Abstract
Type III secretion (T3S) systems are largely used by pathogenic Gram-negative bacteria to inject multiple effectors into eukaryotic cells. Upon cell contact, these bacterial microinjection devices insert two T3S substrates into host cell membranes, forming a so-called ‘translocon' that is required for targeting of type III effectors in the cell cytosol. Here, we show that secretion of the translocon component IpaC of invasive Shigella occurs at the level of one bacterial pole during cell invasion. Using IpaC fusions with green fluorescent protein variants (IpaCi), we show that the IpaC cytoplasmic pool localizes at an old or new bacterial pole, where secretion occurs upon T3S activation. Deletions in ipaC identified domains implicated in polar localization. Only polar IpaCi derivatives inhibited T3S, while IpaCi fusions with diffuse cytoplasmic localization had no detectable effect on T3S. Moreover, the deletions that abolished polar localization led to secretion defects when introduced in ipaC. These results indicate that cytoplasmic polar localization directs secretion of IpaC at the pole of Shigella, and may represent a mandatory step for T3S.
Keywords: IpaC, polarization, secretion, Shigella, type III translocator
Introduction
Type III secretion system (T3SS) are found in a wide variety of Gram-negative bacteria that are pathogenic to vertebrates or plants (Cornelis, 2006; Galan and Wolf-Watz, 2006). These nanomachines allow rapid injection of effectors from the bacterial cytoplasm to the host cell cytosol upon cell contact. Once injected, type III effectors divert various cell functions, acting on regulators of the actin cytoskeleton, intracellular trafficking, or modulating inflammatory responses. Although injected type III effectors might be specific for a given bacterial pathogen, approximately 25 proteins are required for the assembly of T3SS, and they share significant levels of homology among bacterial species and a similar genetic organization, indicating common functional features.
Considerable progress has been made in recent years on the structural characterization of these machineries, mostly through the visualization of T3SS from various bacterial pathogens by electron microscopy. T3SS are related to structures of flagella and are composed of a basal-body spanning the two bacterial membranes and the periplasm, extended by a needle (Kubori et al, 1998; Tamano et al, 2000). In the case of Yersinia pestis, the causative agent of plague, LcrV, a T3S substrate that is not required for the assembly of the T3SS, was shown to be part of a complex located at the tip of the needle (Mueller et al, 2005). The tip complex of Shigella, the causative agent of bacillary dysentery, is composed of IpaB and IpaD (Espina et al, 2006; Veenendaal et al, 2007). Because of its location, the Shigella tip complex was proposed to serve as a sensor that recognizes host cell membranes and triggers T3S (Veenendaal et al, 2007). As opposed to wild-type Shigella for which T3S is repressed under bacterial culture conditions, ipaB or ipaD mutants show constitutive secretion (Menard et al, 1994a), consistent with the role of IpaB and IpaD in controlling the activity of T3SS.
The precise sequence of events leading to the injection of type III effectors from the bacterial to host cell cytoplasm is poorly understood. For all T3SS described to date, contact with the host's cells triggers secretion of two proteins that insert into host cell membranes to form the translocon (Buttner and Bonas, 2002; Coombes and Finlay, 2005). Translocon components are required for the injection of type III effectors and therefore, it is thought that they are the first substrates secreted by the T3SS upon cell contact. In the case of Shigella, these translocon components are the IpaB and IpaC proteins. In the absence of secretion stimuli, IpaB and IpaC are bound to the IpgC chaperone and stored in the cytoplasm, which prevents premature IpaB–IpaC association (Menard et al, 1994b). Upon cell contact, the entire cytoplasmic pool of IpaB and IpaC is secreted within minutes (Enninga et al, 2005).
In addition to its role in translocation, IpaC was also shown to mediate actin polymerization, which, with the concerted action of injected T3S effectors, promotes bacterial invasion of epithelial cells in a triggering or macropinocytic process (Tran Van Nhieu et al, 1999; Kueltzo et al, 2003; Handa et al, 2007). As opposed to zipper-phagocytosis, which implies high-affinity interactions between bacterial surface ligands and cell receptors driving the extension of cellular pseudopods in tight apposition with the bacterial body, trigger-phagocytosis relies on the formation of T3S-dependent localized membrane ruffles (Swanson and Baer, 1995; Cossart and Sansonetti, 2004). Since Shigella T3S effectors of invasion include IpaC or IpgB1, which promote actin polymerization to drive the formation of cellular extensions, but also IpaA, which depolymerizes actin filaments, a controlled spatio-temporal action of these effectors would be required to coordinate the cytoskeletal responses leading to bacterial invasion.
Here, we show that upon cell contact, secretion of IpaC, and therefore T3S, does not occur diffusely over the bacterial surface, but occurs at the level of one bacterial pole. We show that polar T3S is determined by a unipolar cytoplasmic localization of IpaC before secretion, which appears to determine not only the localization but also the efficiency of secretion.
Results
IpaC localizes at one pole in Shigella cells
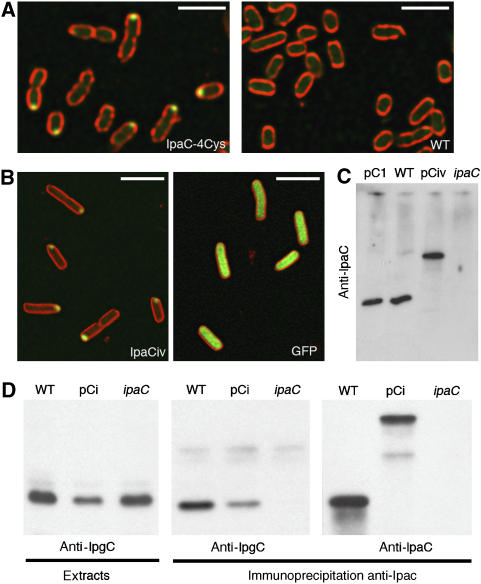
To study the localization of IpaC inside the bacterial cytoplasm, we analyzed the localization of IpaC-4Cys, a recombinant form of IpaC that binds to the FlAsH fluorescent derivative, and that was shown to complement a Shigella ipaC-mutant strain SF621 for cell invasion (Materials and methods; Enninga et al, 2005). IpaC-4Cys in strain SF621 labeled with the FlAsH compound showed a unipolar localization (Figure 1A, IpaC-4Cys). In control experiments and as previously described, labeling of the wild-type Shigella flexneri 5a strain M90T with the FlAsH compound did not result in significant staining (Enninga et al, 2005; Figure 1A, WT).
Figure 1.
Unipolar localization of fluorescently labeled recombinant IpaC. (A) Confocal micrograph of SF621/pIpaC-4Cys (IpaC-4Cys) or M90T (WT) labeled with the FlAsH derivative (Materials and methods). Green: FlAsH fluorescence; red: membrane staining with FM 4-64. Scale bar=5 μm. (B) Confocal micrographs of SF621/pCiv (left panel) and M90T/pFPV25.1 (right panel). Green: IpaCiv and eGFP fluorescence; red: membrane staining with FM 4-64. Scale bar=3 μm. (C) Anti-IpaC western blot analysis of bacterial lysates. (D) Western blot analysis on anti-IpaC immunoprecipitates from bacterial lysates. Bacterial lysates (left panel) are used as controls. Western blot using anti-IpgC antibody (left and middle panels); anti-IpaC antibody (right panels). Strains pC1: SF621/pC1; WT: wild type; pCi: SF621/pCic; ipaC: ipaC mutant.
Because the low quantum yield of the FlAsH reagent does not allow the detection of low protein levels, to further analyze the localization of IpaC, we constructed recombinant IpaCi proteins in which eGFP (IpaCig), mCFP (IpaCic), or ‘Venus', a fast-maturing YFP variant (IpaCiv), were inserted at position 57 of IpaC so as to preserve the amino-terminal secretion signal and the carboxy-terminal effector domain (Materials and methods; Nagai et al, 2002; Harrington et al, 2003; Hoppe and Swanson, 2004). When IpaCiv was produced in the Shigella ipaC-mutant SF621 strain, the recombinant protein showed a unipolar localization that was very similar to the one observed for IpaC-4Cys (Figure 1B, left panel). In most cases, a fluorescent spot was detected at one bacterial pole, although in a few marginal cases, fluorescence could be detected at both poles. This was in contrast to GFP alone showing diffuse fluorescence throughout the bacterial body (Figure 1B, right panel). In control experiments, western blot analysis of bacterial extracts showed that IpaCiv was produced in equal amounts as the endogenous IpaC protein in the wild-type M90T strain, indicating that polar localization was not linked to IpaCiv overproduction (Figure 1C). To test whether IpaCi, as endogenous IpaC, could associate with the IpgC chaperone, we performed co-immunoprecipitation assays. Soluble extracts from wild-type, SF621, and SF621/pCig strains were subjected to anti-IpaC immunoprecipitation and analyzed by western blotting. As shown in Figure 1D, IpgC associated with IpaCig as efficiently as the endogenous IpaC, indicating that GFP insertion in IpaC did not interfere with IpgC binding. These results indicate that like functional IpaC-4Cys, IpaCi shows a predominant localization at one bacterial pole.
IpaC localizes to the same pole as IcsA and targets the old or the new pole upon septation with the same frequency
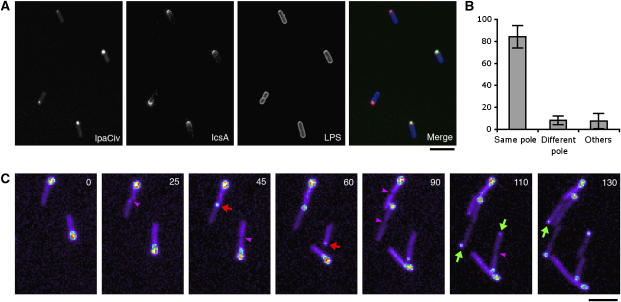
IscA is a Shigella surface protein that is localized at one bacterial pole and mediates actin-based motility in host cells (Goldberg et al, 1993). The basis for IcsA polar localization has been the subject of many studies, and is one of the best-characterized models of polarity in enterobacteria (Ebersbach and Jacobs-Wagner, 2007). Because the polar localization of IpaC was reminiscent of that of IcsA, we investigated their respective localization by immunofluorescence staining of IcsA at the surface of SF621/pCiv strain. As shown in Figure 2A, IpaCiv and IcsA were found at the same pole in the vast majority of the cells, with 84.2±10.3% displaying staining of IcsA and IpaCiv at the same pole, whereas only 8.1±4% of bacteria showing IcsA staining at the pole opposite IpaCiv (Figure 2B, 1304 bacteria, n=5).
Figure 2.
IpaCiv rapidly localizes at the old or new pole during division. (A) Confocal micrographs of SF621/pCiv. Red: anti-IcsA immunofluorescent staining (Materials and methods); blue: LPS staining; green: IpaCiv fluorescence. (B) Quantitative analysis of the respective localization of IpaCiv and IcsA (1304 individual bacteria, n=5). Percentages were normalized on the total number of bacteria showing a fluorescent signal for IcsA and IpaCiv. (C) Time-lapse fluorescence microscopy of SF621/pCiv growing on an agarose pad (Materials and methods). Time points are indicated in minutes. Fluorescence intensity is represented in pseudo-color code. Red and green arrows point to the formation of a fluorescent dot at the newborn or the old pole, respectively. Purple arrowheads indicate septum formation. Scale bar=5 μm.
To identify whether IpaCi targeted the old or the newly formed pole, we performed time-lapse experiments on Shigella strain SF621/pCiv growing on an agar pad during several division periods (Materials and methods). As shown in Figure 2C, fluorescent dots were detected as they emerged directly at a pole or at the bacterial septal area. Moreover, dot appearance generally correlated with septum formation (Supplementary movie 1). After septation, the inherited dot usually remained at the same bacterial pole in one of the daughter cells during several divisions. The new dot, which appeared rapidly before or after septation, was localized at the newborn pole or the old pole of the second cell. When quantified, the appearance of fluorescent dots occurred in 50.3% of the cases at the new pole and 49.7% of the cases at the old pole of the daughter cell (161 septation events, n=3). This result indicates that IpaCiv is targeted with equal frequency to the old or new pole.
IpaC polarization is conserved in Escherichia coli
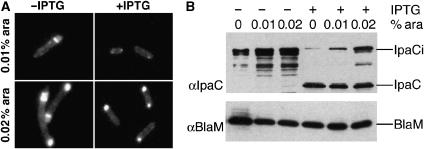
IcsA polarization implies mechanisms that are shared between Shigella and Escherichia coli, since for both bacteria an internal domain of IcsA fused to GFP is localized at the pole in the cytoplasm (Charles et al, 2001). To analyze the localization of IpaCi in E. coli cells, the expression of ipaCi under the control of the pBAD promoter, was induced in the presence of low concentrations of arabinose and bacteria were observed by fluorescence microscopy. As shown in Figure 3A, IpaCig was detected at the bacterial pole, indicating that, like for IcsA, IpaC polarization is not restricted to Shigella and is independent of the T3SS. To test if IpaCi could be chased from the pole by wild-type IpaC, we introduced a compatible plasmid expressing the ipaC gene under the control of the plac promoter in the same cells. Upon induction of ipaC with IPTG, the level of fluorescence at the poles was significantly reduced, compared with levels observed in the absence of IpaC induction (Figure 3A, right panel). Anti-IpaC western blot analysis of these bacteria showed that the loss of polar fluorescence was correlated with a reduction in the amounts of IpaCig protein in total cell extracts (Figure 3B). These experiments indicate that the levels of IpaCig at the pole can be reduced by simultaneous production of IpaC, arguing that polar localization does not result from protein aggregation.
Figure 3.
IpaC polarization is observed in E. coli. E. coli TOP10F' containing plasmid pBadCi and pCHAP4500 grown in the presence of 0.01 or 0.02% arabinose were incubated in the absence (−IPTG) or presence of 1 mM IPTG (+IPTG) to induce ipaC expression (Materials and methods). (A) Fluorescence microscopy analysis of representative samples grown the absence (left panels) or presence (right panels) of IPTG. (B) Extracts from cultures were analyzed by anti-IpaC (top panel) and anti-BlaM (bottom panel) western blotting. The presence or absence of IPTG, and the percentage of arabinose are indicated above each lane. The migration of IpaCi, IpaC, and the β-lactamase (BlaM) used as a control, is indicated.
Residues 170–302 are dispensable for IpaC polar localization
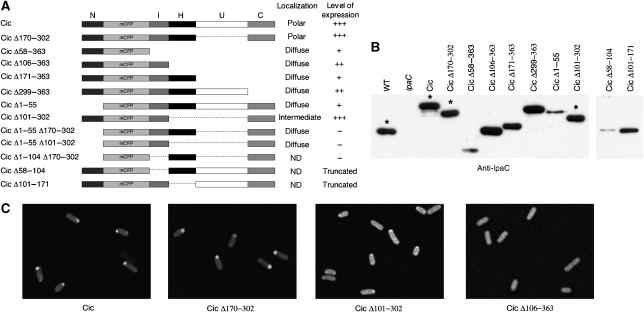
Functional studies on IpaC have led to the identification of various domains (Page et al, 2001; Picking et al, 2001; Harrington et al, 2003; Kueltzo et al, 2003). We used this domain organization as a basis to generate truncated derivatives of IpaCic, to identify regions involved in polar localization. Deletions were performed to remove five regions of IpaC: (1) an amino-terminal domain N, which consists of the first 57 residues and includes the secretion signal; (2) an intermediate domain I (residues 57–100) containing the IpgC-binding region; (3) a hydrophobic domain H (residues 101–170), which allows membrane anchoring of IpaC after secretion; (4) a domain U with no assigned function (residues 171–298); and (5) a carboxy-terminal domain C (residues 299–363), which includes the oligomerization and effector regions (Figure 4A).
Figure 4.
Determination of domains in IpaC required for polar localization. (A) Schematic representation of IpaCic constructs and summary of their localization and expression levels in ipaC. The positions of residues corresponding to deletions in IpaC are mentioned. (B) Anti-IpaC western blot analysis of the IpaCic constructs in ipaC. Asterisks indicate samples of the most expressed constructs, which were diluted for the representation. (C) Representative fluorescence micrographs of SF621/pCic, SF621/pCicΔ170–302, SF621/pCicΔ101–302, and SF621/pCicΔ106–363.
Some of these constructs showed very low levels of expression or were subjected to proteolysis and their localization was not analyzed further (Figure 4A). In particular, deletion of the I domain (IpaCic Δ58–104) led to production of a polypeptide showing unexpected migration, when analyzed by SDS–PAGE (Figure 4B, right panel). This polypeptide of an apparent molecular weight of 45 kDa probably corresponded to a truncated derivative resulting from an endoproteolytic cleavage within the U domain, since it was recognized by monoclonal antibodies directed against the amino-terminal but not the carboxy-terminal part of IpaC (data not shown). Also, deletion of the H domain (IpaCic Δ101–171) led to detection of a polypeptide cleaved within the U domain (Figure 4B, right panel). Deletion of the I domain in combination with other deletions (Δ1–104 or Δ170–302) generated proteins produced in very low amounts (not shown). Deletions Δ1–55, Δ58–104, and Δ58–363 generated proteins with reduced levels of expression (Figure 4B), but which still showed a significant fluorescent signal. Deletion of the N domain (IpaCic Δ1–55) led to a complete loss of polar localization. Fusion of the N domain with CFP (IpaCic Δ58–363) also led to diffuse fluorescence, indicating that the N domain was necessary but not sufficient for polar localization. Other ipaCic deletion mutants were expressed at levels comparable to those of ipaCic or endogenous ipaC. The I domain was not sufficient to confer polar localization, because its fusion to the N domain (resulting in IpaCic Δ106–363) led to diffuse fluorescence. In contrast, deletion of the U domain (IpaCic Δ170–302) did not affect polar localization, indicating that this domain was dispensable for targeting IpaC to the pole (Figure 4C). An IpaCic deleted for both the H and the U domains showed a mixed pattern, with a diffuse localization and some reinforcement at the pole, indicating that the H domain was partially required for polar localization (Figure 4C, Cic Δ101–302). In these bacteria, quantification of the fluorescence intensity indicated a statistically significant 35% increase at one pole over the global fluorescence intensity of the bacteria (Supplementary Figure S1). IpaCic Δ1–55 Δ170–302 and IpaCic Δ1–55 Δ101–302 were diffusely localized, confirming the roles of the N and H domains in polar localization. However, the presence of these two domains was still not sufficient for polar targeting, since IpaCic Δ171–302 showed a diffuse localization. Finally, all the IpaCic variants lacking the C domain showed diffuse localization, indicating that the C domain was also required (Figure 4C). Similar findings were obtained when these IpaCic constructs were expressed in E. coli (Supplementary Figure S2), confirming that polar localization determinants are not restricted to Shigella. Taken together, these results indicate that with the exception of the U domain, all the other identified domains of IpaC are required for polarization, although it is possible that some of these domains are indirectly required by assisting the proper folding or presentation of a polar determinant.
Type III secretion is inhibited by polar but not diffuse IpaCic fusions
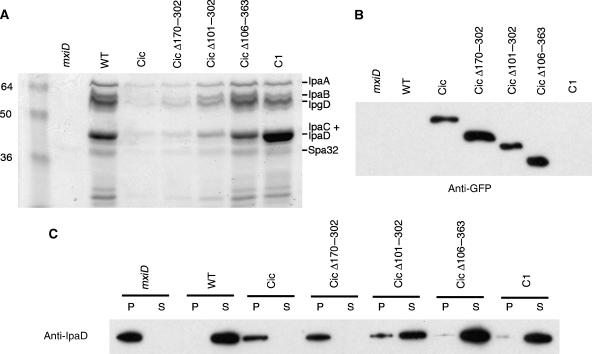
It has been reported that type III substrates fused to GFP were not secreted by the T3SS, probably because type III ATPase is unable to unfold the GFP β-barrel structure (Akeda and Galan, 2005; Enninga et al, 2005). To test if there was a correlation between their localization and secretion via the T3SS, we set to analyze the effects of IpaCic and its deletion derivatives on type III secretion (Figure 5A). As previously described (Enninga et al, 2005), wild-type Shigella producing IpaCic showed a dramatic defect in secretion of type III substrates upon Congo red induction (Figure 5A, Cic). Strikingly, only the polarized hybrid IpaCic Δ170–302 inhibited secretion to the same extent as full-length IpaCic. IpaCic Δ101–302, which showed a mixed localization, partially inhibited T3S. In contrast, all the truncated derivatives showing diffuse localization had no detectable effect on T3S (Figure 5A and data not shown). In control experiments, western blot analysis of bacterial cell extracts indicated that the effects on T3S were not due to difference in the expression levels of the various ipaCic derivatives (Figure 5B). Moreover, we observed that IpaCic, IpaCic Δ101–302, IpaCic Δ170–302, and IpaCic Δ106–363 were not secreted (Supplementary Figure S3). To confirm that the absence of protein in the supernatant was due to a secretion defect and not to variations in the expression of type III substrates, protein contents of bacterial pellets and supernatants were analyzed by anti-IpaD western blotting (Figure 5C). In M90T/pCic or M90T/pCic Δ170–302, IpaD was produced but was not secreted into the bacterial supernatant, whereas it was virtually completely secreted in M90T/pCic Δ101–302 or M90T/pC1 (Figure 5C). In keeping with the observed partial effects on T3S, the majority of the IpaD pool was secreted in M90T/pCic Δ101–302, but some IpaD could still be detected in the bacterial pellet (Figure 5C). These results indicate that only the IpaCic Δ170–302 or the IpaCic Δ101–302 fusions that show polar localization inhibit type III secretion, whereas all tested IpaCic hybrids with diffuse cytoplasmic localization have no detectable effect on type III secretion.
Figure 5.
Polar IpaCic fusions inhibit type III secretion. (A) SDS–PAGE and Coomassie blue staining of proteins secreted upon Congo red induction in the supernatant of the indicated strains (Materials and methods). The expected migration of T3S substrates is mentioned. (B) Anti-GFP western blot analysis of bacterial lysates. (C) Anti-IpaD western blot analysis of bacterial pellets (P) and supernatants (S) following Congo red induction and fractionation (Materials and methods). mxiD: T3S-deficient SF401 mutant; WT: wild-type Shigella strain M90T. Full-length IpaC (C1) and the IpaCic constructs are expressed in M90T.
Deletions in IpaC that affect polar localization, reduce its secretion
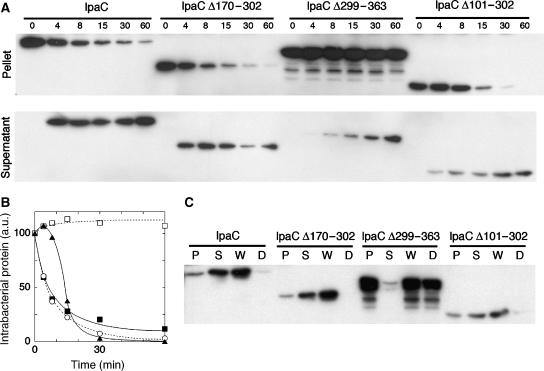
Since IpaCic fusions were not secreted, deletions were introduced in the wild-type ipaC gene to study their impact on secretion. Deletions Δ299–363 and Δ101–302 were chosen for their effect on polar localization, whereas Δ170–302 was used as control. These constructs were introduced into the ipaC-mutant strain and the kinetics of secretion of the resulting IpaC variants were analyzed. As shown in Figure 6A, the entire pool of IpaC was secreted, with half-maximal secretion occurring within 6 min following Congo red induction. The Δ170–302 deletion that did not affect polar localization, did not interfere with secretion, since similar kinetics were observed for IpaCΔ170–302. The entire pool of IpaCΔ101–302 was also secreted, but with kinetics which appeared delayed, since it took 14 min to secrete 50% of the pool of this derivative (Figure 6A). In contrast, secretion of IpaCΔ299–363 was dramatically affected, since only a small fraction of the protein was secreted after 60 min. For all the strains, proteinase K digestion assays indicated that proteins present in the bacterial pellet were protected from degradation in contrast to proteins present in the supernatant, confirming the secretion defect of IpaCΔ299–363 and IpaCΔ101–302 variants (Figure 6B). These results show that deletion Δ170–302 that does not affect, or deletion Δ101–302 that partially affects, polar localization, resulted in constructs which were efficiently secreted. In contrast, deletion Δ299–363, which resulted in diffuse localization, led to significant secretion defects, suggesting that polar localization of IpaC is required for its efficient secretion via the T3SS.
Figure 6.
Kinetics of secretion of IpaC and truncated derivatives. (A) ipaC-mutant strains producing IpaC or IpaC derivatives were grown to exponential phase and incubated with Congo red to induce type III secretion (Materials and methods). Samples were centrifuged at the time points indicated (in minutes) above each lane. Bacterial pellets (top panels) and supernatants (bottom panels) were normalized to the total protein amounts and equivalent aliquots were analyzed by anti-IpaC western blot analysis (Materials and methods). (B) Quantification of the amounts of IpaC and IpaC derivatives in the bacterial pellets by scanning the band intensities in (A). Solid squares: IpaC; empty circles: IpaC Δ170–302; solid triangles: IpaC Δ101–302; empty squares: IpaC Δ299–363. The values are normalized to the initial values obtained at t=0, arbitrarily set up to 100. (C) Proteinase K digestion was performed on whole samples after induction of secretion for 30 min (Materials and methods). Bacterial pellets (P), supernatants (S), whole sample (W), and digested samples (D) were normalized and equivalent aliquots were analyzed by anti-IpaC western blotting.
T3S is predominantly polar and occurs at the pole where IpaC is stored
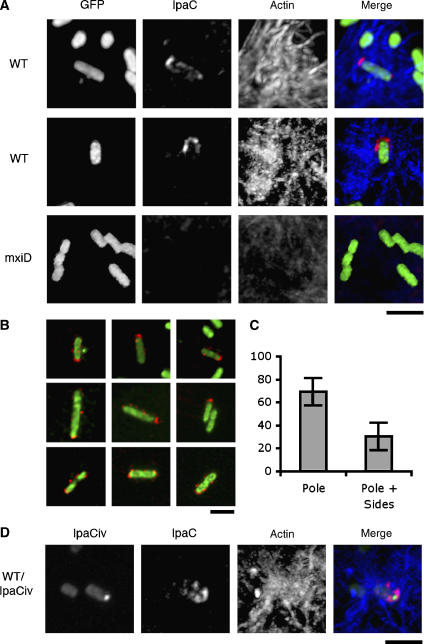
The fact that polar IpaC fusions, but not fusions with diffuse cytoplasmic localization, significantly inhibit T3S, suggests that secretion occurs mostly at one bacterial pole. Since real-time analysis indicated that IpaC has high diffusion dynamics following secretion into host cell membranes (Enninga et al, 2005), the localization of secreted IpaC needed to be analyzed shortly after cell contact. IpaC secreted in wild-type Shigella upon invasion was visualized by immunofluorescence, using a technique that allows permeabilization of epithelial cells but not of bacteria (Materials and methods). After 3 min following cell contact, IpaC staining could be detected in association with wild-type bacteria in nascent entry foci (Figure 7A, WT). IpaC staining was particularly strong at one pole of the bacteria, often forming a U-shaped structure (Figure 7A, WT). Although for most bacteria, IpaC staining was predominantly detected at the pole (Figure 7B, top and middle panels), some bacteria showed staining as distinct spots along the bacterial sides (Figure 7B, bottom panels). When quantified, polar staining of secreted IpaC was clearly more frequent, with 70±12% of the bacteria showing staining predominantly associated with one pole (Figure 7C, 125 bacteria, n=3). No signal was observed for an ipaC mutant (not shown) or for the secretion-defective mxiD mutant (Figure 7A, mxiD), indicating that staining was specific for secreted IpaC. In control experiments, labeling performed in the absence of permeabilization did not result in significant staining (data not shown), indicating that IpaC labeling corresponded to the IpaC inserted into host cell membranes. Consistent with the dynamic distribution of IpaC after secretion, labeling of IpaC could be detected in association with bacteria during the first 7 min following cell contact. At later time points of bacterial invasion, a diffuse IpaC fluorescent signal was generally observed throughout infected cells.
Figure 7.
Secretion of IpaC at the pole labeled by IpaCiv during HeLa cells infection. Bacteria were incubated with HeLa cells at 37°C for 3–5 min, fixed, and processed for anti-IpaC and F-actin fluorescence staining (Materials and methods). Red: IpaC staining; blue: F-actin; green: GFP or IpaCiv fluorescence. (A) Fluorescent micrographs of HeLa cells infected by WT: wild-type Shigella/pFPV25.1; mxiD: SF401/pFPV25.1. (B) Confocal micrographs of WT showing a predominant IpaC staining at the pole (top and middle panels), or also showing staining at the bacterial sides (bottom panels). Scale bar=3 μm. (C) Individual bacteria were scored and classified as: bacteria presenting a predominant IpaC staining at one bacterial pole (pole), or bacteria also presenting a significant staining at the bacterial sides (pole+sides). The results are expressed as percentage of the total bacteria showing IpaC staining±s.d. (125 bacteria, n=3). (D) Fluorescent micrographs of HeLa cells infected by wild-type Shigella/pCiv/pMM100.
The M90T/pCiv strain is deficient for T3S and, consistently, is defective for entry foci formation and cell invasion (data not shown).
To correlate the polar secretion of IpaC to its polar intra-bacterial localization, we reasoned that reducing the levels of IpaCiv may allow to partially relieve its inhibitory effect on T3S, while preserving sufficient fluorescence to label the pole. In order to modulate the expression of ipaCiv driven by the lac promoter, we introduced the lacI repressor on pMM100, a multi-copy plasmid compatible with pCiv (Laviña et al, 1986), by varying the concentrations of the inducer IPTG. As expected, M90T/pCiv/pMM100 bacteria grown in the absence of IPTG were not fluorescent and showed active T3S upon Congo red induction, indicating that LacI repression was highly efficient (Supplementary Figure S4). When M90T/pCiv/pMM100 were grown in the presence of 1 mM IPTG, however, individual bacteria showing a fluorescent dot at one pole could be observed albeit with a much lower intensity and frequency than in M90T/pCiv (Supplementary Figure S4). At these IPTG concentrations, repression via LacI was efficient since western blot analysis indicated that the amounts of IpaCiv were significantly reduced compared to growth in the absence of pMM100 (Supplementary Figure S4). These levels of IpaCiv did not allow full inhibition of T3S, since M90T/pCiv/pMM100 secreted significant amounts of type III substrates upon Congo red induction (Figure 7C; Supplementary Figure S4). Consistent with functional T3S, when immunofluorescence staining was performed under these growth conditions, M90T/pCiv/pMM100 was observed to induce entry foci, albeit at a lower frequency than M90T. In addition, bacteria associated with these foci showed staining for secreted IpaC at one bacterial pole, similar to that observed with M90T. Whenever detected, IpaC staining localized at the pole labeled by IpaCiv and could never be detected at the opposite pole (12 bacteria, n=2; Figure 7C). These results indicate that type III secretion of IpaC is predominantly polar and occurs at the pole corresponding to the pool pre-accumulated in the cytoplasm.
Discussion
Studies on protein localization during bacterial division have revealed the complexity of processes that were long thought to occur in a single compartment defined by the bacterial cytoplasm. In this article, we show that in the vast majority of the cells secretion of IpaC via the T3SS occurs at one pole of the Shigella cell during epithelial cell invasion (Figure 7). This predominant localization of IpaC secretion might have an important impact on the coordination of cytoskeletal rearrangements required for invasion. Insertion of a translocon component such as IpaC at a specific location in host cell membranes would be an efficient manner to locally target injection of T3S effectors, and to spatially coordinate cytoskeletal responses during invasion. According to the translocon model, polarized secretion of IpaC would imply that early T3SS effectors that are constitutively expressed and secreted upon cell contact, such as those involved in bacterial invasion, are injected at the bacterial pole. Moreover, we observed that a polar IpaCic fusion inhibits the secretion of all pre-stored T3SS substrates (Figure 5A), suggesting that early T3SS effectors, as well as substrates such as IpaB and IpaD, are also targeted and secreted at the pole. However, previous observations by electron microscopy indicated that T3SS are diffusely distributed at the surface of Shigella (Blocker et al, 1999). Whether the non-polarly distributed T3SSs are functional and allow the secretion of another subset of T3SS substrates whose function does not require translocon components is unknown. For example, it is possible that these non-polar T3SSs allow intracellular secretion of T3SS effectors that are upregulated after cell contact, by bacteria replicating freely in the host cell cytosol.
Polarized secretion has been reported in other systems, but the mechanisms leading to polarity and providing links of secretion machineries with cytoplasmic substrates remain by and large ill-defined. For example, in Agrobacterium tumefaciens, polar localization of the type IV secretion machinery determines the polarity of secretion. This may be a general feature for type IV secretion systems that translocate substrate proteins and DNA across both bacterial and eukaryotic membranes, since several components of these systems have been reported to be have a polar localization (Judd et al, 2005; Atmakuri et al, 2007). Also, several structural components of the type II secretion system (T2SS) in Vibrio cholerae localize at one pole, and secretion of substrate enzymes to the extracellular medium occurs in a polar manner (Scott et al, 2001). Here, we observed that IpaC localized at one pole in the absence of secretion.
These results suggest that polar secretion of IpaC during epithelial cell invasion is determined before secretion by localization of IpaC in the bacterial cytoplasm. A similar model was proposed for IcsA, another protein of Shigella and member of the auto-transporter family, which shows a polar localization at the cell surface (Goldberg et al, 1993; Brandon et al, 2003). IcsA is exported to the periplasm through the Sec machinery, which is not restricted to the poles and is distributed in a helical pattern within the inner membrane around the bacterial body, at least in E. coli and Bacillus subtilis (Charles et al, 2001; Campo et al, 2004; Shiomi et al, 2006). It has been proposed that polar targeting of the IcsA precursor occurs in the cytoplasm and determines its final localization. Although plausible, such a mechanism is difficult to prove in vivo due to the short-lived nature of IcsA cytoplasmic precursor pool and their rapid and constitutive export through the Sec machinery. This is in contrast to T3SS, whose substrates are stored in the bacterial cytoplasm until secretion is activated. This property of T3SS allowed the visualization of the cytoplasmic polar pool of IpaC before secretion. We found correlations between the cytoplasmic IpaC location and that of IcsA, suggesting that they share a common mechanism of polarization. Only a region corresponding to one-third of the protein between the hydrophobic domain and the effector domain was dispensable for IpaC polar localization. No significant sequence similarity between the polar localization domains of IcsA and IpaC could be detected, suggesting that there are several polarization determinants, possibly functioning as structural motifs.
Here, we also report an intricate correlation between cytoplasmic polar IpaCi location and T3SS, which impacts not only the localization but also on efficiency of T3S. Secretion of IpaC occurs at the pole that is labeled by cytoplasmic IpaCi. IpaCi fusions, which localize at the pole, inhibit T3S, whereas those with a diffuse localization had no detectable effects on T3S. It was demonstrated that substrates have to be unfolded by the ATPase of the T3SS, to allow their secretion but that GFP is resistant to this unfolding, thus explaining the lack of secretion of GFP fusions by the T3SS (Akeda and Galan, 2005). Our observations suggest that IpaCi engages in the Shigella T3SS irreversibly, possibly by physically clogging the apparatus due to the compact folding of the GFP β-barrel structure. Consistent with this, membrane fractionation experiments on sucrose flotation gradients showed that an IpaC-GFP fusion associated with membrane fractions containing the T3SS, but not with membranes isolated from a T3S-defective mxiD-mutant strain (data not shown). This T3SS-associated fraction of IpaCi, however, corresponded to a minor fraction of the total pool, since the majority of the pool of IpaC-GFP fusion is soluble. This indicates that the majority of the IpaCi pool at the pole is not engaged in the T3SS.
How the polar cytoplasmic localization of IpaC determines its efficient secretion is a highly interesting issue that will deserve more investigation. An estimation for bacterial rods of Shigella with an average of 2–5 μm length and 0.9 μm diameter, indicate that polar localization in a sphere ranging from 100 to 500 nm could increase the local concentration of IpaC by 25- to 7400-fold. The high local concentration of IpaC allowed by polarization may significantly accelerate recognition events between the substrate and the T3SS, and therefore accelerate the secretion rate.
Materials and methods
Bacterial strains, cell line, and plasmids
HeLa cells from ATCC were grown in DMEM containing 10% fetal calf serum at 37°C, in an incubator containing air supplemented with 10% CO2. The S. flexneri serotype 5a strain M90T was used as a wild-type strain (Sansonetti et al, 1982). The ipaC-mutant (SF621) and mxiD-mutant (SF401) isogenic derivatives of this strain have been described previously (Allaoui et al, 1993; Menard et al, 1993). The E. coli strain used in this study was MC1000 F'tet (araD139, Δ[ara-leu]7679, galU, galK, Δ[lac]174, rpsL, thi-1 F'∷Tn10 proA+B+lacIq Δ(lacZ)M15 TetR). Bacteria were grown in trypticase soy (TCS). Carbenicillin was used at 100 μg/ml and tetracycline was used at 5 μg/ml. The pIpaC-4Cys plasmid was described previously (Enninga et al, 2005). Plasmid pFPV25.1 was used to produce GFP in bacteria (Valdivia and Falkow, 1996). Plasmid pMM100 containing the lacIQ gene was used to repress the plac promoter-driven expression of ipaCic (Laviña et al, 1986). IpaC fusions were made with enhanced green fluorescent protein (IpaCig), monomeric cyan fluorescent protein (IpaCic) and fast-maturing yellow fluorescent protein Venus (IpaCiv) (Nagai et al, 2002; Hoppe and Swanson, 2004). The procedures to construct plasmids pCic and pCiv, expressing ipaC fused to mCFP and venus genes, respectively, the ipaCic truncated derivatives, as well as pBadCi are given in the Supplementary data.
Antibodies and reagents
The anti-IpaC, -IpaD, -IpgC, -IcsA, and -LPS polyclonal antibodies, and the anti-IpaC monoclonal antibodies J22 have been described previously (Phalipon et al, 1992; Menard et al, 1994a). Polyclonal antiserum against β-lactamase was purchased from 5′–3′. The lipophilic styryl dye FM 4-64, the FlAsH reagent (Lumio green in cell), anti-rabbit IgG antibodies coupled to Alexa Fluor 568, anti-mousse IgG coupled to Cy5, and phalloidin coupled to Alexa Fluor 633 were from Invitrogen Corporation.
Immunoprecipitation
To test the association between IpaCic and the IpgC chaperone, bacterial strains were grown to OD600 0.3. Bacteria were centrifuged at 5000 g for 10 min at at 4°C, and bacterial pellets were washed in lysis buffer containing 50 mM NaCl, 0.1 mM EDTA, 0.1 mM DTT, and 50 mM Tris, pH 7.4, in the presence of protease inhibitors (CompleteTM; Roche Diagnostics GmBh, Mannheim, Germany). Bacterial pellets were resuspended in 1/20th of the initial volume in lysis buffer. Samples were transferred in a glass tube in an ice-bath and subjected to sonication using a microtip and a Branson Sonifier 250. Each sample was sonicated at output 3 for 4 times 20 seconds. All the subsequent steps were carried out at 4°C. Samples were centrifuged at 2000 g for 10 min, and the supernatant was transferred to an ultracentrifuge tube and centrifuged for 30 min at 45 K in a 55-Ti rotor. The supernatant (S1) was used as soluble fraction of the bacterial lysates for immunoprecipitation. A 500-μl volume of S1 aliquots was incubated with the polyclonal anti-IpaC antibody at 1:200 dilution for 30 min on ice, followed by incubation with protein G–Sepharose 4B freshly equilibrated in the lysis buffer (30 μl beads per sample) with end-over-end rolling for 60 min. After three washes in lysis buffer, samples were resuspended in Laemmli loading buffer, boiled for 5 min, and analyzed by western blotting using the J22 anti-IpaC monoclonal antibody (2 μg/ml final concentration) or anti-IpgC (diluted 1:1000), as previously described (Menard et al, 1994a).
Bacterial immunofluorescence labeling
To label IcsA at the bacterial surface, overnight pre-cultures of bacteria were diluted 100 times in TCS broth and incubated at 37°C until the cultures reached OD600 of 0.5. Bacteria were allowed to bind for 10 min to glass coverslips, which were previously coated with poly-L-lysine at a final concentration of 50 μg/ml, and fixed for 15 min with 4% of paraformaldehyde in phosphate-buffered saline (PBS). Samples were washed three times in PBS and incubated 20 min with 2% of bovine serum albumin (BSA) in PBS. Samples were first incubated for 1 h with anti-IcsA rabbit polyclonal antibody (diluted 1:100) and anti-LPS mouse polyclonal antibody (1:1000). Samples were washed three times with PBS and incubated for 1 h with anti-rabbit IgG antibody coupled to Alexa Fluor 568 (diluted 1:400) and anti-mouse IgG antibody coupled to Cy5 (diluted 1:200). Samples were washed three times with PBS and mounted with Prolong (Molecular Probes). Bacterial labeling with the FlAsH reagent was essentially performed as described previously (Enninga et al, 2005), except that bacteria were incubated with the FlAsH reagent at a final concentration of 5 μM for 3 h at 21°C.
Chase assay
E. coli bacteria of strain TOP10F' (Invitrogen), containing plasmids pCHAP4500 and pBadCi or pBadCi alone, were grown in LB medium in the absence or presence of 0.01 or 0.02% L-arabinose for 2 h at 37°C. Cultures were diluted four-fold and IPTG (1 mM final concentration) was added to induce the expression of wild-type ipaC, and growth was continued for another 2 h. Before and after 2 h of IPTG induction, bacteria from 0.5 ml of cultures at OD600=0.5 were collected by centrifugation and subjected to two consecutive washes in PBS, and then allowed to bind to coverslips previously coated with poly-L-lysine at a final concentration of 50 μg/ml. Samples were processed for fluorescence microscopy. In parallel, samples of the same cultures were centrifuged, resuspended in Laemmli loading sample buffer, and analyzed by western blotting using rabbit polyclonal antibodies against anti-IpaC (diluted 1:10 000) (Mounier et al, 1997), or against the β-lactamase (diluted 1:2000) that served as internal control (5′–3′).
HeLa cells infection and immunofluorescence
Infection of HeLa cells by Shigella was performed as described (Clerc and Sansonetti, 1987). Sixteen hours before the experiment, HeLa cells were plated at a density of 4 × 105 cells per well onto a six-well plate containing 22 × 22 mm glass coverslips. Briefly, bacteria were grown in TCS until mid-exponential phase, centrifuged, and resuspended in DMEM containing 20 mM HEPES pH 7.3. Bacteria were added to cells and centrifuged at 700 g for 10 min at 21°C, using a Rotixa/RP swinging-bucket centrifuge equipped with tray holders (Hettich). Samples were incubated during various times at 37°C to allow infection, and then fixed for 15 min with 4% paraformaldehyde in PBS. They were washed three times in PBS and permeabilized for 5 min with 0.1% of Triton X-100 in PBS. Samples were washed three times and blocked with 2% BSA in PBS for 30 min. Samples were first incubated with the rabbit polyclonal anti-IpaC antibody (diluted 1:500) for 1 h. Then samples were washed three times with PBS and incubated with anti-rabbit IgG antibody coupled to Alexa Fluor 568 (diluted 1:400) and phalloidin coupled to Alexa Fluor 633 (diluted 1:100) for another hour. Samples were washed three times with PBS and mounted with Prolong (Invitrogen Corporation).
Secretion assay and proteinase K digestion
Overnight precultures of bacteria were diluted 1:200 in TCS broth and incubated at 37°C until they reached OD600 of 0.5. For each sample and each time point, 2 ml of the bacterial culture were centrifuged for 30 s at 16 100 g in an Eppendorf microcentrifuge, and the bacterial pellet was resuspended in 500 μl PBS containing 0.01% of Congo red. Samples were incubated at 37°C and at the specified time points, and then were centrifuged for 30 s at 16 100 g, and the supernatant was immediately transferred to a fresh tube. The supernatant was subjected to precipitation using trichloroacetic acid at 5% final concentration, as described previously (Menard et al, 1994a), and the precipitated proteins were resuspended 40 μl Laemmli loading sample buffer. Samples were analyzed in SDS–PAGE, followed by Coomassie blue staining. Alternatively, following centrifugation of the culture, the bacterial pellet was resuspended in the same volume of PBS as the supernatant, and 10 μl of each fraction were analyzed by rabbit western blotting using polyclonal anti-IpaC antibody, followed by anti-rabbit IgG coupled to horse radish peroxidase and detection using the ECL+system (GE Healthcare).
To control that IpaD detected in the pellet fractions after induction of secretion by Congo red, did not correspond to secreted but aggregated material, proteinase K digestion experiments were conducted (Menard et al, 1994b). In these experiments, following bacterial growth and induction of secretion with Congo red as described above, samples were chilled at various time points in an ice-cold bath for 5 min to stop the reaction. Proteinase K was added at a final concentration of 10 μg/ml and the samples were incubated for 10 min at 20°C. Proteinase K was inactivated by addition of phenylmethylsulfonyl fluoride (2 mM), and samples were analyzed by SDS–PAGE and anti-IpaD western blotting.
Microscopy
Analysis of fixed samples was performed using a LSM 510 Meta (Zeiss) and an SP5 (Leica Microsystems) confocal laser scanning microscopes with a × 63/1.4 Apochromat objective. Confocal sections were deconvolved using Huygens 2 and a theoretically calculated point spread function, and images were processed using the Image J software. Time-lapse microscopy of IpaCiv-expressing bacteria growing on a pad containing 1% agarose in TCS was performed as previously described (Stewart et al, 2005), using a wide-field fluorescence inverted DMRIBe microscope (Leica Microsystems), with a × 63/1.32 HCX PL APO objective and fluorescence band pass filters (excitation 480±20 nm, emission 527±30 nm), equipped with a Cascade 512B CCD camera (Roper Instruments). In these experiments, IpaCiv, which consists of IpaC fused to the Venus variant of GFP, was used because Venus is characterized by a short maturation time that limits drawbacks linked to the time required for GFP folding (Nagai et al, 2002). Images of bacteria were captured every minute during 3–5 h. Image acquisition and analysis were performed with Metamorph (Universal Imaging Corp.).
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Movie
Supplementary Figure Legends
Supplementary References
Acknowledgments
We thank Drs Debabrata RayChaudhuri and Anthony Pugsley and members of the Unité de Pathogénie Microbienne Moléculaire for helpful discussions. We also thank Dr Atsushi Miyawaki for the gift of the Venus construct. We are grateful to Drs Jost Enninga and Stéphane Romero for technical advices, and Emmanuelle Perret and Pascal Roux at the PFID of the Institut Pasteur for help in image acquisition. PJS is a Howard Hugues fellow. This work was supported in part by a grant from the ACI Microbiologie 2003 to OF.
References
- Akeda Y, Galan JE (2005) Chaperone release and unfolding of substrates in type III secretion. Nature 437: 911–915 [DOI] [PubMed] [Google Scholar]
- Allaoui A, Sansonetti PJ, Parsot C (1993) MxiD, an outer membrane protein necessary for the secretion of the Shigella flexneri Ipa invasions. Mol Microbiol 7: 59–68 [DOI] [PubMed] [Google Scholar]
- Atmakuri K, Cascales E, Burton OT, Banta LM, Christie PJ (2007) Agrobacterium ParA/MinD-like VirC1 spatially coordinates early conjugative DNA transfer reactions. EMBO J 26: 2540–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blocker A, Gounon P, Larquet E, Niebuhr K, Cabiaux V, Parsot C, Sansonetti P (1999) The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J Cell Biol 147: 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon LD, Goehring N, Janakiraman A, Yan AW, Wu T, Beckwith J, Goldberg MB (2003) IcsA, a polarly localized autotransporter with an atypical signal peptide, uses the Sec apparatus for secretion, although the Sec apparatus is circumferentially distributed. Mol Microbiol 50: 45–60 [DOI] [PubMed] [Google Scholar]
- Buttner D, Bonas U (2002) Port of entry—the type III secretion translocon. Trends Microbiol 10: 186–192 [DOI] [PubMed] [Google Scholar]
- Campo N, Tjalsma H, Buist G, Stepniak D, Meijer M, Veenhuis M, Westermann M, Muller JP, Bron S, Kok J, Kuipers OP, Jongbloed JD (2004) Subcellular sites for bacterial protein export. Mol Microbiol 53: 1583–1599 [DOI] [PubMed] [Google Scholar]
- Charles M, Perez M, Kobil JH, Goldberg MB (2001) Polar targeting of Shigella virulence factor IcsA in enterobacteriaceae and Vibrio. Proc Natl Acad Sci USA 98: 9871–9876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc P, Sansonetti PJ (1987) Entry of Shigella flexneri into HeLa cells: evidence for directed phagocytosis involving actin polymerization and myosin accumulation. Infect Immun 55: 2681–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes BK, Finlay BB (2005) Insertion of the bacterial type III translocon: not your average needle stick. Trends Microbiol 13: 92–95 [DOI] [PubMed] [Google Scholar]
- Cornelis GR (2006) The type III secretion injectisome. Nat Rev Microbiol 4: 811–825 [DOI] [PubMed] [Google Scholar]
- Cossart P, Sansonetti PJ (2004) Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 304: 242–248 [DOI] [PubMed] [Google Scholar]
- Ebersbach G, Jacobs-Wagner C (2007) Exploration into the spatial and temporal mechanisms of bacterial polarity. Trends Microbiol 15: 101–108 [DOI] [PubMed] [Google Scholar]
- Enninga J, Mounier J, Sansonetti P, Tran Van Nhieu G (2005) Secretion of type III effectors into host cells in real time. Nat Methods 2: 959–965 [DOI] [PubMed] [Google Scholar]
- Espina M, Olive AJ, Kenjale R, Moore DS, Ausar SF, Kaminski RW, Oaks EV, Middaugh CR, Picking WD, Picking WL (2006) IpaD localizes to the tip of the type III secretion system needle of Shigella flexneri. Infect Immun 74: 4391–4400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JE, Wolf-Watz H (2006) Protein delivery into eukaryotic cells by type III secretion machines. Nature 444: 567–573 [DOI] [PubMed] [Google Scholar]
- Goldberg MB, Barzu O, Parsot C, Sansonetti PJ (1993) Unipolar localization and ATPase activity of IcsA, a Shigella flexneri protein involved in intracellular movement. J Bacteriol 175: 2189–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa Y, Suzuki M, Ohya K, Iwai H, Ishijima N, Koleske AJ, Fukui Y, Sasakawa C (2007) Shigella IpgB1 promotes bacterial entry through the ELMO-Dock180 machinery. Nat Cell Biol 9: 121–128 [DOI] [PubMed] [Google Scholar]
- Harrington AT, Hearn PD, Picking WL, Barker JR, Wessel A, Picking WD (2003) Structural characterization of the N terminus of IpaC from Shigella flexneri. Infect Immun 71: 1255–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe AD, Swanson JA (2004) Cdc42, Rac1, and Rac2 display distinct patterns of activation during phagocytosis. Mol Biol Cell 15: 3509–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd PK, Kumar RB, Das A (2005) Spatial location and requirements for the assembly of the Agrobacterium tumefaciens type IV secretion apparatus. Proc Natl Acad Sci USA 102: 11498–11503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galan JE, Aizawa SI (1998) Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280: 602–605 [DOI] [PubMed] [Google Scholar]
- Kueltzo LA, Osiecki J, Barker J, Picking WL, Ersoy B, Picking WD, Middaugh CR (2003) Structure–function analysis of invasion plasmid antigen C (IpaC) from Shigella flexneri. J Biol Chem 278: 2792–2798 [DOI] [PubMed] [Google Scholar]
- Laviña N, Pugsley AP, Moreno MF (1986) Identification, mapping, cloning and characterization of a gene (SbmA) required for microcin B17 action on Escherichia coli K-12. J Gen Microbiol 132: 1685–1693 [DOI] [PubMed] [Google Scholar]
- Menard R, Sansonetti P, Parsot C (1994a) The secretion of the Shigella flexneri Ipa invasins is activated by epithelial cells and controlled by IpaB and IpaD. EMBO J 13: 5293–5302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard R, Sansonetti P, Parsot C, Vasselon T (1994b) Extracellular association and cytoplasmic partitioning of the IpaB and IpaC invasins of S flexneri. Cell 79: 515–525 [DOI] [PubMed] [Google Scholar]
- Menard R, Sansonetti PJ, Parsot C (1993) Non-polar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol 175: 5899–5906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounier J, Bahrani F, Sansonetti PJ (1997) Secretion of Shigella flexneri Ipa invasins on contact with epithelial cells and subsequent entry of the bacterium into cells are growth stage dependent. Infect Immun 65: 774–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller CA, Broz P, Muller SA, Ringler P, Erne-Brand F, Sorg I, Kuhn M, Engel A, Cornelis GR (2005) The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science 310: 674–676 [DOI] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A (2002) A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20: 87–90 [DOI] [PubMed] [Google Scholar]
- Page AL, Fromont-Racine M, Sansonetti P, Legrain P, Parsot C (2001) Characterization of the interaction partners of secreted proteins and chaperones of Shigella flexneri. Mol Microbiol 42: 1133–1145 [DOI] [PubMed] [Google Scholar]
- Phalipon A, Arondel J, Nato F, Rouyre S, Mazie JC, Sansonetti PJ (1992) Identification and characterization of B-cell epitopes of IpaC, an invasion-associated protein of Shigella flexneri. Infect Immun 60: 1919–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picking WL, Coye L, Osiecki JC, Barnoski Serfis A, Schaper E, Picking WD (2001) Identification of functional regions within invasion plasmid antigen C (IpaC) of Shigella flexneri. Mol Microbiol 39: 100–111 [DOI] [PubMed] [Google Scholar]
- Sansonetti PJ, Kopecko DJ, Formal SB (1982) Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect Immun 35: 852–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott ME, Dossani ZY, Sandkvist M (2001) Directed polar secretion of protease from single cells of Vibrio cholerae via the type II secretion pathway. Proc Natl Acad Sci USA 98: 13978–13983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi D, Yoshimoto M, Homma M, Kawagishi I (2006) Helical distribution of the bacterial chemoreceptor via colocalization with the Sec protein translocation machinery. Mol Microbiol 60: 894–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart E, Madden R, Paul G, Taddei F (2005) Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol 2: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson JA, Baer SC (1995) Phagocytosis by zippers and triggers. Trends Cell Biol 5: 89–93 [DOI] [PubMed] [Google Scholar]
- Tamano K, Aizawa S, Katayama E, Nonaka T, Imajoh-Ohmi S, Kuwae A, Nagai S, Sasakawa C (2000) Supramolecular structure of the Shigella type III secretion machinery: the needle part is changeable in length and essential for delivery of effectors. EMBO J 19: 3876–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran Van Nhieu G, Caron E, Hall A, Sansonetti PJ (1999) IpaC induces actin polymerization and filopodia formation during Shigella entry into epithelial cells. EMBO J 18: 3249–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia RH, Falkow S (1996) Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol Microbiol 22: 367–378 [DOI] [PubMed] [Google Scholar]
- Veenendaal AK, Hodgkinson JL, Schwarzer L, Stabat D, Zenk SF, Blocker AJ (2007) The type III secretion system needle tip complex mediates host cell sensing and translocon insertion. Mol Microbiol 63: 1719–1730 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Movie
Supplementary Figure Legends
Supplementary References