Abstract
TRIM5 proteins constitute a class of restriction factors that prevent host cell infection by retroviruses from different species. TRIM5α restricts retroviral infection early after viral entry, before the generation of viral reverse transcription products. However, the underlying restriction mechanism remains unclear. In this study, we show that during rhesus macaque TRIM5α (rhTRIM5α)–mediated restriction of HIV-1 infection, cytoplasmic HIV-1 viral complexes can associate with concentrations of TRIM5α protein termed cytoplasmic bodies. We observe a dynamic interaction between rhTRIM5α and cytoplasmic HIV-1 viral complexes, including the de novo formation of rhTRIM5α cytoplasmic body–like structures around viral complexes. We observe that proteasome inhibition allows HIV-1 to remain stably sequestered into large rhTRIM5α cytoplasmic bodies, preventing the clearance of HIV-1 viral complexes from the cytoplasm and revealing an intermediate in the restriction process. Furthermore, we can measure no loss of capsid protein from viral complexes arrested at this intermediate step in restriction, suggesting that any rhTRIM5α-mediated loss of capsid protein requires proteasome activity.
Introduction
The TRIM5 family of proteins has recently been identified as a class of restriction factors that can act as a barrier to the cross-species transmission of retroviruses, which is exemplified by the ability of TRIM5α from rhesus macaques (rhesus macaque TRIM5α [rhTRIM5α]) to restrict HIV-1 infection (Stremlau et al., 2004). TRIM5α-mediated restriction of retroviral infection involves the specific recognition of determinants present on the capsid of the targeted retrovirus (Bieniasz, 2004; Goff, 2004; Stremlau et al., 2004). These determinants in retroviral capsids are recognized by the C-terminal B30.2 domain of TRIM5 proteins, and the amino acids that dictate antiviral specificity have been identified (Stremlau et al., 2005; Yap et al., 2005). Moreover, in vitro studies demonstrate that TRIM5 proteins associate with viral capsid structures (Sebastian and Luban, 2005; Stremlau et al., 2006). Importantly, these studies show that TRIM5α does not recognize or bind free capsid protein but rather binds capsid protein in the context of an intact, mature viral core (Sebastian and Luban, 2005; Stremlau et al., 2006).
Although the regions within the B30.2 domain that contribute to retroviral capsid binding have been mapped, less is known about the mechanism by which TRIM5 proteins restrict retroviral infection after capsid recognition. It is known that virions encounter this replicative block before the accumulation of viral reverse transcription (RT) products (Towers et al., 2000; Cowan et al., 2002; Stremlau et al., 2004).
Our laboratory has recently found that proteasome inhibition abrogates the ability of TRIM5 proteins to prevent the accumulation of RT products but does not relieve the ability of TRIM5 proteins to restrict viral infection (Anderson et al., 2006; Wu et al., 2006). In the presence of proteasome inhibitor, virions complete RT and form functional preintegration complexes (PICs), but infection and 2–long terminal repeat circle formation remains impaired (Anderson et al., 2006; Wu et al., 2006). As 2–long terminal repeat circles can act as surrogate markers of the nuclear entry of retroviral cDNA, this suggests that incoming viral PICs are unable to access the nucleus under these conditions, although the mechanism by which nuclear access is inhibited is currently unknown.
TRIM5 proteins belong to a larger family of TRIM proteins that were originally observed to oligomerize into high-order structures localizing to specific cellular compartments in the cytoplasm and nucleus (Reymond et al., 2001). rhTRIM5α and its primate counterparts are known to form discreet concentrations in the cytoplasm referred to as cytoplasmic bodies (Stremlau et al., 2004). A second population of TRIM5α exists with a diffuse cytoplasmic localization. Formation of the cytoplasmic bodies is concentration dependent. We have recently reported that TRIM5α is continuously exchanging between the cytoplasmic pool and the subset of rhTRIM5α protein present in cytoplasmic bodies (Campbell et al., 2007a). The potential importance of TRIM5α cytoplasmic bodies remains controversial, primarily because there are no antibodies available that can determine whether these structures are formed by endogenous expression levels of the protein. Preliminary examination of the role of these bodies in restriction showed that preexisting cytoplasmic bodies were not required for retroviral restriction by TRIM5 proteins (Perez-Caballero et al., 2005b; Song et al., 2005). Importantly, however, these studies only examined the requirement for preexisting cytoplasmic bodies in restriction and did not examine TRIM5 localization after virus addition. For clarity, we define a cytoplasmic body as being a discrete, observable accumulation of rhTRIM5α signal above background.
In this study, we use fluorescently labeled HIV-1 particles to examine the fate of these viral complexes during restriction. We find that HIV-1 viral complexes associate with rhTRIM5α cytoplasmic bodies in live and fixed cell experiments. We also find that proteasome inhibition prevents the clearance of HIV-1 viral complexes from the cytoplasm and leads to the stable sequestration of these complexes in cytoplasmic bodies, where they still contain readily detectable amounts of p24. Collectively, these results suggest a model in which the association of discreet concentrations of rhTRIM5α (currently known as cytoplasmic bodies) with cytoplasmic HIV-1 viral complexes is a key intermediate in the restriction process leading to the proteasome-dependent destruction of these viral complexes.
Results
HIV-1 viral complexes accumulate in rhTRIM5α cytoplasmic bodies after proteasome inhibition
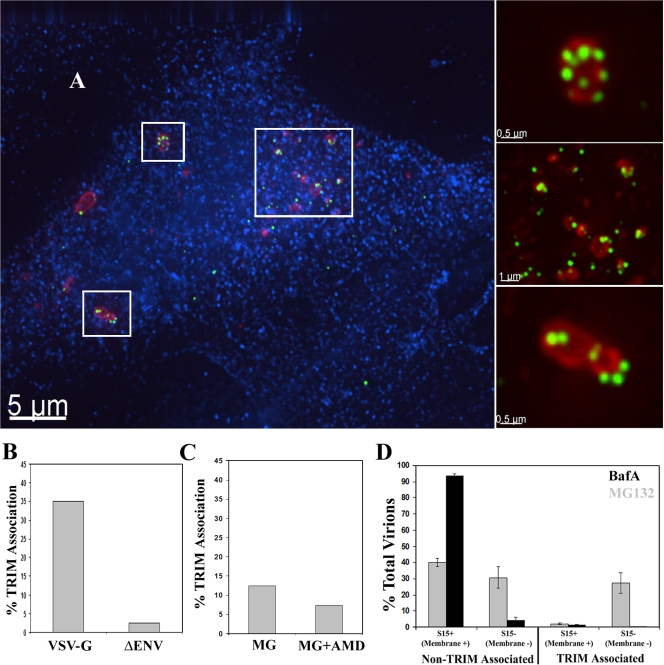
We first examined the fate of HIV-1 particles in restricted cells after proteasome inhibition. To this end, HeLa cells stably expressing HA-tagged rhTRIM5α, which have been shown to potently restrict HIV-1 infection (Stremlau et al., 2004), were incubated with GFP-Vpr–labeled HIV-1 virions (McDonald et al., 2002) in the presence of the proteasome inhibitor MG132 for 6 h, and the localization of TRIM5α and HIV-1 viral complexes was examined. As we previously reported, rhTRIM5α localized into large cytoplasmic bodies after 6 h of MG132 treatment (Fig. 1 A; Wu et al., 2006). HIV-1 viral complexes clearly localized to these cytoplasmic bodies, and accumulations of viral complexes were often observed in these large cytoplasmic bodies (Fig. 1 A, insets), where they appear to be sequestered.
Figure 1.
Proteasome inhibition induces the accumulation of HIV-1 in rhTRIM5α cytoplasmic bodies. (A) rhTRIM5α-HA–expressing HeLa cells were continuously infected with GFP-Vpr–labeled HIV-1 virions (green) in the presence of 1 μg/ml MG132 proteasome inhibitor for 6 h. Cells were then fixed and immunostained with antibodies to HA (red) or the 20s proteasomal subunit (blue) and imaged. The boxed regions are enlarged in the insets to the right for better visualization of the described interaction. The provided image is one z section from a deconvolved z-stack image. (B) The percentage of HIV-1 virions associated with rhTRIM5α cytoplasmic bodies was calculated for virions lacking envelope protein (ΔEnv) or pseudotyped with the VSV-G envelope protein (VSV-G). The mean of two independent experiments is shown in which a total of 698 virions (VSV-G) and 837 virions (ΔEnv) were counted. (C) rhTRIM5α-HA–expressing HeLa cells were spinoculated with GFP-Vpr–labeled NL43 virions in the presence of 1 μg/ml MG132, and the percentage of HIV-1 virions associated with rhTRIM5α cytoplasmic bodies was calculated for virions infected in the presence of absence of AMD3100. The mean of two independent experiments is shown in which a total of 369 virions (MG132) and 418 virions (MG132 + AMD3100) were counted. (D) The ability of fused and unfused HIV-1 virions to associate with rhTRIM5α cytoplasmic bodies was analyzed using HIV-1 dually labeled with S15-mCherry and GFP-Vpr. Cells were infected in the presence of 1 μg/ml MG132 (gray bars) or 20 nM BafA (black bars). Error bars represent the SEM from three experiments in which a total of 2,578 or 1,445 virions were counted for cells treated with MG132 or BafA, respectively.
TRIM5 proteins restrict retroviruses by interacting with incoming retroviral cores that are released into the cell cytoplasm after fusion of the virion and cell membranes (Bieniasz, 2004; Goff, 2004). Therefore, we investigated whether the association of HIV-1 viral complexes and rhTRIM5α cytoplasmic bodies during proteasome inhibition (Fig. 1 A) was specific to viral complexes that had fused and entered the cytoplasm.
First, rhTRIM5α cells were incubated for 6 h in MG132 with HIV-1 lacking a functional envelope protein (ΔEnv) to block fusion or were pseudotyped with vesicular stomatitis virus G (VSV-G) envelope protein to allow the productive entry of virions into target cells. HIV-1 localization with rhTRIM5α bodies was compared (Fig. 1 B and Table I). In data compiled from two independent experiments, 35.1% of 698 VSV-G–pseudotyped virions localized to rhTRIM5α cytoplasmic bodies. Conversely, only 2.5% of 837 viral complexes lacking envelope protein associated with rhTRIM5α under similar conditions, indicating that it is the fused viral complexes that have entered the cytoplasm and are susceptible to restriction that interact with rhTRIM5α cytoplasmic bodies formed in the presence of the proteasome inhibitor.
Table I. Comparative analysis of HIV-1 association with rhTRIM5α.
| Viral envelope |
Cell type | Treatment | Total TRIM association |
TRIM association
|
|
|---|---|---|---|---|---|
| S15+
(unfused) |
S15−
(fused) |
||||
| % | % | % | |||
| VSV-G | rhTRIM-HA HeLa | MG132 | 35.1 (Fig. 1 B) | ND | ND |
| None | rhTRIM-HA HeLa | MG132 | 2.5 (Fig. 1 B) | ND | ND |
| HIV | rhTRIM-HA HeLa | MG132 | 12.4 (Fig. 1 C) | ND | ND |
| HIV | rhTRIM-HA HeLa | MG132 + AMD3100 | 7.3 (Fig. 1 C) | ND | ND |
| VSV-G | rhTRIM-HA HeLa | BafA | 1.7a | 1.5 (Fig. 1 D) | 5.1 (Fig. 1 D) |
| VSV-G | rhTRIM-HA HeLa | MG132 | 28.6a | 5.0 (Fig. 1 D) | 44.7 (Fig. 1 D) |
| VSV-G | rhTRIM-HA CRFK | No drug | 17.8a | 4.7 (Fig. 3 B) | 32.1 (Fig. 3 B) |
| VSV-G | rhΔpatch-HA CRFK | No drug | 5.1a | 3.2 (Fig. 3 B) | 7.5 (Fig. 3 B) |
| VSV-G | rhTRIM-HA CRFK | MG132 | 13.7a | 5.4 (Fig. 3 C) | 26.5 (Fig. 3 C) |
| VSV-G | rhΔpatch-HA CRFK | MG132 | 6.9a | 6.2 (Fig. 3 C) | 7.9 (Fig. 3 C) |
Data from the experiments averaged in Figs. 1 and Figs.3 were pooled to examine the percentage of all viral particles associating with rhTRIM5α under all conditions examined. The figure from which the pooled data were derived is indicated in parentheses.
In the case of experiments examining S15-mCherry, data were pooled to determine the degree of rhTRIM5α associations in both S15+ and S15− populations.
We also examined whether this association occurred when viral entry into target cells was mediated by the HIV-1 envelope protein. To allow for entry of these wild-type HIV-1 virions into rhTRIM5α-expressing HeLa cells, the cells were transfected with a CFP-CD4 fusion protein that has been previously shown to be functional in mediating HIV-1 envelope–mediated fusion (Steffens and Hope, 2003). These transfected cells were then spinoculated (O'Doherty et al., 2000) at 17°C with GFP-Vpr–labeled wild-type NL43 virus in the presence of MG132. The CXCR4-specific fusion inhibitor AMD3100 was included as a control in these experiments. When the degree of TRIM5α association of these viral complexes was determined, 12.4% of 418 viral complexes were observed to associate with TRIM5α signal 6 h after infection (Fig. 1 C and Table I), whereas only 7.3% of 369 viral complexes were observed to associate with TRIM5α signal when the infection was performed in the presence of AMD3100. This difference in the degree of association observed between viral complexes pseudotyped with the VSV-G envelope protein and those possessing native HIV-1 envelope is likely because VSV-G mediates the cytoplasmic entry of viral complexes much more efficiently than HIV-1 envelope (Luo et al., 1998). However, given the relatively smaller degree of association observed in this system, and as restriction by TRIM5α is known to occur independently of the envelope protein used (Hofmann et al., 1999; Stremlau et al., 2004), we used virions pseudotyped with the VSV-G envelope protein in all subsequent experiments.
To gain further insight into the association of HIV-1 and rhTRIM5α, we also examined this interaction using HIV-1 virions dually labeled with GFP-Vpr and S15-mCherry to identify virions that had productively entered the target cell. We have recently demonstrated that this S15-mCherry protein is efficiently incorporated into HIV-1 virions (Campbell et al., 2007b). As this fluorescent membrane label is lost from the virion core after fusion, this system can effectively discriminate between virions that remain intact and unfused (S15+ and Vpr+) from those that have fused and productively entered the cell cytoplasm (S15− and Vpr+). rhTRIM5α-expressing cells were incubated with these dually labeled virions in the presence of MG132 for 6 h, and virion association with rhTRIM5α bodies was assessed. As a control, the experiment was also performed with bafilomycin A (BafA), which blocks endosomal acidification by inhibiting the vacuolar ATPase and thus blocks the pH-dependent fusion of VSV-G–pseudotyped virions.
In the control experiment with the BafA fusion inhibitor, the viral population was almost exclusively dually labeled unfused virus that did not appreciably associate with rhTRIM5α (Fig. 1 D, black bars; and Table I). In data compiled from three independent experiments examining a total of 1,445 viral complexes, 93.1% of viral complexes were S15+ (unfused) and not associated with rhTRIM5α in the presence of BafA. We also observed 5.1% of viral complexes that were S15− and not associated with rhTRIM5α bodies (Fig. 1 D and Table I), although these viral complexes likely represent virions inefficiently labeled with the S15-mCherry protein (Campbell et al., 2007b). Importantly, only 1.7% of these viral complexes (S15+ and S15−) associated with rhTRIM5α bodies (Fig. 1 D and Table I) in the presence of this fusion inhibitor.
In the MG132-treated cells, a greater percentage of HIV-1 associated with rhTRIM5α signal compared with the BafA control (Fig. 1 C and Table I). Moreover, the population of HIV-1 that associated with rhTRIM5α signal during MG132 treatment had fused and lost their membrane (S15−). In 2,578 viral complexes counted from four independent experiments, 26.6% of viral complexes were S15− (fused) and associated with rhTRIM5α bodies compared with only 2.0% of S15+ (unfused) viral complexes associated with rhTRIM5α bodies (Fig. 1 C and Table I). Therefore, these data collectively reveal that incoming HIV-1 viral complexes that have fused and entered the cell cytoplasm can associate with rhTRIM5α cytoplasmic bodies in cells treated with proteasome inhibitors. Sequestration within these bodies provides a plausible mechanistic explanation for our previous observation that proteasome inhibitors allow RT and PIC formation by restricted viral complexes but that these complexes are unable to enter the nucleus (Anderson et al., 2006; Wu et al., 2006).
HIV-1 viral complexes associate with rhTRIM5α bodies in the absence of drug
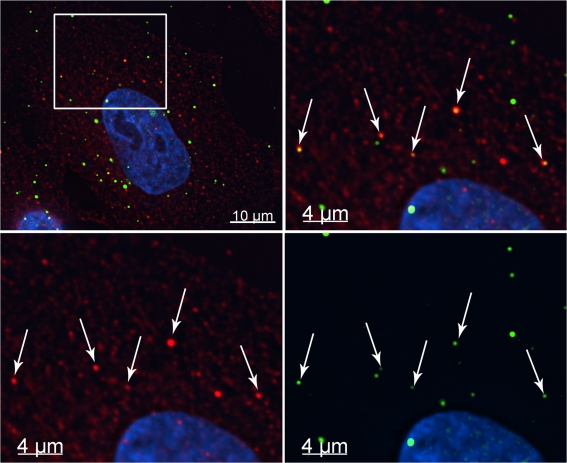
As HIV-1 interacted with rhTRIM5α cytoplasmic bodies during proteasome inhibition, we examined whether HIV-1 also interacted with rhTRIM5α bodies in the absence of drug. When rhTRIM5α-expressing cells were continuously infected with GFP-Vpr–labeled virions for 2 h in the absence of drug, it was possible to observe HIV-1 viral complexes associating with rhTRIM5α signal, although rhTRIM5α accumulated in smaller cytoplasmic bodies (Fig. 2) compared with the relatively larger bodies observed after MG132 treatment (Fig. 1 A). This association was only detectable at earlier times after infection as opposed to the case of proteasome inhibition, which induces the stable accumulation of viral complexes within enlarged rhTRIM5α cytoplasmic bodies. Moreover, although the association between HIV-1 and rhTRIM5α was readily observed in the presence of proteasome inhibitor, this interaction in the absence of drug was not frequently observable. This is consistent with the notion that in the absence of drug, restriction occurs rapidly after the entry of viral complexes into the target cell as previously reported (Perez-Caballero et al., 2005b).
Figure 2.
HIV-1 interacts with rhTRIM5α cytoplasmic bodies in the absence of drug. (A) rhTRIM5α-HA–expressing HeLa cells were continuously infected with GFP-Vpr–labeled HIV-1 virions (green) for 2 h. Cells were fixed and immunostained with anti-HA antibody (red) or Hoechst to visualize cellular DNA (blue). Arrows indicate HIV-1 virions associated with rhTRIM5α cytoplasmic bodies. The boxed area in the top left panel has been enlarged in the other three panels, and the fluorescent signals for either rhTRIM5α, GFP-Vpr HIV-1, or both (merge) are shown for clarity. The provided image is one z section from a deconvolved z-stack image.
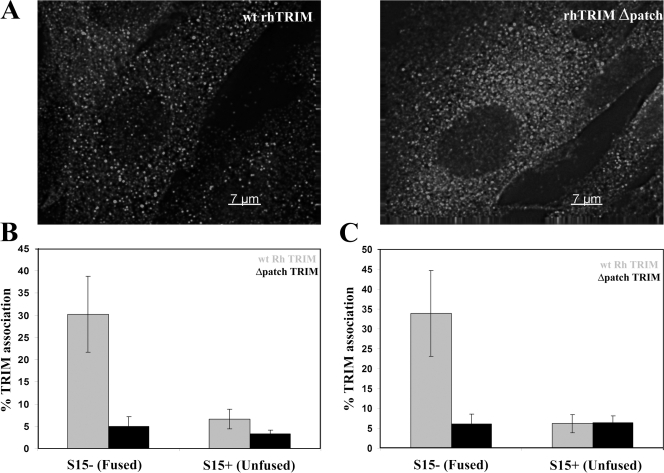
HIV-1 viral complex association with rhTRIM5α bodies requires the rhTRIM5α B30.2 domain that recognizes HIV-1 capsid and is essential for HIV-1 restriction. Recent studies show that elements in the C-terminal B30.2 domain of TRIM5 proteins mediate their ability to recognize specific retroviral capsids (Sawyer et al., 2005; Stremlau et al., 2005; Yap et al., 2005). Moreover, removal of a positively selected 13–amino acid patch of this domain (Δpatch) abrogates the ability of rhTRIM5α to restrict HIV-1 infection (Sawyer et al., 2005). Therefore, we next tested whether this region in the rhTRIM5α B30.2 domain, which is critical for recognizing retroviral capsid and restricting HIV-1, was also required for HIV-1 association with rhTRIM5α cytoplasmic bodies.
To this end, we used CRFK feline cells stably expressing HA-tagged versions of either wild-type rhTRIM5α or rhTRIM5α containing the Δpatch mutations in the B30.2 domain (Sawyer et al., 2005). Microscopic examination of both cell lines revealed that both the wild-type and Δpatch rhTRIM5α formed cytoplasmic bodies (Fig. 3 A). These cells were then infected with S15-mCherry and GFP-Vpr dually labeled HIV-1 virions, and association of these viral complexes with the wild-type or Δpatch rhTRIM5α bodies was examined. For these experiments, cells were synchronously infected as previously described (Campbell et al., 2007b). Cells were infected without drug and fixed 90 min after the infection was initiated by raising the temperature to 37°C (Fig. 3 B). For the cells expressing wild-type rhTRIM5α, 32.2% of 432 fused (S15−) viral complexes associated with rhTRIM5α bodies over three independent experiments. In contrast, only 4.7% of 473 unfused (S15+) viral complexes associated with rhTRIM5α cytoplasmic bodies, again demonstrating that HIV-1 interacts with rhTRIM5α bodies after fusion and entry into the cytoplasm (Fig. 3 B and Table I). Conversely, for the cells expressing rhTRIM5αΔpatch, only 7.1% of 379 fused (S15−) viral complexes associated with cytoplasmic bodies. This degree of association likely represents the background degree of association in these experiments, as a similar degree of association was observed for unfused viral complexes (S15+) in both cell lines.
Figure 3.
HIV-1 association with rhTRIM5α cytoplasmic bodies requires the rhTRIM5α B30.2 domain essential for recognizing HIV-1 capsids and restriction. (A) CRFK cells stably expressing HA-tagged wild-type (wt) rhTRIM5α or rhTRIM5αΔpatch were immunostained with anti-HA antibody, and protein localization was visualized microscopically. The provided image is one z section from a deconvolved z-stack image. (B and C) Each cell line was also infected with VSV-G–pseudotyped HIV-1 virions dually labeled with S15-mCherry and GFP-Vpr in a synchronized infection. Cells were fixed either 90 min after infection (B) or 4 h after infection in the presence of MG132 (C), immunostained with anti-HA antibody, and imaged. The association of both unfused (S15+) and fused (S15−) HIV-1 virions with the HA-positive cytoplasmic bodies was examined in both cell lines. The mean results from three independent experiments are graphed, counting a total of 1,716 (B) and 3,381 virions (C). Error bars represent the SEM from three experiments.
Similar experiments were performed in the presence of MG132 using cells analyzed 4 h after synchronized infection (Fig. 3 C and Table I). For the cells expressing wild-type rhTRIM5α, 26.4% of 654 fused (S15−) viral complexes associated with rhTRIM5α bodies, whereas only 5.4% of 1,014 unfused (S15+) viral complexes associated with rhTRIM5α bodies (Fig. 3 C and Table I). Moreover, for the cells expressing rhTRIM5αΔpatch, only 7.9% of 693 fused (S15−) viral complexes and 6.2% of 1,020 unfused (S15+) viral complexes associated with the rhTRIM5αΔpatch bodies with MG132 treatment (Fig. 3 C and Table I). Again, this level likely represents the background amount of association observed in these experiments. Therefore, both with and without MG132, rhTRIM5α requires the patch region in its B30.2 domain for rhTRIM5α cytoplasmic bodies to efficiently interact with fused HIV-1 viral complexes (Fig. 3, B and C) as well as mediate restriction (Sawyer et al., 2005). This indicates that rhTRIM5α recognition and restriction of HIV-1 viral complexes correlates with and involves HIV-1 interaction with rhTRIM5α cytoplasmic bodies.
Proteasome inhibition causes rhTRIM5α-specific accumulation of cytoplasmic HIV-1 viral complexes in target cells
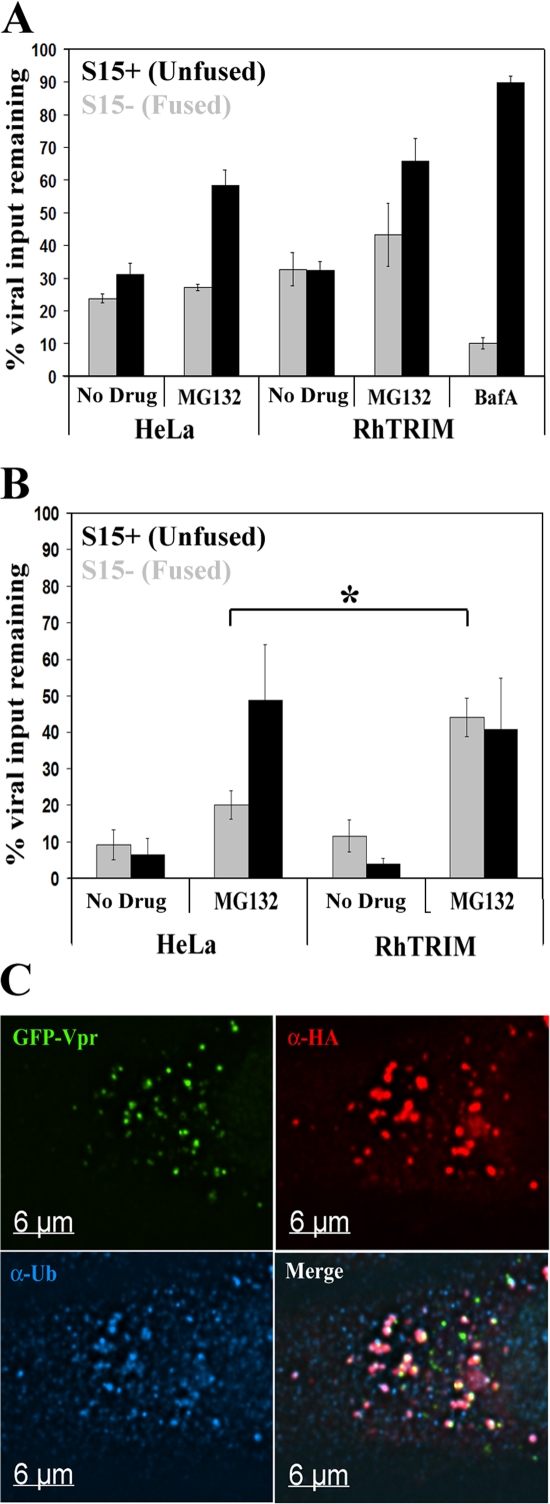
To examine viral populations under conditions of restriction and proteasome inhibition, we performed a synchronized infection with S15-mCherry– and GFP-Vpr–labeled HIV-1 virions and compared the viral populations present in HeLa cells stably expressing rhTRIM5α or control HeLa cells at 1 or 4 h after infection, both in the presence or absence of MG132. The number of S15− (fused) and S15+ (unfused) viral complexes per cell was counted for each sample. To normalize for differences in viral input between experiments, these values were expressed as percentages of the total number of viral complexes per cell counted in the control sample containing BafA, which blocks VSV-G–mediated fusion and viral degradation mediated by endosomal acidification (Fig. 4 A).
Figure 4.
Proteasome function is required for rhTRIM5α-mediated loss of HIV-1 virions. HeLa cells or rhTRIM5α-HA–expressing HeLa cells were synchronously infected with S15-mCherry– and GFP-Vpr–labeled virions in the presence of 1 μg/ml MG132 or 20 nM BafA or in the absence of drug as indicated. (A and B) Cells were fixed for 1 (A) or 4 h (B) after the initiation of infection, and the number of fused (S15+; black bars) and unfused (S15−; gray bars) virions per cell was calculated. All values are expressed as a percentage of the total virions per cell (fused and unfused) present in the BafA sample at 1 h. The mean results from three independent experiments examining a total of 493 cells and 11,727 virions are graphed. Error bars represent the SEM from three independent experiments. The difference in values indicated by the asterisk are statistically significant (*, P = 0.0152). (C) HeLa cells stably expressing rhTRIM5α were continuously infected with GFP-Vpr–labeled HIV-1 virions (green) in the presence of 1 μg/ml MG132 for 6 h. Cells were then fixed and immunostained with anti-HA antibody (red) and antiubiquitin antibody (blue). The provided image is one z section from a deconvolved z-stack image.
At 1 and 4 h after infection, similar numbers of fused viral complexes were observed in the control HeLa and rhTRIM5α-expressing HeLa cells in the absence of MG132 (Fig. 4, A and B), suggesting that virions productively enter both cell types at similar rates. MG132 treatment increased the number of unfused virions present in both cell types 1 and 4 h after infection. This is likely caused by the fact that monoubiquitination regulates trafficking within the endosomal/lysosomal pathway (for review see d'Azzo et al., 2005). Therefore, depletion of the ubiquitin pool by proteasome inhibitors alters vesicular trafficking. This would change the rate at which these pseudotyped virions are able to productively enter cells using the VSV-G envelope protein and subsequently increase the amount of virus within the endosomal compartment.
Importantly, in addition to this global effect of delayed entry kinetics, we also observed a specific increase in the number of fused viral complexes present in rhTRIM5α-expressing cells relative to HeLa control cells in the presence of MG132 (Fig. 4 B). This trend is apparent at 1 h after infection and becomes more pronounced at 4 h after infection. At 4 h after infection, this difference is statistically significant (P = 0.0152). This indicates that MG132 is specifically increasing the number of fused viral complexes in the rhTRIM5α-expressing cells compared with the HeLa cells by 4 h after infection. This observation correlates well with our previous report that MG132 treatment allows the accumulation of HIV-1 RT products and formation of functional PICs but does not abrogate the ability of rhTRIM5α to restrict infection (Anderson et al., 2006; Wu et al., 2006). Here, the increased number of fused cytoplasmic HIV-1 viral complexes observed in the presence of MG132 indicates that rhTRIM5α can effectively interrupt the normal trafficking of viral complexes under these conditions.
rhTRIM5α cytoplasmic bodies containing HIV-1 viral complexes after proteasome inhibition stain positively for ubiquitin
As proteasome-mediated degradation requires ubiquitination, we examined whether the rhTRIM5α bodies containing sequestered HIV-1 viral complexes also contained ubiquitin. In the presence of MG132, the cytoplasmic bodies stained positively for ubiquitin (Fig. 4 C). Ubiquitin localized to entire cytoplasmic body structures, similar to a previous study (Diaz-Griffero et al., 2006), but did not intensify on or around viral complexes (Fig. 4 C). This demonstrates that rhTRIM5α cytoplasmic bodies containing HIV-1 viral complexes also contain ubiquitin and further suggests that these complexes could be degraded in a proteasome-mediated manner in the absence of MG132.
The viral intermediate associated with cytoplasmic bodies after proteasome inhibition contains abundant capsid protein
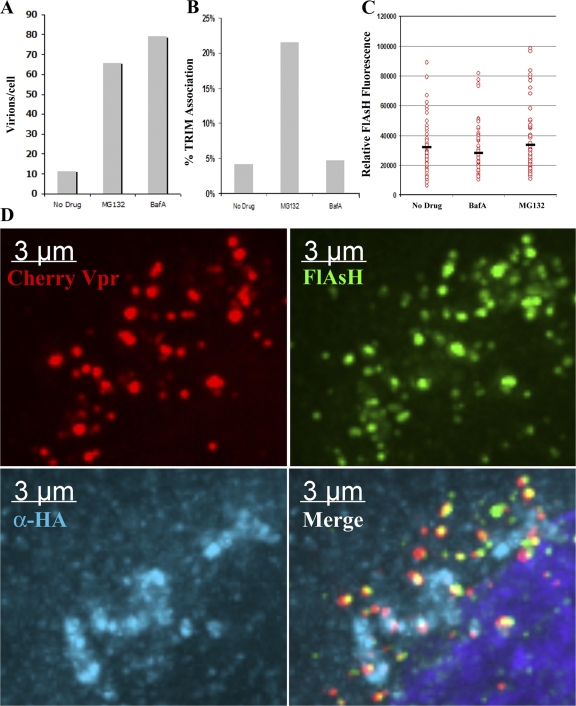
HIV-1 capsid protein (p24CA) forms the conical capsid structure containing the viral genome in mature virions. Interaction of rhTRIM5α with HIV-1 is mediated through interaction with this structure (Stremlau et al., 2006). This conical capsid is released into the cytoplasm after fusion between virion and target cell membranes. It has recently been suggested by other groups that TRIM5α restriction may occur via a mechanism that involves either the targeted degradation of p24CA from viral cores (Chatterji et al., 2006) or the accelerated loss of p24CA from the viral core during uncoating (Stremlau et al., 2006). Previously, we have been able to identify the association of p24CA with RT complexes using a p24CA-specific monoclonal antibody (McDonald et al., 2002). However, because of the protein-dense nature of the cytoplasmic bodies induced after proteasome inhibition, we were concerned that antibodies may have a limited ability to penetrate these structures during immunostaining, potentially confounding our analysis. Therefore, to investigate the effect of rhTRIM5α on p24CA association with cytoplasmic HIV-1 viral complexes, we developed an HIV-1 strain containing a tetracysteine tag within the capsid protein (HIVFC). This tag can be effectively labeled with the fluorescent FlAsH reagent (Adams et al., 2002). By combining this technology with our ability to fluorescently label virions with mCherry-Vpr, we sought to determine the presence or absence of capsid protein on HIV-1 viral complexes sequestered within rhTRIM5α cytoplasmic bodies in the presence of proteasome inhibitor.
Initial characterization of this HIVFC virus indicated that insertion of the tag into the capsid protein severely impaired the ability of the virus to infect unrestricted HeLa target cells (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200706154/DC1). However, using an approach described by others previously (Muller et al., 2004), we found that virions produced by transfecting an equal amount of wild-type HIV-1 and modified HIVFC plasmids generated mixed particles (HIVFCM) that could now efficiently infect unrestricted HeLa target cells (Fig. S1 A) as well as be effectively labeled by the FlAsH reagent (Fig. S1 B). This virus was also still completely restricted when used to infect HeLa cells stably expressing rhTRIM5α-HA (Fig. S1 A). Therefore, we used this virus to examine whether HIV-1 viral complexes sequestered in rhTRIM5α cytoplasmic bodies contained or lacked p24CA.
To determine whether HIV-1 viral complexes sequestered in rhTRIM5α cytoplasmic bodies contained or lacked p24CA, we synchronously infected rhTRIM5α-HA HeLa cells with this FlAsH- p24CA– and mCherry-Vpr–labeled virus in the presence of MG132 and BafA or in the absence of drug. When these cells were fixed 6 h after infection, there was more virus visible in cells that had been infected in the presence of MG132 or BafA relative to the infection performed in the absence of drug (Fig. 5 A), which is similar to the pattern observed in the case of wild-type virus (Fig. 4, A and B). Association of these viral complexes with rhTRIM5α signal was also readily apparent in MG132-treated cells, whereas there were only background levels of association visible in cells that were infected in the absence of drug or in the presence of BafA (Fig. 5, B and D). Therefore, we examined the intensity of the FlAsH capsid signal in viral complexes that were associated with rhTRIM5α signal and compared that intensity to viral complexes observed in the other control infections. We did not observe any reduction in the FlAsH capsid signal in viral complexes associated with rhTRIM5α relative to viral complexes observable after the BafA control infection or the infection performed in the absence of drug (Fig. 5 D). This indicates that if rhTRIM5α does act to induce the loss of capsid protein from the viral ribonucleoprotein complex, this loss of capsid is a proteasome-sensitive step in rhTRIM5α-mediated restriction, which is consistent with another recent study (Diaz-Griffero et al., 2007).
Figure 5.
HIV-1 virions sequestered within rhTRIM5α cytoplasmic bodies after proteasome inhibition contain p24CA protein. (A and B) HeLa cells stably expressing HA-tagged rhTRIM5α were spinoculated with VSV-G–pseudotyped HIVFCM containing FlAsH-labeled capsid and mCherry-Vpr for 2 h at 17°C. Unbound virus was removed and replaced with media, and cells were incubated for 6 h at 37°C in the presence of MG132. Cells were then fixed, stained with an HA-specific antibody, and analyzed by fluorescent microscopy to determine the total number of mCherry-Vpr+ virions/cell and the percentage of virions that associated with rhTRIM5α signal (B). (C) The fluorescent FlAsH intensity of mCherry-Vpr virions associated with rhTRIM5α signal in the MG132-treated sample was compared with the intensity of virions observed in the untreated and BafA infections. The mean FlAsH intensity of each population is indicated by black bars. (D) HIVFCM particles positive for both FlAsH-labeled capsid (green) and mCherry-Vpr (red) within rhTRIM5α cytoplasmic bodies (blue). The provided image is one z section from a deconvolved z-stack image.
Analyzing the dynamics of rhTRIM5α and HIV-1 interactions
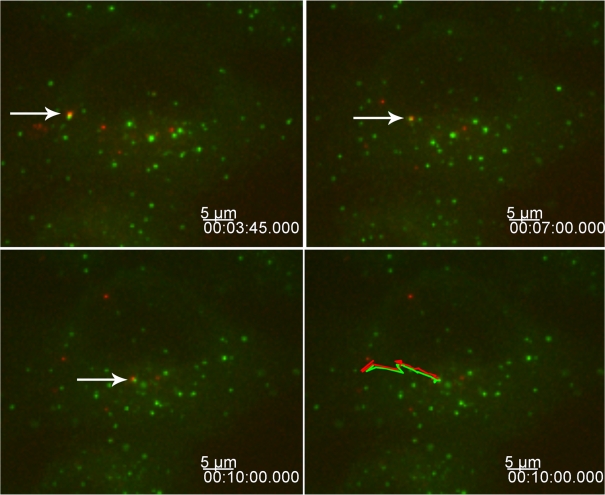
We also used live cell microscopy to monitor the interaction between HIV-1 and rhTRIM5α interaction in real time to gain a better understanding of this rapidly occurring dynamic interaction. To this end, we infected a HeLa cell line stably expressing YFP-rhTRIM5α, which effectively restricts HIV-1 infection (Campbell et al., 2007a), with mCherry-Vpr–labeled HIV-1 virions and monitored the interaction between YFP-rhTRIM5α and HIV-1 viral complexes. Z stacks of images were acquired every 10–15 s for 10 min or more. These experiments revealed three distinct types of interactions between HIV-1 viral complexes and YFP-rhTRIM5α protein. In the first type, viral complexes could be seen associating with and trafficking with preexisting cytoplasmic bodies (Fig. 6 and Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200706154/DC1). In this example, the YFP-rhTRIM5α body and HIV-1 complex was maintained for 6 min while trafficking a considerable distance within the cell, demonstrating the stability of this association (Fig. 6).
Figure 6.
Stable interaction between rhTRIM5α cytoplasmic bodies and HIV-1 virions. HeLa cells expressing YFP-rhTRIM5α (green) were infected with mCherry-Vpr–labeled HIV-1 virions (red). Images were acquired every 15 s for 10 min. Arrows indicate one YFP-rhTRIM5α cytoplasmic body associating and trafficking with one mCherry-Vpr–labeled virus over time. The bottom right panel shows the path traveled by the YFP-rhTRIM5α– and mCherry-Vpr–labeled virion throughout the experiment. The provided image is one z section from a deconvolved z-stack image showing the z section in which the interaction is currently trafficking.
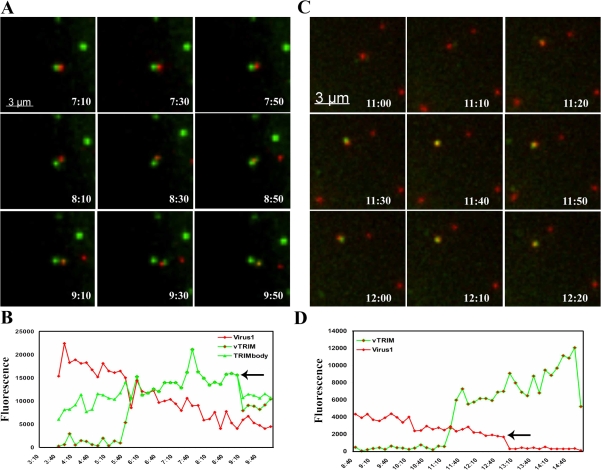
In the second type of interaction, HIV-1 viral complexes again encountered a preexisting rhTRIM5α cytoplasmic body but, this time, dissociated from this interaction apparently coated with protein derived from the original preexisting body (Fig. 7 A and Videos 2 and 3, available at http://www.jcb.org/cgi/content/full/jcb.200706154/DC1). To confirm this hypothesis, we quantified the total fluorescence intensity of the indicated cytoplasmic body over the course of its interaction with virus. We observed a measurable loss of fluorescent signal in the original YFP-rhTRIM5α body coinciding with dissociation of the viral complex now interacting with a newly formed YFP-TRIM5α body (Fig. 7 B and Fig. S2 A). This demonstrates that the new body observed around the indicated viral complex was derived from protein originally present in the first preexisting body.
Figure 7.
Dynamic interactions between rhTRIM5α cytoplasmic bodies and HIV-1 virions. (A and C) HeLa cells expressing YFP-rhTRIM5α (green) were infected with mCherry-Vpr–labeled HIV-1 virions (red). Images were acquired every 10 s for 15 min. Virus could induce the formation of new cytoplasmic bodies by becoming coated with YFP-rhTRIM5α protein derived from a preexisting cytoplasmic body (A) or by the de novo formation of a cytoplasmic body around a virion (C). (B and D) Quantifying these interactions confirms that the YFP-rhTRIM5α signal (vTRIM) becoming associated with an mCherry-Vpr–labeled virion (virus 1) in C derives from a preexisting rhTRIM5α body (TRIMbody) at the time point indicated by the arrows. The provided images were derived from a volume compression of a z stack of 10 images taken at a 0.5-μm step size at each time point.
In the third type of interaction, we observed the de novo formation of YFP-rhTRIM5α cytoplasmic bodies on or around mCherry-Vpr–labeled viral complexes (Fig. 7 C and Videos 4 and 5, available at http://www.jcb.org/cgi/content/full/jcb.200706154/DC1). Quantifying this interaction revealed that body formation in this type of interaction is very rapid, with body intensity reaching its maximal intensity in <1 min (Fig. 7 D and Fig. S2 B). Unfortunately, it was not possible to reliably quantify the fate of these YFP-rhTRIM5α body–associated viral complexes, as the outcomes of most interactions could not be ascertained before the end of the acquisition time. However, during the short period of analysis, we did observe two different outcomes in the cases in which the interaction appeared to terminate. In some cases (Videos 4 and 5), the fluorescent signal of mCherry-Vpr–labeled HIV-1 viral complexes disappeared during the acquisition time. Alternatively, in other examples, the YFP-rhTRIM5α–associated viral complexes separated from the cytoplasmic body without detectable YFP-rhTRIM5α remaining with the virus (Video 3). Therefore, these time-lapse images suggest that the dynamic nature of rhTRIM5α cytoplasmic bodies allows this protein to rapidly recognize and respond to cytoplasmic HIV-1 viral complexes during restriction.
Discussion
A proteasome-sensitive intermediate of restriction
In this study, we demonstrate that rhTRIM5α-mediated restriction in the presence of proteasome inhibitors induces the accumulation of HIV-1 viral complexes within rhTRIM5α cytoplasmic concentrations termed cytoplasmic bodies (Stremlau et al., 2004), where they are apparently sequestered (Fig. 1 A). In the presence of proteasome inhibitors, we could measure a statistically significant retention of HIV-1 viral complexes in the cytoplasm of restricted cells relative to control cells (Fig. 4 B). The finding that proteasome inhibitors induce the stable accumulation of viral complexes in cytoplasmic bodies also indicates that proteasome inhibition allows an intermediate step in the restriction process to be observed. Our previous work has demonstrated that proteasome inhibition allows HIV-1 RT and PIC formation in restricted cells but that these complexes are apparently unable to enter the nucleus (Anderson et al., 2006; Wu et al., 2006). Therefore, viral sequestration within cytoplasmic bodies represents a mechanistic explanation for these findings, as the reverse transcribing genome trapped within these structures would likely be unable to interact with cellular factors required for entry into the nucleus (Fig. 1 A).
We did not observe every cytoplasmic viral complex associating with rhTRIM5α cytoplasmic bodies during MG132 treatment (Fig. 1 C and Table I). The relatively high intensity of fluorescent signal associated with large rhTRIM5α cytoplasmic bodies after MG132 treatment necessitated very short exposure times to obtain unsaturated signal in these images (Fig. 1 A). This short exposure time may have been insufficient to detect weaker signals associated with small amounts of rhTRIM5α associating with HIV-1 capsids.
The observed association of rhTRIM5α and HIV-1 cytoplasmic viral complexes had the expected specificity. The association of GFP-Vpr–labeled viral complexes with rhTRIM5α was only observed to be above background levels when the HIV capsid surface became accessible to the cytoplasmic rhTRIM5α protein after fusion (Table I). Furthermore, viral cores shown to have entered the cytoplasm, as indicated by loss of the S15 membrane label, preferentially associated with rhTRIM5α (Figs. 1 and Figs.3 and Table I). Moreover, we also find that mutations within the B30.2 domain that abrogate restriction (Sawyer et al., 2005) also reduce the association between HIV-1 and rhTRIM5α cytoplasmic bodies to background levels (Fig. 3, B and C), demonstrating that the interactions between rhTRIM5α cytoplasmic bodies and HIV-1 reflect the ability of rhTRIM5α to restrict HIV-1 infection.
Using live cell microscopy, we observed three types of interactions leading to an association of rhTRIM5α with cytoplasmic HIV-1 during infection, underscoring the dynamic nature of these complexes. First, HIV-1 cytoplasmic complexes were observed interacting with preexisting cytoplasmic bodies and trafficking with these bodies over time within cells (Fig. 6 and Video 1). Second, we also observed that interaction between rhTRIM5α bodies and HIV-1 could lead to the dynamic transfer of rhTRIM5α protein from a preexisting cytoplasmic body onto a viral complex in close proximity (Fig. 7 A and Videos 2 and 3). This is consistent with our recent observation showing that cytoplasmic bodies are dynamic structures capable of rapidly separating and condensing to form smaller and larger cytoplasmic bodies, respectively (Campbell et al., 2007a). Third, we also observed the rapid association of large amounts of rhTRIM5α with HIV-1 viral complexes, forming a structure indistinguishable from a cytoplasmic body (Fig. 7 B and Videos 4 and 5). The ability of rhTRIM5α to dynamically reorganize its localization upon encountering a cytoplasmic viral complex represents a mechanism by which rhTRIM5α protein associates with and concentrates around the viral capsid structure during restriction. In all cases, the outcome is a viral capsid associated with a discrete localized accumulation of rhTRIM5α that meets the definition of a cytoplasmic body. Collectively, these results reveal that diverse types of interactions between rhTRIM5α and HIV-1 cytoplasmic complexes can lead to the formation of a complex of rhTRIM5α protein bound to an HIV-1 capsid in the context of a cell line expressing YFP-rhTRIM5α (Fig. 7 B and Videos 4 and 5). In a situation in which rhTRIM5α expression is lower and cytoplasmic bodies are not readily apparent, the de novo formation of rhTRIM5α on the HIV core would no doubt predominate.
Moreover, in vitro studies demonstrate that TRIM5 proteins associate with incoming viral capsids (Sebastian and Luban, 2005; Stremlau et al., 2006). Importantly, these studies show that TRIM5α does not recognize or bind free capsid protein but rather only binds capsid protein in the context of the larger viral core (Sebastian and Luban, 2005; Stremlau et al., 2006). TRIM proteins are also reported to form trimers and higher order oligomers (Reymond et al., 2001; Mische et al., 2005). This ability to oligomerize, as measured by biochemical cross-linking, is reported to be critical for the ability of rhTRIM5α to recognize purified capsid cores and restrict retroviral infection (Javanbakht et al., 2006). These studies cumulatively suggest that the ability of TRIM5α to concentrate around a viral core is a requisite step in retroviral restriction, as TRIM mutants that cannot oligomerize cannot restrict infection. Our observation that rhTRIM5α can induce the de novo nucleation of rhTRIM5α protein around a cytoplasmic viral complex further supports a mechanism in which rhTRIM5α could restrict HIV-1 infection by multimerizing into higher order structures to increase the localized concentration of restriction factor around a cytoplasmic viral core. Cytoplasmic bodies may therefore represent the ability of rhTRIM5α to form localized concentrations of proteins in specific locations, such as around viral cores, as this ability to multimerize into higher order structures around a core would increase the avidity of the interaction between rhTRIM5α and the viral complex.
Fate of capsid during restriction
Our assay for the presence of p24CA within the intermediate of restriction observed with proteasome inhibition reveals that there are similar amounts of p24CA present in the complexes of GFP-Vpr–labeled HIV complexes as is found in intact viral cores (Fig. 5). At first glance, this observation would seem to be in conflict with reports that rhTRIM5α restricts HIV infection by mediating premature disassociation of the HIV conical capsid as proposed by the Sodroski laboratory (Perron et al., 2004; Stremlau et al., 2006). However, a recent paper from that laboratory showing that the facilitated uncoating mediated by rhTRIM5α is proteasome dependent (Diaz-Griffero et al., 2007) is consistent with the two-step model of restriction that we have proposed previously (Anderson et al., 2006) and is consistent with the results presented here, which suggest that a proteasome-independent structure consisting of an HIV capsid coated with large amounts of rhTRIM5α is a key intermediate in the mechanism of restriction of HIV infection by this protein.
We also observed that rhTRIM5α cytoplasmic bodies containing sequestered HIV-1 viral complexes contain ubiquitin (Fig. 4 C). This suggests that in the absence of proteasome inhibitor, proteasome activity would play a role in the disruption of cytoplasmic viral complexes. We have shown that rhTRIM5α is rapidly turned over (Wu et al., 2006), and others have shown that this rapid turnover can be influenced by polyubiquitination and proteasome-mediated degradation of rhTRIM5α (Diaz-Griffero et al., 2006). These data collectively suggest that in the absence of drug, rhTRIM5α binds HIV-1 capsid determinants in the cytoplasm and mediates restriction by a mechanism that involves proteasome-dependent degradation of the rhTRIM5α protein.
The precise mechanism by which proteasome activity mediates this second step in the restriction process requires further investigation. Our data have failed to demonstrate a concentration of ubiquitin on viral complexes sequestered within cytoplasmic bodies, although the bodies sequestering these viral complexes do contain ubiquitin (Fig. 4). With no evidence showing that the viral capsid is polyubiquitylated, perhaps proteasomes target polyubiquitylated rhTRIM5α during restriction and not the viral capsid protein. rhTRIM5α insertion into the capsid lattice of incoming viral cores (Mische et al., 2005) and the subsequent dislocation of rhTRIM5α during its degradation by the proteasome could induce significant strain on the capsid structure, perturbing the integrity of the viral capsid lattice and causing the capsid lattice to fall apart in the absence of direct capsid degradation. Moreover, this model would also explain the loss of HIV viral complexes observed during restriction via imaging here, where dissociation of the viral capsid lattice into individual capsid proteins would allow diffusion of the concentrated GFP-Vpr signal used to identify viral complexes. Therefore, a model invoking the proteasome-mediated turnover of TRIM5 proteins inserted into incoming retroviral capsid lattices and subsequent dissociation of the capsid lattice could reconcile our observations that TRIM5 restriction involves proteasomes (Anderson et al., 2006; Wu et al., 2006) with the Sodroski laboratory findings that restriction occurs without any apparent degradation of the viral capsid protein (Stremlau et al., 2006).
The data presented here collectively reveal the importance of rhTRIM5α cytoplasmic bodies for rhTRIM5α restriction of HIV-1 and further inspire a novel view of the restriction process in which the ability of rhTRIM5α protein to concentrate around a cytoplasmic viral core likely plays an important role in the mechanism to restrict invading retroviruses. Future work is required to better understand how proteasome activity mediates the destruction of the restriction intermediate observed when proteasome activity is inhibited.
Materials and methods
Cells and pharmaceuticals
293T and HeLa cells were obtained from the American Type Culture Collection. rhTRIM5α-HA–expressing HeLa cells were gifts from J. Sodroski (Harvard Medical School, Boston, MA). CRFK cells stably expressing rhTRIM5α-HA or rhTRIM5αΔpatch-HA were provided by M. Emerman (Fred Hutchinson Cancer Center, Seattle, WA). Cells were cultured as previously described (Wu et al., 2006). BafA1 (Sigma-Aldrich) was prepared in DMSO and diluted to a final concentration of 20 nm in DME. MG132 (Sigma-Aldrich) was prepared in ethanol and diluted to a final concentration of 1 μg/ml.
Virus generation
The tetracysteine motif was introduced into the cyclophilin-binding loop of capsid (H87C/A88C/I91C/A92C) of the proviral strain R7Δenv GFP by segment overlap PCR and using BssHII–SpeI restriction enzyme sites for cloning to create R7Δenv GFPFC (HIVFC in the text).
To produce virus, 10-cm plates of 293T virus-producing cells were transfected with the appropriate plasmids using polyethylenimine (mol wt of 25,000 kD; Polysciences). GFP-Vpr– or mCherry-Vpr–labeled virus and GFP-Vpr and S15-mCherry dually labeled virus were prepared as previously described (McDonald et al., 2002; Campbell et al., 2007b). GFP-Vpr–labeled NL43 virions were produced by cotransfecting 17.5 μg NL43 proviral plasmid DNA with 2.5 μg GFP-Vpr. HIVFC virus was generated by cotransfecting 12 μg HIVFC, 8 μg VSV-G, and 1.5 μg mCherry-Vpr, whereas mixed HIVFC (HIVFCM) was generated by cotransfecting 6 μg R7Δenv GFP, 6 μg pHIVFC, 8 μg VSV-G, and 1.5 μg mCherry-Vpr. Virus was harvested 38 h after transfection and filtered through a 0.22-μm filter. Virus concentration was determined by p24CA ELISA (PerkinElmer).
FlAsH labeling solution was prepared by incubating 0.2 μM FlAsH reagent (Invitrogen) with 1 μl DMSO for 10 min at room temperature. 200 μl HBSS supplemented with 1 mg/ml glucose was then added to the labeling solution and incubated for 15 min at room temperature. Labeling solution was then added to 2 ml of virus and incubated for 2 h at 37°C. Unbound FlAsH reagent was removed by ultracentrifugation of the virus through a 30% sucrose cushion. Virus was resuspended in DME and assayed for p24 concentration by ELISA (PerkinElmer).
Infections
Synchronized infections were performed as previously described (O'Doherty et al., 2000; Campbell et al., 2007b). Cells were spinoculated with virus at 17°C for 2 h at 1,200 g. Unbound virus was removed and replaced with fresh 37°C media to initiate infection. Cells were fixed at the indicated times after the initiation of infection. Unsynchronized infections were performed at 37°C in the constant presence of virus. Glass coverslips were fixed with 3.7% formaldehyde (Polysciences) in 0.1 M Pipes buffer, pH 6.8. Coverslips were stained with mouse or rabbit α-HA antibody (Sigma-Aldrich), α-p24 mAb AG3.0 (National Institutes of Health AIDS Research and Reference Reagent Program), α-ubiquitin monoclonal antibody P4D1 (Santa Cruz Biotechnology, Inc.), or α-20S proteasome α-subunit polyclonal antibody (EMD) followed by fluorescently labeled secondary antibodies (Jackson ImmunoResearch Laboratories).
Imaging
Images were collected on a digital camera (CoolSNAP HQ; Photometrics) using a 100× NA 1.4 objective lens and were deconvolved with a microscope (DeltaVision) and DeltaVision software (Applied Precision).
Virus infectivity was analyzed by adding equal amounts of virus normalized by p24 ELISA (PerkinElmer) onto control HeLa cells or HeLa cells stably expressing HA-tagged rhTrim5α. GFP expression was measured 48 h later using a flow cytometer (FACSCalibur; Becton Dickinson) analyzing 5,000 events per infection.
Live cell microscopy
Cells stably expressing YFP-rhTRIM5α (Campbell et al., 2007a) were plated in delta DPG dishes (Thermo Fisher Scientific) in DME/F-12 50/50 (Mediatech, Inc.) containing 20% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 292 μg/ml l-glutamine, and 10 μg/ml ciprofloxacin the night before imaging. Virus was allowed to bind the cells for 1 h at 17°C. Virus was then removed and replaced with fresh 37°C DME/F-12 50/50 media and incubated for 5 min at 37°C before imaging. Cells were imaged at 37°C on a DeltaVision microscope, and images were captured in a z series on an electron multiplied charge coupled device digital camera (Cascade 2; Photometrics) and deconvolved using SoftWoRx deconvolution software (Applied Precision). In the presented images, the threshold fluorescent intensity was set to allow the visualization of all virions above background. Because virion labeling is heterogeneous, this led to postacquisition saturation of extremely fluorescent virions, giving them the appearance of being larger. This is only a result of the threshold required to view poorly labeled virions, and these virions can be observed to be punctate virions of appropriate size when a threshold more appropriate for an intensely labeled virion is used.
Data analysis
Deconvolved data were corrected for channel-specific phase shifts in the z axis and analyzed manually using Imaris software (Bitplane). Individual GFP-Vpr–positive virions were assessed for the presence of S15-mCherry stain and localization within an area of cytoplasmic rhTRIM5α concentration. For virion quantification, any punctate signal in the GFP channel above background was counted as a virion. Any signal above background in the rhTRIM5α channel was counted as rhTRIM5α association. The data quantifications shown reflect the mean results from two (Fig. 1 B only) or three independent experiments. Error bars represent the SEM of the individual calculated means. In Fig. 4 (A and B), the number of virions present in the 1-h BafA slide was set to 100% to normalize differences in viral input between experiments.
Live cell analysis was performed using the data inspector function on SoftWoRx software. The total intensity of a 6 × 6–pixel region of interest was measured, and the final total intensity at each time point was calculated by subtracting the mean total intensity of three similar regions without visible fluorescent signal.
Online supplemental material
Fig. S1 shows validation of the HIVFCM virus, demonstrating that the virus is labeled by FlAsH reagent, which was infectious and appropriately restricted by rhTRIM5α. Fig. S2 shows additional fluorescence quantification of the interactions observed in Videos 3 and 5 as performed in Fig. 7 (C and D). Video 1 shows an mCherry-Vpr–labeled HIV-1 virion trafficking with a preexisting YFP-rhTRIM5α cytoplasmic body over the course of 6 min. Videos 2 and 3 show the association of an mCherry-Vpr–labeled virion and YFP-rhTRIM5α cytoplasmic body terminating with an amount of YFP-rhTRIM5α protein remaining associated with the virion. Videos 4 and 5 show the de novo formation of a YFP-rhTRIM5α cytoplasmic body around an mCherry-Vpr–labeled virion. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200706154/DC1.
Supplemental Material
Acknowledgments
We thank Dr. Joseph Sodroski and Dr. Michael Emerman for providing cell lines used in this study. We also thank Wes Sundquist for helpful discussions and Cindy Danielson for a thoughtful critique of this manuscript.
This work was supported by National Institutes of Health grants R01 AI47770 and 1P50GM082545-01 to T.J. Hope and by the James B. Pendleton Charitable Trust. T.J. Hope is an Elizabeth Glaser Scientist.
E.M. Campbell and O. Perez contributed equally to this paper.
Abbreviations used in this paper: BafA, bafilomycin A; PIC, preintegration complex; rhTRIM5α, rhesus macaque TRIM5α; RT, reverse transcription; VSV-G, vesicular stomatitis virus G.
References
- Adams, S.R., R.E. Campbell, L.A. Gross, B.R. Martin, G.K. Walkup, Y. Yao, J. Llopis, and R.Y. Tsien. 2002. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. J. Am. Chem. Soc. 124:6063–6076. [DOI] [PubMed] [Google Scholar]
- Anderson, J.L., E.M. Campbell, X. Wu, N. Vandegraaff, A. Engelman, and T.J. Hope. 2006. Proteasome inhibition reveals that a functional preintegration complex intermediate can be generated during restriction by diverse TRIM5 proteins. J. Virol. 80:9754–9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz, P.D. 2004. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 5:1109–1115. [DOI] [PubMed] [Google Scholar]
- Campbell, E.M., M.P. Dodding, M.W. Yap, X. Wu, S. Gallois-Montbrun, M.H. Malim, J.P. Stoye, and T.J. Hope. 2007. a. TRIM5 alpha cytoplasmic bodies are highly dynamic structures. Mol. Biol. Cell. 18:2102–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, E.M., O. Perez, M. Melar, and T.J. Hope. 2007. b. Labeling HIV-1 virions with two fluorescent proteins allows identification of virions that have productively entered the target cell. Virology. 360:286–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterji, U., M.D. Bobardt, P. Gaskill, D. Sheeter, H. Fox, and P.A. Gallay. 2006. Trim5alpha accelerates degradation of cytosolic capsid associated with productive HIV-1 entry. J. Biol. Chem. 281:37025–37033. [DOI] [PubMed] [Google Scholar]
- Cowan, S., T. Hatziioannou, T. Cunningham, M.A. Muesing, H.G. Gottlinger, and P.D. Bieniasz. 2002. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. USA. 99:11914–11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Azzo, A., A. Bongiovanni, and T. Nastasi. 2005. E3 ubiquitin ligases as regulators of membrane protein trafficking and degradation. Traffic. 6:429–441. [DOI] [PubMed] [Google Scholar]
- Diaz-Griffero, F., X. Li, H. Javanbakht, B. Song, S. Welikala, M. Stremlau, and J. Sodroski. 2006. Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology. 349:300–315. [DOI] [PubMed] [Google Scholar]
- Diaz-Griffero, F., A. Kar, M. Lee, M. Stremlau, E. Poeshla, and J. Sodroski. 2007. Comparative requirements for the restriction of retrovirus infection by TRIM5alpha and TRIMCyp. Virology. 369:400–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff, S.P. 2004. Retrovirus restriction factors. Mol. Cell. 16:849–859. [DOI] [PubMed] [Google Scholar]
- Hofmann, W., D. Schubert, J. LaBonte, L. Munson, S. Gibson, J. Scammell, P. Ferrigno, and J. Sodroski. 1999. Species-specific, postentry barriers to primate immunodeficiency virus infection. J. Virol. 73:10020–10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javanbakht, H., W. Yuan, D.F. Yeung, B. Song, F. Diaz-Griffero, Y. Li, X. Li, M. Stremlau, and J. Sodroski. 2006. Characterization of TRIM5alpha trimerization and its contribution to human immunodeficiency virus capsid binding. Virology. 353:234–246. [DOI] [PubMed] [Google Scholar]
- Luo, T., J.L. Douglas, R.L. Livingston, and J.V. Garcia. 1998. Infectivity enhancement by HIV-1 Nef is dependent on the pathway of virus entry: implications for HIV-based gene transfer systems. Virology. 241:224–233. [DOI] [PubMed] [Google Scholar]
- McDonald, D., M.A. Vodicka, G. Lucero, T.M. Svitkina, G.G. Borisy, M. Emerman, and T.J. Hope. 2002. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159:441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mische, C.C., H. Javanbakht, B. Song, F. Diaz-Griffero, M. Stremlau, B. Strack, Z. Si, and J. Sodroski. 2005. Retroviral restriction factor TRIM5alpha is a trimer. J. Virol. 79:14446–14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, B., J. Daecke, O.T. Fackler, M.T. Dittmar, H. Zentgraf, and H.G. Krausslich. 2004. Construction and characterization of a fluorescently labeled infectious human immunodeficiency virus type 1 derivative. J. Virol. 78:10803–10813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Doherty, U., W.J. Swiggard, and M.H. Malim. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74:10074–10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Caballero, D., T. Hatziioannou, F. Zhang, S. Cowan, and P.D. Bieniasz. 2005. b. Restriction of Human Immunodeficiency Virus type 1 by TRIM-CypA occurs with rapid kinetics and independently of cytoplasmic bodies, ubiquitin, and proteasome activity. J. Virol. 79:15567–15572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron, M.J., M. Stremlau, B. Song, W. Ulm, R.C. Mulligan, and J. Sodroski. 2004. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. USA. 101:11827–11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond, A., G. Meroni, A. Fantozzi, G. Merla, S. Cairo, L. Luzi, D. Riganelli, E. Zanaria, S. Messali, S. Cainarca, et al. 2001. The tripartite motif family identifies cell compartments. EMBO J. 20:2140–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer, S.L., L.I. Wu, M. Emerman, and H.S. Malik. 2005. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. USA. 102:2832–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian, S., and J. Luban. 2005. TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology. 2:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, B., F. Diaz-Griffero, D.H. Park, T. Rogers, M. Stremlau, and J. Sodroski. 2005. TRIM5alpha association with cytoplasmic bodies is not required for antiretroviral activity. Virology. 343:201–211. [DOI] [PubMed] [Google Scholar]
- Steffens, C.M., and T.J. Hope. 2003. Localization of CD4 and CCR5 in living cells. J. Virol. 77:4985–4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau, M., C.M. Owens, M.J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 427:848–853. [DOI] [PubMed] [Google Scholar]
- Stremlau, M., M. Perron, S. Welikala, and J. Sodroski. 2005. Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J. Virol. 79:3139–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau, M., M. Perron, M. Lee, Y. Li, B. Song, H. Javanbakht, F. Diaz-Griffero, D.J. Anderson, W.I. Sundquist, and J. Sodroski. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc. Natl. Acad. Sci. USA. 103:5514–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers, G., M. Bock, S. Martin, Y. Takeuchi, J.P. Stoye, and O. Danos. 2000. A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. USA. 97:12295–12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X., J.L. Anderson, E.M. Campbell, A.M. Joseph, and T.J. Hope. 2006. Proteasome inhibitors uncouple rhesus TRIM5alpha restriction of HIV-1 reverse transcription and infection. Proc. Natl. Acad. Sci. USA. 103:7465–7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap, M.W., S. Nisole, and J.P. Stoye. 2005. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr. Biol. 15:73–78. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.