
| YK15 | S288C | MATa can1Δ∷MFA1pr-HIS3 leu2Δ0 his3Δ1 ura3Δ1 lys2Δ0 NOP56-3FLAG∷KAN hsp82ts-URA3 hsc82Δ∷LEU2 | This study |
RVB1, RVB2, and PIH1 were found to genetically interact with 34, 91, and 124 ORFs, respectively (Fig. 5 A). No useful data were obtained from the tah1Δ strain. The hits that overlapped among the three genes are listed in Fig. 5 B. Several of those hits code for proteins involved in pre-rRNA processing or PolII DNA transcription. Consistent with our previous results (Zhao et al., 2005), RVB1 and RVB2 were found to genetically interact with PIH1 (Fig. 5 B). More interestingly, PIH1, RVB1, and RVB2 were all found to genetically interact with a pseudo-ORF, YOR309C. YOR309C was originally identified as a separate gene but subsequently was found to be the 3′ end fragment of the essential NOP58 gene. Thus, yor309cΔ is a deletion of ∼252 bp from the 3′ end of NOP58, which we will refer to as the nop58-3′Δ mutant. Therefore, PIH1, RVB1, and RVB2 are genetically linked to NOP58, which encodes a core protein component of the box C/D snoRNP complex (Lafontaine and Tollervey, 2000). This strongly suggests that the R2P complex and, consequently, the R2TP complex are involved in box C/D snoRNP biogenesis/assembly/maintenance (see next section). This observation is consistent with a previous study that showed a direct interaction between Pih1 and Nop58 (Gonzales et al., 2005).
To gain further insights into the in vivo functions of Tah1 and Pih1, we performed microarray analysis of tah1Δ and pih1Δ cells and compared the mRNA profiles obtained from those cells with those from WT cells (Fig. S3 A, available at http://www.jcb.org/cgi/content/full/jcb.200709061/DC1). Deletion of PIH1 caused up-regulation of a cluster of genes involved in ribosome biogenesis and pre-rRNA processing and metabolism, some of which were also up-regulated in the tah1Δ strain. The transcript levels for some genes were down-regulated, but they had various functions. The microarray expression profiles from tah1Δ and pih1Δ cells were then clustered with 224 microarray expression profiles obtained from cells in which the expression of essential genes was placed under the TetO7 promoter (Mnaimneh et al., 2004). Strikingly, the profile of pih1Δ cells clustered with those of genes involved in pre-rRNA processing such as RRP46, UTP23, and ESF2 (Fig. S3 B).
Our SGA and microarray results clearly support previous observations (Gonzales et al., 2005; Granato et al., 2005) that Pih1 plays a role in pre-rRNA processing. Importantly, it is suggested that R2TP as a complex is involved in pre-rRNA processing.
Hsp90 and R2TP proteins affect pre-rRNA processing and snoRNA accumulation/maintenance
The aforementioned data establish a link between the R2TP complex and pre-rRNA processing. Because Hsp90 and Tah1 modulate the stability of Pih1, we expected that Hsp90 will also have an effect on pre-rRNA processing. To this end, heat shock experiments were performed on cells knocked out of HSC82 and expressing a temperature-sensitive mutant allele of HSP82, R0013 (hsp82ts hsc82Δ; Zhao et al., 2005). As can be seen in Fig. 6 A, when cells grown to log or stationary phase at 30°C are shifted to the nonpermissive temperature of 37°C for the indicated time periods, there is an increased accumulation of 35S pre-rRNA in hsp82ts hsc82Δ versus WT cells. The levels of the U2 snRNA (a component of the spliceosome) are shown as a loading control. This seemed to indicate that Hsp90 acts on upstream components that result in 35S accumulation.
Figure 6.
Northern blot analysis of pre-rRNA and snoRNA in different strains. All Northern blot analyses shown in this figure have been repeated at least three times. (A) Accumulation of 35S pre-rRNA in WT and hsp82ts hsc82Δ cells grown to log or stationary phase at 30°C upon temperature upshift to 37°C for the indicated time periods. The levels of the U2 snRNA (a component of the spliceosome) are shown as a loading control. (B) Comparison of 35S pre-rRNA and snoRNAs levels in hsp82ts hsc82Δ cells to those in WT cells. Cells were grown at 30°C to log or stationary phase. Numbers next to each panel give the relative intensity of the bands in the mutant strain compared with WT levels after normalization to loading controls of scR1 RNA (involved in signal recognition particle–dependent cotranslational protein targeting to the membrane). Bottom panels show Western blot analysis using αHsp90 antibodies of total yeast lysates obtained from cells with equal OD600. (C) 35S pre-rRNA and snoRNA accumulation in different strains grown at 30°C during log and stationary phases was assessed by Northern blot analysis. U2 snRNA and scR1 were used as internal controls for RNA loading of rRNA and snoRNA, respectively. (D) Quantification of the bands from Northern analysis experiments similar to those shown in C normalized to loading controls. WT snoRNA levels are arbitrarily set to 1. The errors in these measurements are based on three separate experiments.
The aforementioned observations together with the observed effect of Hsp90 on Pih1 stability and the genetic interaction of R2P with Nop58 led us to speculate that Hsp90 affects 35S processing by affecting snoRNA accumulation. To formally test this possibility, further analysis was performed on WT, hsp82ts hsc82Δ, pih1Δ, tah1Δ, rvb1-DAmP, rvb2-DAmP, and nop58-3′Δ cells (Fig. 6, B–D). It should be noted that when grown at the permissive temperature of 30°C, the expression levels of Hsp82ts in the R0013 strain are reduced to 50% and 20% of the levels of WT Hsp90 (Hsp82 + Hsc82) in WT cells grown at log and stationary phases, respectively (Fig. 6 B). However, we confirmed that the cell viability of the hsp82ts hsc82Δ strain is similar to that of the WT strain at 30°C (unpublished data). Thus, the hsp82ts hsc82Δ strain was used to examine the effect of reduction in Hsp90 levels on rRNA/snoRNA accumulation at permissive temperatures. Furthermore, instead of analyzing the hsp82ts hsc82Δ cells under heat shock conditions, which might result in cell death, the analysis was performed at the permissive temperature of 30°C at log phase and stationary phase, which is a defined stress phase of the cell. It should be noted that the transcription of ribosomal DNA in stationary-phase cells occurs at a reduced rate, ∼10% compared with that in log-phase cells (Fahy et al., 2005; Hiley et al., 2005).
First, 35S pre-rRNA levels were analyzed by Northern analysis at 30°C. At log phase, pih1Δ, rvb1-DAmP, and rvb2-DAmP cells showed more 35S pre-rRNA accumulation compared with WT cells, but in hsp82ts hsc82Δ and tah1Δ cells, 35S pre-rRNA levels were similar to those in WT cells (Fig. 6, B and C). At stationary phase, however, 35S pre-rRNA accumulation was detected in all mutants, including hsp82ts hsc82Δ (Fig. 6 B) and tah1Δ (Fig. 6 C) cells. These results indicate that Pih1, Rvb1, and Rvb2 function in pre-rRNA processing at both log and stationary phases, but the effect of Tah1 and Hsp90 on pre-rRNA processing can be observed only at stationary phase, a stress phase for the cell. However, it should be noted that the levels of the chaperone might not be reduced enough in the R0013 strain to see an effect of Hsp90 in log-phase cells.
Second, the accumulation of box C/D and H/ACA snoRNAs was examined. Northern analysis showed that rvb1- and rvb2-DAmP cells have defects in box C/D and H/ACA accumulation at log and stationary phases (Fig. 6, C and D). In pih1Δ and nop58-3′;Δ cells, box C/D snoRNA accumulation is significantly reduced at both growth phases, whereas box H/ACA snoRNA accumulation is generally not affected (Fig. 6, C and D). As expected, levels of both box C/D and H/ACA snoRNAs in log-phase tah1Δ and hsp82ts hsc82Δ cells were similar to those in WT cells; however, accumulation of those snoRNAs was reduced in stationary-phase cells (Fig. 6, B–D). This is consistent with the results for 35S pre-rRNA.
Subsequently, microarrays were used to gain a more global view of snoRNA accumulation in hsp82ts hsc82Δ, pih1Δ, and tah1Δ stationary-phase cells. As shown in Fig. 7 A, there was generally a reduction in the accumulation of several box C/D snoRNAs in all three strains, with the greatest reduction evident in pih1Δ cells. The pih1Δ strain showed an overall accumulation of box H/ACA snoRNAs, whereas in tah1Δ and hsp82ts hsc82Δ cells, levels of these snoRNAs varied (Fig. 7 B). There is general agreement between the trends obtained from the microarray analysis and the Northern analysis (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200709061/DC1).
Figure 7.
Microarray and pull-down experiments link Hsp90 and R2TP to snoRNA accumulation. (A and B) Microarray analysis of changes in the levels of snoRNAs in pih1Δ, hsp82ts hsc82Δ, and tah1Δ strains compared with levels in the WT strain. Cells were grown to stationary phase at 30°C. The figures include all of the probes on the arrays showing statistically significant changes with respect to WT. Colors reflect intensity changes expressed as log2(mutant/WT), with up-regulated probes in red and down-regulated probes in green. (C) Nop56–3FLAG protein complexes were purified from WT, pih1Δ, and hsp82ts hsc82Δ strains grown to stationary phase. The proteins were separated on SDS-PAGE gels and were visualized by silver staining (top) or immunoblotted with αHsp90 and αFLAG antibodies (middle and bottom, respectively). Nop58, Nop56, Nop1, and Snu13 were identified by mass spectrometry.
The deletion or depletion of Rvb1, Rvb2, Tah1, Pih1, and Hsp90 results in the same general effect on box C/D snoRNA accumulation, which is similar to the effect seen in nop58-3′;Δ cells. This is clearly evident in stationary-phase cells. Therefore, further experiments were performed to determine how Hsp90 and R2TP proteins affect box C/D snoRNA. Four core proteins are associated with the mature box C/D snoRNAs: namely, Nop1, Nop56, Nop58, and Snu13 (Lafontaine and Tollervey, 2000). Snu13 directly binds to snoRNA, whereas Nop1 has methyltransferase activity (Galardi et al., 2002). Chromosomal NOP56 was tagged to express a C-terminal 3xFLAG-tagged Nop56 protein, which was immunoprecipitated using anti-FLAG antibodies from stationary-phase cells. As shown in Fig. 7 C, Nop1, Nop58, and Snu13 copurified with Nop56–3FLAG; however, in both pih1Δ and Hsp90-depleted hsp82ts hsc82Δ cells, the levels of Snu13 were reduced compared with WT cells. Snu13 is a known factor needed for the initiation of snoRNP core protein assembly (Watkins et al., 2002). Thus, the isolated Nop56 complex includes proteins associated with free Nop56 as well as with Nop56 bound to mature box C/D snoRNP. In pih1Δ cells, there was also a decrease in the levels of Nop1 immunoprecipitated with the Nop56–3FLAG complex (Fig. 7 C).
Surprisingly, there was significantly increased levels of Hsp90 associated with the core box C/D protein complex in pih1Δ and hsp82ts hsc82Δ strains (Fig. 7 C). This might indicate that Hsp90 compensates for the absence of Pih1 in the pih1Δ strain, whereas in the hsp82ts hsc82Δ strain, Hsp82ts might not be as active as WT Hsp82, and, therefore, more of the chaperone has to bind to the box C/D core protein complex. For tah1Δ, rvb1-DAmP, and rvb2-DamP cells, the levels of the core box C/D proteins immunoprecipitated with Nop56–3FLAG seem to be unaffected (Fig. S5 B); however, there was a moderate increase in the levels of Hsp90 associated with the complex, possibly indicating that the chaperone is playing a compensatory role. Western blot analysis of all strains used in these pull downs indicated that all strains have similar levels of Nop1, Nop56, Nop58, Snu13, and Hsp90 (unpublished data).
The aforementioned microarray, Northern, and pull-down assays suggest an effect of Hsp90 and R2TP proteins on rRNA biogenesis and snoRNA accumulation. We propose that this effect is mediated, in part, by their common effect on box C/D snoRNP assembly.
Discussion
The Hsp90 chaperone has been known to regulate multiple signaling pathways. However, until now, there has been no evidence that Hsp90 might modulate rRNA biogenesis through an effect on snoRNP accumulation/assembly/maintenance. In this study, we have provided detailed in vitro and in vivo biochemical data that suggest such a role for the chaperone by affecting snoRNA accumulation. This novel function of Hsp90 seems to be partially mediated by the chaperone interactors Tah1 and Pih1 that associate tightly with the essential helicases Rvb1 and Rvb2 to form the R2TP complex (Figs. 1 and Figs.2). Tah1 is a small protein containing TPR motifs, whereas Pih1 has no known motifs. Rvb1, Rvb2, Tah1, and Pih1 form a tight complex, as shown by in vivo and in vitro assays (Figs. 1 and Figs.2). Although Hsp90 was shown to directly bind to Tah1 by in vitro pull-down and yeast 2H experiments (Fig. 1; Zhao et al., 2005) and to Pih1 by yeast 2H, Hsp90 does not seem to interact significantly with the R2TP complex in log-phase cells (Fig. 3 A). However, more of the chaperone interacts with the R2TP complex in stationary-phase cells, a stress condition, or if Pih1 is destabilized by the deletion of Tah1 (Fig. 3 A). This implies that Hsp90 and Tah1 maintain the R2P complex by stabilizing Pih1. Here, Tah1 plays a classical role as a cochaperone for Hsp90 by conferring a specialized function to the chaperone aimed at stabilizing Pih1.
Pih1 is likely the key component of the R2TP complex. Pih1 binds directly to the Rvb1–Rvb2 complex, whereas Tah1 does not (Fig. 2 E). Also, quantification of the amount of Rvb1, Rvb2, Tah1, and Pih1 in living yeast cells revealed that the relative abundance of each of these proteins is Rvb1 ≈ Rvb2 >> Tah1 >> Pih1 (unpublished data). This indicates that Pih1 is the limiting protein in forming the in vivo R2TP complex, and, therefore, Pih1 plays a critical role in the regulation of R2TP complex levels. Paradoxically, Pih1 seems to be the most unstable component in the R2TP complex and requires stabilization by Hsp90 and Tah1.
Based on our data, the R2P complex functions in the proper assembly of the box C/D core protein complex composed of Nop1, Nop56, Nop58, and Snu13. Several of our experiments support this conclusion. (1) SGA array indicated that RVB1, RVB2, and PIH1 have many genetic interactions, suggesting that they are involved in a variety of cellular functions. However, all three genes genetically interact with NOP58 (Fig. 5, A and B). (2) Cells deleted or depleted of Rvb1, Rvb2, or Pih1 have a reduced accumulation of box C/D snoRNAs in log and stationary phases (Fig. 6, C and D). (3) The box C/D core protein complex is destabilized in rvb1, rvb2, and pih1 mutant backgrounds as indicated by enhanced binding of Hsp90 to this complex (Figs. 7 C and S5 B) under stationary-phase growth conditions. Our observation of the binding of Hsp90 to the box C/D core protein complex has not been reported before and further implicates Hsp90 in playing a role in rRNA biogenesis or maintenance. This role of Hsp90 is most evident under stress conditions such as heat shock or in stationary phase. Thus, the chaperone might either be required for the biogenesis of snoRNA or for the maintenance of snoRNA under these stress conditions. This effect of Hsp90 partly results from its effect on the stability of Pih1 (Fig. 8).
Figure 8.
A model of Hsp90 and R2TP function in the modulation of snoRNP assembly. In vivo, Pih1 is degraded in the absence of Hsp90/Tah1, whereas in vitro, Pih1 can misfold and form large soluble aggregates. Hsp90 and Tah1 bind and stabilize Pih1, probably resulting in a transient formation of an Hsp90–Tah1–Pih1 complex. Hsp90, Tah1, and Pih1 associate with the Rvb1/2 complex, resulting in formation of the R2TP complex and Hsp90, both of which are then involved in proper assembly/maintenance of box C/D snoRNPs. Hsp90, Tah1, Pih1, and Rvb1/2 are proposed to also affect the accumulation of other noncoding RNAs (not depicted).
We suggest that the role played by Hsp90/R2TP in modulating or maintaining the stability of snoRNPs might be similar to that played by other protein factors that have been found to promote the proper assembly of snoRNPs. For example, Naf1 has been shown to be recruited to RNA polymerase II and to be involved in the assembly of a stable box H/ACA preRNP that is inactive until Naf1 is exchanged for Gar1, a core protein component of the final mature box H/ACA snoRNP (Dez et al., 2002; Fatica et al., 2002).
The effect of Hsp90 on snoRNA accumulation and rRNA processing is most evident in stationary-phase cells. This phase is a stress phase of the cell, in which molecular chaperones are expected to play a more prominent role. In this phase, cells face nutrient deprivation, and, therefore, proteins might be more prone to rapid degradation by the proteasome or autophagy to provide a source of amino acids (Gray et al., 2004). Therefore, in stationary phase, if Pih1, an unstable protein, is not stabilized by Hsp90 and its cofactor Tah1, it would be rapidly degraded as observed in Fig. 3 B, resulting in improper box C/D snoRNA biogenesis/assembly/maintenance and in 35S rRNA accumulation. On the other hand, in log-phase cells, Pih1 would not be prone to rapid degradation (Fig. 3 B), and, thus, the effect of Hsp90 on rRNA processing will not be as evident. Alternatively, one or more other chaperones might stabilize Pih1 in log-phase cells.
Interestingly, in a recent proteomic study, the human Rvb1 and Rvb2 orthologues, Tip49A and Tip49B, and the human Pih1 orthologue, Nop17, were found to coimmunoprecipitate with human Hsp90 (Te et al., 2007). This indicates that the R2P complex is highly conserved in eukaryotes. There is currently no known human orthologue of yeast Tah1; however, other TPR domain–containing proteins might serve as an alternative to Tah1 in human cells. We propose that human Hsp90 will have a similar effect on rRNA biogenesis as that shown for yeast Hsp90.
In conclusion, our study has elucidated a molecular basis in which Hsp90 could modulate the assembly, maturation, or maintenance of box C/D snoRNP complexes by maintaining the stability of Pih1. An effect of Hsp90, Tah1, Pih1, and Rvb1/2 on box H/ACA snoRNPs is also suggested by our data (Figs. 6, B–D; and 7 B). However, the molecular basis of this effect requires further investigation. It is reasonable to predict that other chaperone systems might have similar activities directed toward noncoding RNAs. Studying the molecular details of these activities will be an important future endeavor.
Materials and methods
Molecular biology
DNA fragments of Tah1, Tah1-TPR (residues 1–75), Tah1-C (residues 76–111), Sti1 TPR2a domain (residues 260–386), and Rvb2 were amplified from the yeast S. cerevisiae S288C genome, cloned into p11 expression vector (Savchenko et al., 2003), and expressed as N-terminal His6–tobacco etch virus (TEV) protease fusion proteins. DNA fragments of Hsp82-NM (residues 1–560), Hsp82-MC (residues 260–709), and Rvb1 were amplified by PCR from the yeast S. cerevisiae S288C genome, cloned into pProEX HTb expression vector (Invitrogen), and expressed as N-terminal His6-TEV fusion proteins. C-terminal His6-tagged full-length Tah1 cloned into pET22b (EMD), full-length untagged Pih1 cloned into pET22b, and N-terminal His6-tagged Hsp82 cloned into pProEX HTb have been previously described (Zhao et al., 2005). The yeast Tah1 overexpression vector, p414Tah1, was constructed by cloning the TAH1 gene into yeast overexpression vector p414 (Mumberg et al., 1995), which contains a glyceraldehyde-3-phosphate dehydrogenase promoter.
To construct a plasmid suitable for the expression of ORFs with C-terminal 3xFLAG tag in yeast, a short DNA fragment, GCTTAATTAAATGGATTACAAGGATGACGACGATAAGGACTATAAGGACGATGATGACAAGGACTACAAAGATGATGACGATAAATAGGGCGCGCCAC, encoding the 3xFLAG sequence MDYKDDDDKDYKDDDDKDYKDDDDK was synthesized and inserted into the PacI–AscI sites of plasmid pFA6a-GFP(S65T)-kanMX6 (Longtine et al., 1998), generating pF6a-3FLAG-kanMX6. To make deg-TAH1, the degron cassette was obtained from the plasmid pRS315-KanMX-URA3pr-Ubi-Arg-DHFRts.
Protein purification
All proteins except Pih1 were expressed in E. coli BL21(DE3) gold (pRIL) and purified using Ni–nitrilotriacetic acid resin (QIAGEN) according to the manufacturer's protocols. The purified proteins were dialyzed and stored in buffer A (25 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10% glycerol, and 1 mM DTT) unless otherwise noted. TEV protease was used to remove the His6 tag to generate untagged proteins when required. Rvb1 and Rvb2 proteins were further purified on a Bio-Scale CHT5-I hydroxyapatite column (Bio-Rad Laboratories) and a MonoQ column (GE Healthcare) according to the manufacturers' protocols. Hsp82 and Tah1 were further purified on a Superdex 200 SE column (GE Healthcare).
Untagged Pih1 was purified as follows. 1 liter of culture of BL21(DE3) gold (pRIL) cells overexpressing untagged Pih1 was collected by centrifugation and lysed by sonication. Soluble proteins were precipitated with 40% ammonium sulfate. Protein pellet was resuspended in 2.5 ml of buffer B (50 mM potassium phosphate, pH 7.4, 100 mM KCl, 10% glycerol, and 1 mM DTT). The sample was desalted using the PD10 column, diluted to 50 ml, filtered through 0.45-μm syringe filters (Pall), and loaded onto an SP-Sepharose column equilibrated with buffer B. Pih1 was eluted using a gradient of KCl. Fractions containing Pih1 were dialyzed against buffer A, concentrated, and reloaded onto a Superdex 200 column. Soluble Pih1 eluted as a monomer at the corresponding position in the column and in the void volume of the column as an aggregate, Pih1agg, which was also partly bound to the 0.2-μM precolumn filter. The concentration of purified proteins was determined using the Bradford method (Bradford, 1976). All molar concentrations are for monomers except as indicated.
SE chromatography
SE chromatography was performed using a calibrated Superdex 200 HR 10/30 column (GE Healthcare) attached to a fast performance liquid chromatography system (AKTA; GE Healthcare). For samples containing Rvb1 and/or Rvb2 proteins, the column was equilibrated with buffer C (25 mM Tris-HCl, pH 7.5, 200 mM NaCl, 5 mM MgCl2, 10% glycerol, and 1 mM DTT). Otherwise, the column was equilibrated with buffer D (25 mM Tris-HCl, pH 7.5, 100 mM KCl, 5 mM MgCl2, 10% glycerol, and 1 mM DTT). The molecular mass standards (Sigma-Aldrich) used were thryoglobulin (669 kD), apoferritin (443 kD), β-amylase (200 kD), alcohol dehydrogenase (150 kD), BSA (66 kD), carbonic anhydrase (21 kD), and cytochrome c (12.4 kD). All experiments were performed at 4°C, and absorbance was monitored at 280 nm. Nucleotides were added to the buffer when needed.
Light scattering
Disaggregation of Pih1agg by Hsp82 and Tah1 was monitored by changes in the intensity of the scattered light of protein solution as previously described (Gribun et al., 2005). Measurements were performed at room temperature using a spectrofluorometer (Fluorolog 3-222; Horiba Jobin Yvon) with the excitation and emission wavelengths set at 350 nm and slits set to 2 nm. All solutions were filtered through 0.2-μm syringe filters (Gelman Laboratories) except for Pih1agg. Typically, 6 μM Pih1agg was added to buffer E (25 mM Tris-HCl, pH 7.5, 10 mM KCl, 5 mM MgCl2, and 10% glycerol) in the absence or presence of 4 mM ATP followed by the addition of 2 μM Hsp82 and 2 μM Tah1 in buffer E. Proteins were incubated for 5 min, and the intensity change of 90° scattered light was monitored over 60 min at room temperature. The background signal intensities were subtracted from all experiments.
AFM
Protein samples containing 5 μM Hsp82 and 15 μM Pih1agg in buffer E containing 5 mM ATP (Sigma-Aldrich) were incubated at 30°C for 60 min. Aliquots of 10 μl were taken at different time points and diluted 10 times with buffer E containing 20 mM geldanamycin. Samples were processed as previously described (Yip et al., 2000). In brief, solution tapping mode AFM images were acquired on a scanning probe microscope (Multimode; Veeco Metrology) with a Nanoscope IIIa controller (Veeco Metrology) and E scanner (13.6 × 13.6–μm maximum lateral scan area) using 120-μm V-shaped cantilevers with oxide-sharpened silicon nitride tips. The scan rate for imaging was 2 Hz, and the typical driving frequency was ∼7 kHz. 100 μl of incubation mixture containing geldanamycin was injected into a contact/tapping mode fluid cell sealed with a Teflon O-ring against freshly cleaved mica substrate and imaged at a resolution of 512 × 512 pixels. The images were flattened at order 0, and the plane was fitted at order 2. The particle diameter and height were determined using Nanoscope software version 4.42r9 (Veeco Metrology). Particle volumes were calculated assuming that the particle shape is a partially filled sphere.
Electron microscopy
Protein samples containing 5 μM Hsp82 and 15 μM Pih1agg in buffer F (25 mM Tris-HCl, pH 7.5, 10 mM KCl, 5 mM MgCl2, and 10% glycerol) containing 5 mM ATP (Sigma-Aldrich) were incubated at 30°C for 60 min. 10-μl aliquots were taken at different time points, diluted 10 times with buffer F containing 20 mM geldanamycin, applied on a carbon-coated copper grid, and negatively stained using uranyl acetate. The samples were examined and photographed using a transmission electron microscope (H-600; Hitachi) at an accelerating voltage of 75 kV. Images were captured with a CCD camera (Advantage HR; Advanced Microscopy Techniques).
Antibodies
Rabbit αTah1 and rabbit αHsp82 antibodies were generated as previously described (Zhao et al., 2005). Rabbit αPih1 antibody was raised using GST-Pih1 fusion protein and was a gift from M. Tyers (University of Toronto, Toronto, Canada). Rabbit αRvb1 and rabbit αRvb2 antibodies were raised using Rvb1 and Rvb2 proteins expressed and purified from E. coli. Monoclonal αFLAG antibody (F1804) was purchased from Sigma-Aldrich.
To prepare total crude protein samples from yeast for immunoblotting, cells were suspended in 200 μl of 20% TCA and broken by vortexing with 100 mg of 0.5-mm glass beads for 15 min. The lysates were spun at 20,000 g for 5 min. The protein pellet was then resuspended in 100 μl of 200 mM Tris-HCl buffer, pH 8.0, and subsequently analyzed by Western blotting.
Yeast strains
PCR-based gene deletion and tagging procedures were used (Longtine et al., 1998) to construct specific gene deletion mutants to make strains with genes that have a C-terminal 3xFLAG tag and to construct the deg-TAH1 strain. Hypomorphic DAmP alleles were constructed using PCR-based homologous recombination as previously described (Schuldiner et al., 2005). All yeast strains used in this work are listed in Table I.
For experiments using the deg-TAH1 strain, cells were grown in YPD medium to log phase (OD600 ≈ 0.6) or stationary phase (OD600 ≈ 3.0) at 30°C. Cycloheximide (C4859; Sigma-Aldrich) was added at a final concentration of 40 μg/ml, and the culture was then incubated at 38°C while shaking. Aliquots equivalent to 1 OD600 were withdrawn at 0 and 2 h after heat shock, and the cells were then collected by centrifugation. Microarray expression profiling of deletion strains was performed as previously described (Mnaimneh et al., 2004).
Protein complex purification and identification
Yeast cells containing specific ORFs having a C-terminal 3xFLAG tag were grown in YPD medium to log phase (OD600 ≈ 0.6) or stationary phase (OD600 > 3.0) at 30°C. Cell pellets were frozen and then broken using a coffee grinder cooled with dry ice. Protein complexes were purified using αFLAG antibody resin (A2220; Sigma-Aldrich) according to established protocols (Shen, 2004). For the isolation of Nop56–3FLAG complexes, only low salt washes were used.
Interaction screens
Yeast 2H analysis was performed as previously described (Zhao et al., 2005) using WT strains or PJ69-4a-tah1Δ and PJ69-4α-tah1Δ strains. For SGA analysis, MATα tah1Δ, MATα pih1Δ, MATα rvb1-DAmP, and MATα rvb2-DAmP strains were crossed to an array comprising ∼5,000 viable deletion mutant strains as previously described (Tong et al., 2004; Tong and Boone, 2006). Random spore analysis was performed as described previously (Tong and Boone, 2006).
Extraction of total yeast RNA
Yeast cells were grown in YPD media at 30°C with vigorous shaking to log phase (OD600 = 0.5–0.7) or stationary phase (3 d). Cells were harvested by centrifugation in a 50-ml centrifuge tube. Cell pellet was dissolved in buffer G (10 mM Tris-HCl, pH 7.4, 100 mM LiCl, 10 mM EDTA, and 0.2% SDS), and then phenol and glass beads were added. Cells were disrupted with vigorous shaking at 4°C. RNA was extracted with phenol/chloroform and precipitated with 1/10 vol of 5 M LiCl and 2.5 vol ethanol.
Microarray analysis of noncoding RNA
Analysis of noncoding RNA using microarrays was performed as previously described (Leeds et al., 2006). A microarray containing 44,000 70-mer oligonucleotide probes (Agilent Technologies) was used. The array was designed with overlapping probes covering all ribosomal and noncoding RNAs and included 2,300 control probes. Microarray quantification and data analysis were performed as described previously (Leeds et al., 2006) with the exception that normalization was performed by applying a Lowess function to all noncoding RNA features. Features that showed saturation and those that did not exceed background levels (signal intensity ≤ one SD from the mean) were excluded from further analysis.
Northern blot analysis
For pre-rRNA Northern blot analysis, 6 μg of total RNA was separated on 1.5% agarose-formaldehyde gels and transferred to a Biodyne B membrane (Pall) by capillary transfer. For snoRNA Northern blot analysis, 6 μg of total RNA was separated on 5% polyacrylamide-urea gels and electrotransferred to a Biodyne B membrane using 0.5× TBE (45 mM Tris-borate and 1 mM EDTA) as the transfer buffer. The membranes were hybridized in Church buffer (250 mM sodium phosphate, 1 mM EDTA, 1% BSA, and 7% SDS) using 5′-32P–labeled oligonucleotide probes. The hybridized membranes were exposed on x-ray film, and band intensities were quantified using Quantity One software (Bio-Rad Laboratories). Oligonucleotides used for pre-rRNA and snoRNA Northern blots were as follows: D-A2 for 35S (GAAAGAAACTTACAAGCCTAGCAAGACCGCGCACTTAAGCGCAGGCCCGG), U2 (GCGACCAAAGTAAAAGTCAAGAACGACTCCACAAGTGCGAGGGTCGCGAC), snR4 (ACACATGAACTATAACCTATCCTCATCGAC), snR13 (GGTAGCTTGAGTTTTTCCACACCGTTACTGATTTGGCAAAAGCCAAACAG), snR64 (TTGAGAATCTGTTGTCCCTATCTGGTTCCT), U14 (GCGGTCACCGAGAGTACTAACGA), U24 (TCAGAGATCTTGGTGATAATTGGTATGTCTCATTCGGAACTCAAAGTTCC), snR8 (CACTCGCGCAGTACCGATCTGGGCCAATGGGAGACACTAAAAGAAGCACC), snR36 (GTCATCCAGCTCAAGATCGTAATATTG), snR43 (CGAGACGCCGTCTACGGTTGTATC), and scR1 (TGTGCTATCCCGGCC-GCCTCCATCACGGGTCACCTTTGCTGACGCTGGAT).
RT-PCR
Total RNA extracted from WT yeast strain Y3068 and from tah1Δ strain R0061 was treated with the RNeasy Mini kit (QIAGEN) to remove possible DNA contaminants. cDNA was synthesized from total RNA using Reverse Transcription III (Invitrogen) with random primers. Primers P3 (GTGTTCCCGCTCATCATC) and P4 (AGTTGTAGCTCGGAGAGAAC) were used to amplify PIH1 cDNA fragment (Fig. S1). Primers P1 (CTGAATTAACAATGGATTCTG) and P2 (GCCAAATCGATTCTCAAAATG) were used to amplify ACT1 cDNA fragment as an internal control (Fig. S1). The amplified DNA fragments were resolved on a 1.2% agarose gel and stained with ethidium bromide.
Online supplemental material
Fig. S1 shows a semiquantitative RT-PCR of total RNA from WT and tah1Δ cells. Fig. S2 shows the three-dimensional topography and electron microscopy of Pih1agg at different time points during incubation with Hsp82 and ATP. Fig. S3 shows microarray expression profile analysis of tah1Δ and pih1Δ cells. Fig. S4 shows a comparison between the microarray and Northern blot results. Fig. S5 shows the composition of the box C/D core protein complex isolated from different mutant strains. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200709061/DC1.
Supplemental Material
Acknowledgments
We thank Steven Doyle (Microscopy Imaging Laboratory, University of Toronto, Toronto, Canada) for his help in the preparation and analysis of electron microscopy samples. We also thank Jeff Pootoolal, Tomas Babak, and Harm van Bakel for their help in carrying out and analyzing some of the microarray experiments. We also thank Dr. Edouard Bertrand (Institut de Génétique Moléculaire de Montpellier, Centre National de la Recherche Scientifique, Montpellier, France) for sharing the results from his group before publication.
Y. Kakihara and A. Gribun are postdoctoral fellows of the Canadian Institutes of Health Research (CIHR) Training Program Grant in Protein Folding: Principles and Diseases. W.A. Houry is a CIHR New Investigator. C.M. Yip is the recipient of a Canada Research Chair in Molecular Imaging. This work was supported, in part, by grants from the National Cancer Institute of Canada (to W.A. Houry), the CIHR (to C. Boone, C.M. Yip, and T.R. Hughes), the National Science and Engineering Research Council of Canada (C.M. Yip), Genome Canada through the Ontario Genomics Institute (to C. Boone and T.R. Hughes), and Genome Ontario (to C. Boone and T.R. Hughes).
R. Zhao, Y. Kakihara, and A. Gribun contributed equally to this paper.
Abbreviations used in this paper: 2H, two hybrid; AFM, atomic force microscopy; rRNA, ribosomal RNA; SE, size exclusion; SGA, synthetic genetic analysis; snRNA, small nuclear RNA; snoRNA, small nucleolar RNA; snoRNP, small nucleolar RNP; TEV, tobacco etch virus; TPR, tetratricopeptide repeat; WT, wild type.
References
- Ansar, S., J.A. Burlison, M.K. Hadden, X.M. Yu, K.E. Desino, J. Bean, L. Neckers, K.L. Audus, M.L. Michaelis, and B.S. Blagg. 2007. A non-toxic Hsp90 inhibitor protects neurons from Abeta-induced toxicity. Bioorg. Med. Chem. Lett. 17:1984–1990. [DOI] [PubMed] [Google Scholar]
- Auluck, P.K., and N.M. Bonini. 2002. Pharmacological prevention of Parkinson disease in Drosophila. Nat. Med. 8:1185–1186. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. [DOI] [PubMed] [Google Scholar]
- Dez, C., J. Noaillac-Depeyre, M. Caizergues-Ferrer, and Y. Henry. 2002. Naf1p, an essential nucleoplasmic factor specifically required for accumulation of box H/ACA small nucleolar RNPs. Mol. Cell. Biol. 22:7053–7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen, R.J., P. Wu, and A. Varshavsky. 1994. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science. 263:1273–1276. [DOI] [PubMed] [Google Scholar]
- Evans, C.G., S. Wisen, and J.E. Gestwicki. 2006. Heat shock proteins 70 and 90 inhibit early stages of amyloid beta-(1-42) aggregation in vitro. J. Biol. Chem. 281:33182–33191. [DOI] [PubMed] [Google Scholar]
- Fahy, D., A. Conconi, and M.J. Smerdon. 2005. Rapid changes in transcription and chromatin structure of ribosomal genes in yeast during growth phase transitions. Exp. Cell Res. 305:365–373. [DOI] [PubMed] [Google Scholar]
- Fatica, A., M. Dlakic, and D. Tollervey. 2002. Naf1 p is a box H/ACA snoRNP assembly factor. RNA. 8:1502–1514. [PMC free article] [PubMed] [Google Scholar]
- Filipowicz, W., and V. Pogacic. 2002. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 14:319–327. [DOI] [PubMed] [Google Scholar]
- Freeman, B.C., and K.R. Yamamoto. 2002. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science. 296:2232–2235. [DOI] [PubMed] [Google Scholar]
- Galardi, S., A. Fatica, A. Bachi, A. Scaloni, C. Presutti, and I. Bozzoni. 2002. Purified box C/D snoRNPs are able to reproduce site-specific 2'-O-methylation of target RNA in vitro. Mol. Cell. Biol. 22:6663–6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales, F.A., N.I. Zanchin, J.S. Luz, and C.C. Oliveira. 2005. Characterization of Saccharomyces cerevisiae Nop17p, a novel Nop58p-interacting protein that is involved in Pre-rRNA processing. J. Mol. Biol. 346:437–455. [DOI] [PubMed] [Google Scholar]
- Granato, D.C., F.A. Gonzales, J.S. Luz, F. Cassiola, G.M. Machado-Santelli, and C.C. Oliveira. 2005. Nop53p, an essential nucleolar protein that interacts with Nop17p and Nip7p, is required for pre-rRNA processing in Saccharomyces cerevisiae. FEBS J. 272:4450–4463. [DOI] [PubMed] [Google Scholar]
- Gray, J.V., G.A. Petsko, G.C. Johnston, D. Ringe, R.A. Singer, and M. Werner-Washburne. 2004. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 68:187–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribun, A., M.S. Kimber, R. Ching, R. Sprangers, K.M. Fiebig, and W.A. Houry. 2005. The ClpP double ring tetradecameric protease exhibits plastic ring-ring interactions, and the N termini of its subunits form flexible loops that are essential for ClpXP and ClpAP complex formation. J. Biol. Chem. 280:16185–16196. [DOI] [PubMed] [Google Scholar]
- Gribun, A., K.L.Y. Cheung, J. Huen, J. Ortega, and W.A. Houry. 2008. Yeast Rvb1 and Rvb2 are ATP-dependent DNA helicases that form a heterohexameric complex. J. Mol. Biol. In press. [DOI] [PubMed]
- Hiley, S.L., T. Babak, and T.R. Hughes. 2005. Global analysis of yeast RNA processing identifies new targets of RNase III and uncovers a link between tRNA 5′ end processing and tRNA splicing. Nucleic Acids Res. 33:3048–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura, T., V.V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang, M. Horikoshi, R. Scully, J. Qin, and Y. Nakatani. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 102:463–473. [DOI] [PubMed] [Google Scholar]
- Jonsson, Z.O., S.K. Dhar, G.J. Narlikar, R. Auty, N. Wagle, D. Pellman, R.E. Pratt, R. Kingston, and A. Dutta. 2001. Rvb1p and Rvb2p are essential components of a chromatin remodeling complex that regulates transcription of over 5% of yeast genes. J. Biol. Chem. 276:16279–16288. [DOI] [PubMed] [Google Scholar]
- Kim, T.S., C.Y. Jang, H.D. Kim, J.Y. Lee, B.Y. Ahn, and J. Kim. 2006. Interaction of Hsp90 with ribosomal proteins protects from ubiquitination and proteasome-dependent degradation. Mol. Biol. Cell. 17:824–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, T.H., W.A. Decatur, E. Bertrand, E.S. Maxwell, and M.J. Fournier. 2001. A well-connected and conserved nucleoplasmic helicase is required for production of box C/D and H/ACA snoRNAs and localization of snoRNP proteins. Mol. Cell. Biol. 21:7731–7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor, M.S., S. Venkatasubrahmanyam, M.D. Meneghini, J.W. Gin, J.L. Jennings, A.J. Link, H.D. Madhani, and J. Rine. 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2:E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N.J., M.C. Keogh, N. Datta, C. Sawa, O.W. Ryan, H. Ding, R.A. Haw, J. Pootoolal, A. Tong, V. Canadien, et al. 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell. 12:1565–1576. [DOI] [PubMed] [Google Scholar]
- Lafontaine, D.L., and D. Tollervey. 2000. Synthesis and assembly of the box C+D small nucleolar RNPs. Mol. Cell. Biol. 20:2650–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds, N.B., E.C. Small, S.L. Hiley, T.R. Hughes, and J.P. Staley. 2006. The splicing factor Prp43p, a DEAH box ATPase, functions in ribosome biogenesis. Mol. Cell. Biol. 26:513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., A. McKenzie III, D.J. Demarini, N.G. Shah, A. Wach, A. Brachat, P. Philippsen, and J.R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Matera, A.G., R.M. Terns, and M.P. Terns. 2007. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 8:209–220. [DOI] [PubMed] [Google Scholar]
- Mitsui, K., H. Nakayama, T. Akagi, M. Nekooki, K. Ohtawa, K. Takio, T. Hashikawa, and N. Nukina. 2002. Purification of polyglutamine aggregates and identification of elongation factor-1alpha and heat shock protein 84 as aggregate-interacting proteins. J. Neurosci. 22:9267–9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi, G., X. Shen, J. Landry, W.H. Wu, S. Sen, and C. Wu. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 303:343–348. [DOI] [PubMed] [Google Scholar]
- Mnaimneh, S., A.P. Davierwala, J. Haynes, J. Moffat, W.T. Peng, W. Zhang, X. Yang, J. Pootoolal, G. Chua, A. Lopez, et al. 2004. Exploration of essential gene functions via titratable promoter alleles. Cell. 118:31–44. [DOI] [PubMed] [Google Scholar]
- Mumberg, D., R. Muller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 156:119–122. [DOI] [PubMed] [Google Scholar]
- Nathan, D.F., M.H. Vos, and S. Lindquist. 1997. In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc. Natl. Acad. Sci. USA. 94:12949–12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, D.R., J.F. Kuhn, G.M. Shanab, and E.S. Maxwell. 2000. Box C/D snoRNA-associated proteins: two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA. 6:861–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl, L.H., and C. Prodromou. 2006. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu. Rev. Biochem. 75:271–294. [DOI] [PubMed] [Google Scholar]
- Peng, W.T., M.D. Robinson, S. Mnaimneh, N.J. Krogan, G. Cagney, Q. Morris, A.P. Davierwala, J. Grigull, X. Yang, W. Zhang, et al. 2003. A panoramic view of yeast noncoding RNA processing. Cell. 113:919–933. [DOI] [PubMed] [Google Scholar]
- Pratt, W.B., and D.O. Toft. 2003. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. (Maywood). 228:111–133. [DOI] [PubMed] [Google Scholar]
- Qiu, X.B., Y.L. Lin, K.C. Thome, P. Pian, B.P. Schlegel, S. Weremowicz, J.D. Parvin, and A. Dutta. 1998. An eukaryotic RuvB-like protein (RUVBL1) essential for growth. J. Biol. Chem. 273:27786–27793. [DOI] [PubMed] [Google Scholar]
- Radovic, S., V.A. Rapisarda, V. Tosato, and C.V. Bruschi. 2007. Functional and comparative characterization of Saccharomyces cerevisiae RVB1 and RVB2 genes with bacterial Ruv homologues. FEMS Yeast Res. 7:527–539. [DOI] [PubMed] [Google Scholar]
- Savchenko, A., A. Yee, A. Khachatryan, T. Skarina, E. Evdokimova, M. Pavlova, A. Semesi, J. Northey, S. Beasley, N. Lan, et al. 2003. Strategies for structural proteomics of prokaryotes: quantifying the advantages of studying orthologous proteins and of using both NMR and X-ray crystallography approaches. Proteins. 50:392–399. [DOI] [PubMed] [Google Scholar]
- Scheufler, C., A. Brinker, G. Bourenkov, S. Pegoraro, L. Moroder, H. Bartunik, F.U. Hartl, and I. Moarefi. 2000. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 101:199–210. [DOI] [PubMed] [Google Scholar]
- Schlatter, H., T. Langer, S. Rosmus, M.L. Onneken, and H. Fasold. 2002. A novel function for the 90 kDa heat-shock protein (Hsp90): facilitating nuclear export of 60 S ribosomal subunits. Biochem. J. 362:675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner, M., S.R. Collins, N.J. Thompson, V. Denic, A. Bhamidipati, T. Punna, J. Ihmels, B. Andrews, C. Boone, J.F. Greenblatt, et al. 2005. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 123:507–519. [DOI] [PubMed] [Google Scholar]
- Shen, X. 2004. Preparation and analysis of the INO80 complex. Methods Enzymol. 377:401–412. [DOI] [PubMed] [Google Scholar]
- Sittler, A., R. Lurz, G. Lueder, J. Priller, H. Lehrach, M.K. Hayer-Hartl, F.U. Hartl, and E.E. Wanker. 2001. Geldanamycin activates a heat shock response and inhibits huntingtin aggregation in a cell culture model of Huntington's disease. Hum. Mol. Genet. 10:1307–1315. [DOI] [PubMed] [Google Scholar]
- Te, J., L. Jia, J. Rogers, A. Miller, and S.D. Hartson. 2007. Novel subunits of the mammalian Hsp90 signal transduction chaperone. J. Proteome Res. 6:1963–1973. [DOI] [PubMed] [Google Scholar]
- Tong, A.H., and C. Boone. 2006. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 313:171–192. [DOI] [PubMed] [Google Scholar]
- Tong, A.H., G. Lesage, G.D. Bader, H. Ding, H. Xu, X. Xin, J. Young, G.F. Berriz, R.L. Brost, M. Chang, et al. 2004. Global mapping of the yeast genetic interaction network. Science. 303:808–813. [DOI] [PubMed] [Google Scholar]
- Watkins, N.J., A. Dickmanns, and R. Luhrmann. 2002. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol. Cell. Biol. 22:8342–8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins, N.J., I. Lemm, D. Ingelfinger, C. Schneider, M. Hossbach, H. Urlaub, and R. Luhrmann. 2004. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol. Cell. 16:789–798. [DOI] [PubMed] [Google Scholar]
- Winzeler, E.A., D.D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J.D. Boeke, H. Bussey, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 285:901–906. [DOI] [PubMed] [Google Scholar]
- Yip, C.M., M.L. Brader, B.H. Frank, M.R. DeFelippis, and M.D. Ward. 2000. Structural studies of a crystalline insulin analog complex with protamine by atomic force microscopy. Biophys. J. 78:466–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, R., M. Davey, Y.C. Hsu, P. Kaplanek, A. Tong, A.B. Parsons, N. Krogan, G. Cagney, D. Mai, J. Greenblatt, et al. 2005. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 120:715–727. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.