Abstract
The chaperone protein network controls both initial protein folding and subsequent maintenance of proteins in the cell. Although the native structure of a protein is principally encoded in its amino-acid sequence, the process of folding in vivo very often requires the assistance of molecular chaperones. Chaperones also play a role in a post-translational quality control system and thus are required to maintain the proper conformation of proteins under changing environmental conditions. Many factors leading to unfolding and misfolding of proteins eventually result in protein aggregation. Stress imposed by high temperature was one of the first aggregation-inducing factors studied and remains one of the main models in this field. With massive protein aggregation occurring in response to heat exposure, the cell needs chaperones to control and counteract the aggregation process. Elimination of aggregates can be achieved by solubilization of aggregates and either refolding of the liberated polypeptides or their proteolysis. Here, we focus on the molecular mechanisms by which heat-shock protein 70 (Hsp70), Hsp100 and small Hsp chaperones liberate and refold polypeptides trapped in protein aggregates.
Keywords: chaperones, Hsp70, Hsp100, protein disaggregation, protein folding
Introduction
One of the most prominent qualities of proteins is their ability to find their native folded conformation within a vast conformational space. The idea of an ‘energy landscape' (reviewed in Dobson, 2004) helped us to understand how it is possible to overcome the Levinthal paradox in protein folding. However, interactions stabilizing the unique conformation of energetic minimum combined with proper functioning are counterbalanced by a large decrease in entropy. Thus, native proteins have a low margin of stability and are always more or less on the verge of denaturation. The level of expression of human genes shows an astonishingly tight inverse correlation with aggregation rates of the corresponding proteins measured in vitro, suggesting that there is hardly any margin of safety to respond to factors that decrease protein solubility (Tartaglia et al, 2007). The situation inside a living cell is much more complex than under idealized test-tube conditions of low protein concentration and a carefully chosen temperature. Molecular crowding and rapidly changing environmental factors interfere with the process of folding of newly synthesized chains or promote loss of native conformation in mature polypeptides. Loss of native conformation not only leads to depletion of functional proteins, but also leads to another problem in the cell, namely aggregation of polypeptides. A special class of proteins, called molecular chaperones, has evolved to counteract this process. The chaperones are a large and diverse group of unrelated proteins that assist in correct noncovalent assembly and/or disassembly of other polypeptide-containing structures, but which are not permanent components of these structures when they are performing their normal biological functions (Ellis, 1997). Either constitutively expressed or induced under stress conditions, they are essential for proper functioning of the cell and are found in every living organism. Here, we focus on the specific mechanisms by which chaperones from three unrelated families, namely heat-shock protein 70 (Hsp70), Hsp100 and the small Hsps, act on protein aggregates to liberate and refold the polypeptides trapped inside the aggregates.
Chaperones and protein folding
Under physiological conditions, proteins are threatened with aggregation from the very first moments of their existence. In a native protein, the interacting amino-acid residues are often dispersed along the polypeptide sequence, and only folding of the polypeptide chain brings them in proximity. During translation, since all the interacting residues are not yet present prior to chain termination, it is not possible for the new polypeptide chain to form all the proper amino-acid contacts that determine the protein's native structure. Additionally, the hydrophobic stretches that are normally hidden inside the three-dimensional structure of a properly folded protein are not shielded from the environment and increase the tendency to form non-native contacts. To minimize the risk of protein aggregation, the cells evolved a system of protecting nascent proteins during folding. The first line of defense is drawn at the ribosome, where prokaryotic trigger factor, or NAC protein in the case of eukaryotic organisms, associates with a nascent peptide to hold it in a folding-competent state. In prokaryotes, where most proteins are rather small, it is estimated that 65–80% of them fold immediately after synthesis. The remaining polypeptides interact with the Hsp70 chaperone system (Deuerling et al, 1999; Teter et al, 1999). It consists of Hsp70 protein, a cooperating DnaJ protein (Hsp40) and a nucleotide exchange factor. Hsp70 possesses two functional domains, an ATPase domain (nucleotide-binding domain, NBD) and a substrate-binding domain (SBD) capable of interaction with an extended polypeptide chain and equipped with a helical ‘lid'. The movement of the lid is controlled by a nucleotide bound to NBD, resulting in either locking of the substrate chain within the SBD or opening the lid and thus releasing the substrate, allowing a subsequent binding event (Mayer and Bukau, 2005; Vogel et al, 2006). The DnaJ protein influences the cycle by stimulating Hsp70 to hydrolyze ATP, while the nucleotide exchange factor promotes ADP dissociation (Liberek et al, 1991). About 10–20% of nascent prokaryotic proteins (and similar levels of eukaryotic proteins) are able to fold properly due to binding to the Hsp70 system. In more difficult cases (approximately 10–15% of the total), eubacteria use a chaperonin system. It consists of a large, oligomeric, double-ring GroEL protein complex forming a two-chambered barrel-like structure, which encloses the improperly folded protein. In a GroES- (a co-chaperone) and ATP-dependent manner, the GroEL cage provides isolated conditions (Anfinsen cage) for folding of a substrate inside the barrel (for review, see Hartl and Hayer-Hartl, 2002). Eukaryotes utilize the analogous TRiC chaperonin system, in a manner dependent on prefolding protein, which interacts with chains emerging from the ribosome, acting co- or post-translationally on approximately 10% of all proteins (Hartl and Hayer-Hartl, 2002). The difference between the prokaryotic and eukaryotic strategies is that, besides the specialized trigger factor, prokaryotes rely on the universal Hsp70 and Hsp60 systems, which have both housekeeping and stress response duties, while in eukaryotes one may distinguish between CLIPS (chaperones linked in protein synthesis) and HSPs, chaperones induced by stress (Albanèse et al, 2006).
Successful initial folding does not ensure long-term stability of a protein, as it still may be affected by many factors leading to partial protein unfolding and subsequent aggregation. For example, mutations in the corresponding DNA sequence, resulting in an increase in polyglutamine repeats, lead to decreased stability and promotes non-native self-association of polypeptides. The formation of amyloid fibrils, induced by partial protein unfolding and enrichment in beta structure, has been studied thoroughly (for review see Jahn and Radford, 2008) due to their involvement in the pathogenesis of many human diseases (Stefani, 2004). Amyloid fibril generation may also be triggered by contagious factors called prions—proteins that switch from native to an alternate, beta-enriched conformation and induce such transitions in other native molecules of the same protein (for review see Prusiner, 1998). In fact, it is believed that the ability to form amyloid fibres may be a common feature of proteins (Stefani, 2004), and the theoretical analysis suggests that this process is ruled by a nonlinear, bistable model, which explains the possibility of triggering massive aggregation effects by relatively small influencing factors (Rieger et al, 2006).
As mentioned before, proteins evolved to possess a relatively small margin of stability (Tartaglia et al, 2007). Therefore, relatively minor changes in physicochemical intracellular conditions, such as an increase in temperature over the physiological limit, often destabilize proteins and cause their aggregation. These aggregates are often amorphous, in contrast to amyloid structures. This type of aggregation is a common problem, especially for organisms that cannot regulate their internal temperature. Here, we focus on molecular mechanisms that allow plant, bacterial and fungal cells to rescue proteins from aggregates and thus increase their chances to survive heat stress conditions. It should be noted that for a cell to survive, it is important not only to deal with aggregates but first and foremost to secure sufficient amounts of active proteins required for metabolism.
Chaperone proteins in changing environmental conditions
Changes in environmental conditions, such as an increase in temperature, challenge the conformation of proteins. Hydrophobic amino-acid residues, buried inside proteins in their native conformation, are exposed under destabilizing conditions, promoting intracellular protein aggregation. Cell survival under such conditions is strongly enhanced by preconditioning at elevated temperatures. The resulting so-called acquired thermotolerance (and tolerance to other factors as well) is due to an increase in chaperone protein synthesis during the preconditioning heat shock (Li and Werb, 1982). It was shown that yeast cells expressing Hsp104 chaperone survive exposure to high temperatures and other conditions that destabilize protein structure, 1000- to 10 000-fold better than cells lacking Hsp104 (Sanchez and Lindquist, 1990). Similar effects were observed for ClpB and Hsp101, the orthologous gene products in bacterial and plant cells, respectively (Kitagawa et al, 1991; Squires et al, 1991; Queitsch et al, 2000). In vivo studies linked the removal of temperature-induced intracellular aggregates to the chaperones responsible for thermotolerance, for example, Hsp104 in the yeast Saccharomyces cerevisiae (Parsell et al, 1994) and ClpB in Escherichia coli (Mogk et al, 1999), both members of the AAA+ (ATPase associated with diverse cellular activities) family. However, several in vitro studies determined that the ClpB and Hsp104 chaperones are not able to disaggregate substrates effectively on their own. In this process, collaboration with the Hsp70 chaperone system (in E. coli: DnaK and co-chaperones DnaJ, GrpE; in S. cerevisiae: Ssa1p and co-chaperone Ydj1p) is required (Glover and Lindquist, 1998; Goloubinoff et al, 1999; Motohashi et al, 1999; Zolkiewski, 1999). Phenotypic effects related to protein aggregation are observed both in clpB and in dnaK, dnaJ or grpE mutants. However, the phenotype of the latter mutants is a result of multiple factors. In addition to their role in protein disaggregation, the components of the Hsp70 system are involved in several other vital cellular processes, including protein folding accompanying translation (Deuerling et al, 1999; Teter et al, 1999), remodeling of protein complexes (Liberek et al, 1988; Zzaman et al, 2004; Goldfless et al, 2006) and regulation of the heat-shock response (Gamer et al, 1992; Liberek and Georgopoulos, 1993), which makes it difficult to analyze the effects related to intracellular aggregation alone.
Elimination of intracellular aggregates can occur either by disaggregation mediated by the Hsp70–Hsp100 bichaperone system and subsequent refolding process, or by proteolysis. A study analyzing removal of aggregated proteins in E. coli strains deficient for genes encoding proteases or chaperones involved in disaggregation revealed that the most prominent defect was caused by deletion of the clpB gene (Laskowska et al, 1996a; Mogk et al, 1999). The importance of the disaggregation process for the cell was substantiated further by mutagenesis studies in which ClpB was modified so that it acted in the degradation pathway instead of disaggregation. The mutant strain showed a lack of thermotolerance, even though intracellular aggregates were efficiently eliminated (Weibezahn et al, 2004). Similar conclusions on the importance of protein disaggregation and reactivation following heat stress were also drawn in studies of DNA replication and translation in mitochondria of the yeast S. cerevisiae. Mitochondria constitute an attractive model for such studies, since nearly all of the mitochondrial proteins are nuclear-encoded and imported into mitochondria. Thus, blocking the translation of nuclear genes eliminates the import of newly synthesized proteins, and the recovery of mitochondrial functions becomes fully dependent on the status and activity of proteins already present. Such studies assessed that the recovery of mitochondrial DNA replication (Germaniuk et al, 2002), translation (Schmitt et al, 1996) and also proper mitochondrial morphology (Lewandowska et al, 2006) following severe heat stress rely more on the reactivation of thermally inactivated proteins residing inside mitochondria, than on the import of newly synthesized proteins.
The experimental data showing that disaggregation and reactivation of aggregated proteins is favored over proteolysis is not entirely surprising. One can assume that the major energy-consuming step for both disaggregation and proteolysis is the separation of a polypeptide chain from aggregates and its unfolding by either a disaggregating chaperone or a protease-regulatory subunit. However, a complete, properly folded protein is the result of the disaggregation process only, in contrast to proteolysis, which leaves the cell with a pool of amino acids. Both energy input and a certain period of time are required to synthesize new polypeptide chains. Thus, the overall gain is significantly different in these two processes despite the similar initial energy expense required to dissolve the aggregate.
Do the Hsp70 and Hsp100 chaperones play any role other than disaggregation in protein quality control under stress conditions? It was proposed that one of the functions of the Hsp70 chaperones is prevention of protein aggregation (Pelham, 1986). This is easily shown in vitro for all Hsp70 chaperones and very often used as a hallmark of Hsp70 activity. However, it is difficult to assess the importance of this process in vivo, since mutations in Hsp70 potentially influence both the prevention and disaggregation of protein aggregates. Additionally, both processes may result in the association of Hsp70 chaperones with protein aggregates. Therefore, the association of Hsp70 with aggregates cannot serve to discriminate between the two functions. The nature of Hsp70 activity, that is, the binding and release of substrates controlled by its co-chaperones, challenges its role in prevention of aggregation, since that would require a rather stable interaction. On the other hand, it was postulated that GrpE (the nucleotide release factor) may function as a thermosensor and at high temperature change the conformation of DnaK to a substrate-binding state (Groemping and Reinstein, 2001; Grimshaw et al, 2003). There are also reports (Park et al, 2007) that associate Hsp70 chaperone activity with proteolysis and suggest that the Hsp70 system can cooperate with proteases in a manner similar to the way it cooperates with disaggregating Hsp100 chaperones.
Mutations in genes encoding disaggregating chaperones result in massive and stable protein aggregation; however, as Hsp100 chaperones do not prevent protein aggregation in in vitro studies, it seems that the major role of at least this family of chaperones is to disaggregate and not to protect proteins from aggregation. In view of the established cooperation between the two protein families, the major task of the Hsp70–Hsp100 bichaperone system seems to be disaggregation rather than prevention of protein aggregation per se.
Hsp100-disaggregating chaperones
ClpB and its orthologs involved in disaggregation belong to the Hsp100 chaperone family and consist of an N-terminal domain and two ATP-binding domains (AAA domains) essential for hexamerization and chaperone function (Schirmer et al, 1996; Zolkiewski, 2006). Despite their diverse functions, AAA+ superfamily members share important features: oligomeric structure, homologous Walker-type nucleotide-binding domain(s) and the ability to utilize energy derived from ATP hydrolysis for remodeling of their substrates (Vale, 2000; Ammelburg et al, 2006). The crystal structure of the monomeric ClpB subunit from Thermus thermophilus and the cryoelectron microscopy reconstitution of its oligomer defined the structure and orientation of its domains. ClpB forms a two-tiered, hexameric ring, with AAA-1 and AAA-2 domains forming the top and bottom ring, respectively, and surrounding an axial pore, approximately 16 Å in diameter (Lee et al, 2003; Diemand and Lupas, 2006). Studies of structural changes in T. thermophilus ClpB in response to nucleotide binding showed that the top ring undergoes ATP-driven conformational change, while the bottom ring remains unchanged (Lee et al, 2007). The AAA-1 domain contains an additional propeller-shaped coiled-coil region protruding from the hexamer, called the ‘middle domain', and built of four α-helices (Schirmer et al, 1996; Lee et al, 2003). Genetic and biochemical analysis defined the mobility of this domain as essential for the proper functioning of these disaggregating chaperones (Lee et al, 2003). This domain differentiates chaperones involved in disaggregation from other family members (ClpA and ClpX) that possess a dual function, being either regulatory subunits of proteases (Wojtkowiak et al, 1993) or acting as molecular chaperones themselves (Wickner et al, 1994; Wawrzynow et al, 1995). The ClpB N-terminal domain, linked to the AAA-1 domain and located on the top of the ring, was suggested to support the interaction of the chaperone with aggregates (Barnett et al, 2005).
Hsp70/Hsp100-dependent protein disaggregation
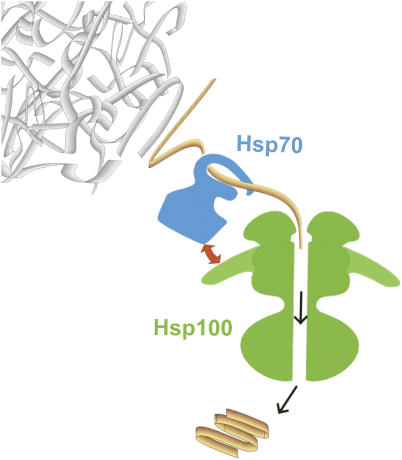
The mechanism of protein disaggregation is not yet fully understood and several models have been proposed. Initially, interaction of ClpB with aggregates at the first step of disaggregation was postulated. This interaction was suggested to change the structure of aggregates into a form recognizable by the Hsp70 chaperone (Ben-Zvi and Goloubinoff, 2001). Another model predicted that ClpB first breaks large aggregates into smaller fragments (Lee et al, 2003) that are further processed by Hsp70. More recently, it was postulated that it is rather the Hsp70 chaperone system that acts on aggregates during the initial step of disaggregation (Weibezahn et al, 2004; Ziętkiewicz et al, 2004) (Figure 1). Bukau and co-workers constructed a ClpB variant capable of participating in proteolysis. The IGL motif, introduced within a short loop of the AAA-2 domain, is characteristic of the regulatory subunits of Clp proteases and is responsible for recognition of the proteolytic subunit. In the presence of the mutated ClpB variant and proteolytic subunit ClpP, aggregated substrates were efficiently digested only in the presence of the Hsp70 chaperone system, highlighting the importance of the threading mechanism of ClpB action and the necessity of Hsp70 action upstream of ClpB (Figure 1). A similar conclusion on the order of events in the disaggregation process was drawn from kinetic studies, which showed that initial incubation of aggregates with the Hsp70 chaperone system but not ClpB changed the physical properties of the aggregates and abolished a delay at the start of the disaggregation reaction (Ziętkiewicz et al, 2004). Furthermore, it was reported that the Hsp70 chaperone system is able to disentangle polypeptides from aggregates in the absence of ClpB (Ziętkiewicz et al, 2006). However, in the absence of ClpB the polypeptides were not able to fold into native structures, suggesting a role for ClpB in the folding process and not necessarily in the separation of polypeptides from aggregates. In a recent mutagenesis study, the propeller-shaped middle domain of ClpB was shown to couple the ClpB threading activity with Hsp70 chaperone activity, suggesting that Hsp70 shuffles polypeptides to the entrance of the ClpB channel through interaction with this domain (Haslberger et al, 2007). The exact mode of cooperation between Hsp70 and Hsp100 remains elusive. However, the cooperation is highly specific, since disaggregation both in vivo and in vitro proceeds only when the Hsp70 and Hsp100 chaperones originate from the same organism (Glover and Lindquist, 1998; Krzewska et al, 2001).
Figure 1.
Model for the mechanism of action of Hsp70 and Hsp100 chaperone system in disaggregation of protein aggregates. First, the Hsp70 chaperone system disentangles the polypeptides from aggregates. The polypeptides are then transferred to ClpB/Hsp104 and unfolded by translocation through the central channel, driven by energy from ATP hydrolysis. The unfolded polypeptides are released following translocation and refold either spontaneously or with the assistance of chaperones. The Hsp70 chaperone system and ClpB/Hsp104 cooperate in the disaggregation process in a specific way (marked with the red arrow). The interaction is linked to the propeller-shaped middle domain of ClpB.
A study by Bukau and co-workers determined that the translocation of a polypeptide chain through the central channel of ClpB is an important step in the disaggregation and reactivation processes (Weibezahn et al, 2004). The importance of channel integrity for the activity of Hsp100 chaperones was additionally shown by mutagenesis studies (Lum et al, 2004; Schlieker et al, 2004). The exact mechanism of translocation was not established for the disaggregating chaperones. However, studies were performed for ClpA, an analogous protein with dual functions, being a protease-regulatory subunit and a chaperone. According to these studies, the binding of a substrate to three loops facing the central channel of ClpA is crucial for substrate unfolding. It was proposed that mechanical force is exerted on the substrate by the movement of one of the loops, triggered by ATP hydrolysis, which results in translocation and unfolding of the substrate (Hinnerwisch et al, 2005).
Recently it was reported that in the absence of the Hsp70 chaperone system both ClpB and Hsp104 are able to perform the disaggregation reaction, albeit with a very low efficiency, provided that the reaction mixture was additionally supplemented with the nonphysiological nucleotide ATPγS (Schaupp et al, 2007; Doyle et al, 2007a, 2007b). This observation raises the possibility that in the disaggregation reaction, the Hsp70 chaperone system not only plays a role in delivering the polypeptide from aggregates to the central pore of Hsp100, but also influences the ATPase cycle of Hsp100 in a way that is mimicked by the artificial nucleotide state obtained by Hsp100 in the presence of both ATPγS and ATP. Recent mutagenesis studies reveal that ATP binding to NBD1 serves as a regulatory switch for Hsp104 activity. It triggers binding of polypeptides and greatly stimulates ATP hydrolysis in NBD2, implying that ATP hydrolysis in this domain is important for disaggregation (Schaupp et al, 2007).
The newly released polypeptide chain regains its native structure downstream of ClpB action. By analogy to the cotranslational folding of a polypeptide chain emerging from a ribosome, the threading mechanism could allow sequential folding of the polypeptide chain emerging from the central channel of ClpB. Therefore, it is very likely that the role of the Hsp70 chaperones is not restricted only to disentangling polypeptides from aggregates and docking of the unfolded polypeptide to ClpB prior to translocation. These chaperones most likely also support folding of a polypeptide chain emerging from the Hsp100 channel, as established for post-translational folding. This explains why the disaggregation reaction in vitro cannot be divided into sequential steps dependent either on Hsp100 or Hsp70 (Goloubinoff et al, 1999; Ziętkiewicz et al, 2004).
The efficiency of disaggregation and reactivation catalyzed by the Hsp70–Hsp100 bichaperone system depends strongly on the physical properties of the protein aggregates. Pioneering in vitro studies performed in the Zylicz laboratory demonstrated that the action of the Hsp70 chaperone system alone is sufficient for disaggregation of certain substrates (Skowyra et al, 1990; Ziemienowicz et al, 1993). Other substrates, however, require bichaperone cooperation in the disaggregation process (Glover and Lindquist, 1998; Goloubinoff et al, 1999; Motohashi et al, 1999; Zolkiewski, 1999). It was also shown that the conditions of aggregation determine the requirement for the presence of Hsp100 in the disaggregation process (Ben-Zvi and Goloubinoff, 2002; Lewandowska et al, 2007). Initially, it was postulated that the requirement for Hsp100 correlates to the size of the aggregates, that is, large aggregates require Hsp100 activity (Goloubinoff et al, 1999; Diamant et al, 2000). Recent studies suggest that the conformational properties of the individual polypeptides forming the aggregates, rather than the size of aggregates, are important factors determining the necessity of the Hsp100 chaperone in the disaggregation process (Lewandowska et al, 2007). It is likely that Hsp70 alone is sufficient for proper folding when polypeptides within the aggregate are close enough to their native conformation and/or not trapped by stable non-native structures. More difficult cases would additionally require unfolding of the polypeptides by Hsp100's translocation activity (Lewandowska et al, 2007).
Small Hsps control aggregate properties
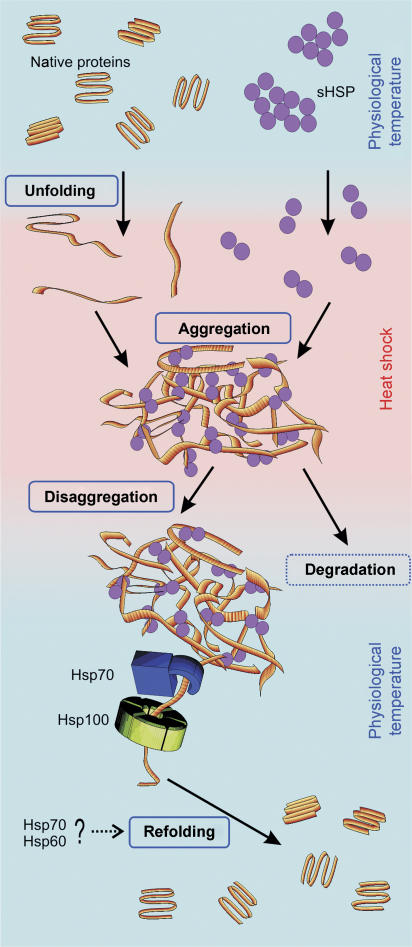
The conformational status of polypeptides in the intracellular aggregates does not depend on the identity of the protein and aggregation conditions alone. Additionally, there exist cellular protein factors—sHsps (Figure 2)—that influence the aggregation process. sHsps are widely distributed in both prokaryotes and eukaryotes. Members of this diverse protein family are characterized by a relatively low monomeric molecular mass (15–43 kDa) and a conserved stretch of approximately 100 amino-acid residues (for reviews see Haslbeck, 2002; Haslbeck et al, 2005). This so-called α-crystallin domain displays sequence similarity to the vertebrate eye lens protein α-crystallin (which prevents protein precipitation and cataract formation in the eye lens). sHsps are distinguished from Hsp100- and Hsp70-disaggregating chaperones and most other chaperones also by the fact that their activity is independent of ATP hydrolysis.
Figure 2.
The Hsp100, Hsp70 and sHsps chaperones control the fate of aggregation-prone proteins under stress conditions.
The deletion of genes encoding sHsps does not usually result in a temperature-sensitive phenotype except in Neurospora crassa (Plesofsky-Vig and Brambl, 1995) and Synechocystis sp. PCC6803 (Giese and Vierling, 2002). However, it has been reported that overproduction of sHsps increases thermotolerance in a number of organisms and cell types, suggesting the involvement of sHsps in the control of protein aggregation and disaggregation upon heat shock (Kitagawa et al, 2000; Nakamoto et al, 2000). In agreement with that, in vivo studies show that in stressed cells sHsps localize mostly in the insoluble protein fraction (Allen et al, 1992; Laskowska et al, 1996b; Basha et al, 2004). Consequently, in E. coli the two genes encoding sHsps were named ‘inclusion body-binding proteins A and B' (IbpA and IbpB) (Allen et al, 1992). Several in vitro studies reported that, in addition to the ability to bind aggregates, sHsps also have a limited ability to protect enzymes from heat denaturation (Jakob et al, 1993; Studer and Narberhaus, 2000).
One of the most striking features of sHsps is their organization in large oligomeric structures, which constantly exchange subunits, presumably dimers (for reviews see Haslbeck, 2002; Haslbeck et al, 2005). An increase in temperature activates the sHsps, thus increasing their affinity for substrates (Haslbeck et al, 1999; Franzmann et al, 2005; Figure 2). The activated subunits of sHsps have the ability to bind protein substrates, both when released from the oligomer and within the oligomeric structure (Franzmann et al, 2005). At the same time, the increase in temperature destabilizes proteins, resulting in aggregation-prone folding intermediates that tend to associate with the temperature-activated sHsps. This leads to the formation of intracellular aggregates containing both destabilized proteins and sHsps. In such cases, the Hsp100/Hsp70-dependent disaggregation and refolding of polypeptides present in these aggregates proceeds more efficiently than in aggregates formed in the absence of sHsps (Lee and Vierling, 2000; Mogk et al, 2003; Matuszewska et al, 2005). The mechanism behind this increase in disaggregation efficiency is elusive. It is believed that the association with sHsps results in changes in the physicochemical properties of individual polypeptides (e.g., the number of hydrophobic contacts with other polypeptides in aggregates, or the number of intramolecular hydrogen bonds within aggregated polypeptides), allowing the Hsp100 and Hsp70 chaperones to separate and unfold the individual polypeptides more efficiently.
Conclusion
There is mounting evidence that the inherent stability of proteins has been thoroughly tailored by evolution (DePristo et al, 2005). Most cellular processes require the structure of a protein to be dynamic and flexible, and increased stability of the protein often is accompanied by a decrease in its activity. This is counterbalanced by the threat of unfolding and/or misfolding, and this low intrinsic stability can sensitize the protein to aggregation. As a result, proteins have evolved to possess a relatively small margin of stability, and moreover, they are tuned to the environmental conditions; for example, temperature specific for the particular species (Zavodszky et al, 1998). Therefore, minor changes in intracellular physicochemical conditions can trigger protein aggregation. Such aggregation is a serious problem for organisms that are constantly challenged by temperature, such as plants, bacteria and fungi. Here, we have discussed the molecular mechanisms that allow chaperones to rescue active proteins from aggregates. sHsps bind to aggregates and influence their properties, while the Hsp70 system is involved in isolation and transfer of polypeptide chains from aggregates to the Hsp100 chaperone, which further unfolds them by translocation through its central channel. The concerted action of chaperones counteracts the aggregation process. Since several vital cellular processes (translation, transcription, DNA replication) are easily blocked due to the collapse of one or more constituent enzymes, the function of chaperones to secure an available pool of active proteins is critical for cellular metabolism. This mechanism of protein quality control (together with the proteolytic machinery necessary for removing an excess of damaged proteins) significantly increases the chances of the cell to survive stress conditions.
Acknowledgments
This work was supported by the Polish Ministry of Science and Higher Education (grant N301 04631/1515). SZ thanks the Foundation for Polish Science for support. We thank Dr Debbie Ang for discussions and critical reading of the paper.
References
- Albanèse V, Yen-Wen Yam A, Baughman J, Parnot C, Frydman J (2006) Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell 124: 75–88 [DOI] [PubMed] [Google Scholar]
- Allen SP, Polazzi JO, Gierse JK, Easton AM (1992) Two novel heat shock genes encoding proteins produced in response to heterologous protein expression in Escherichia coli. J Bacteriol 174: 6938–6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammelburg M, Frickey T, Lupas AN (2006) Classification of AAA+ proteins. J Struct Biol 156: 2–11 [DOI] [PubMed] [Google Scholar]
- Barnett ME, Nagy M, Kedzierska S, Zolkiewski M (2005) The amino-terminal domain of ClpB supports binding to strongly aggregated proteins. J Biol Chem 280: 34940–34945 [DOI] [PubMed] [Google Scholar]
- Basha E, Lee GJ, Breci LA, Hausrath AC, Buan NR, Giese KC, Vierling E (2004) The identity of proteins associated with a small heat shock protein during heat stress in vivo indicates that these chaperones protect a wide range of cellular functions. J Biol Chem 279: 7566–7575 [DOI] [PubMed] [Google Scholar]
- Ben-Zvi AP, Goloubinoff P (2001) Review: mechanisms of disaggregation and refolding of stable protein aggregates by molecular chaperones. J Struct Biol 135: 84–93 [DOI] [PubMed] [Google Scholar]
- Ben-Zvi AP, Goloubinoff P (2002) Proteinaceous infectious behavior in non-pathogenic proteins is controlled by molecular chaperones. J Biol Chem 277: 49422–49427 [DOI] [PubMed] [Google Scholar]
- DePristo MA, Weinreich DM, Hartl DL (2005) Missense meanderings in sequence space: a biophysical view of protein evolution. Nat Rev Genet 6: 678–687 [DOI] [PubMed] [Google Scholar]
- Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B (1999) Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 400: 693–696 [DOI] [PubMed] [Google Scholar]
- Diamant S, Ben-Zvi AP, Bukau B, Goloubinoff P (2000) Size-dependent disaggregation of stable protein aggregates by the DnaK chaperone machinery. J Biol Chem 275: 21107–21113 [DOI] [PubMed] [Google Scholar]
- Diemand AV, Lupas AN (2006) Modeling AAA+ ring complexes from monomeric structures. J Struct Biol 156: 230–243 [DOI] [PubMed] [Google Scholar]
- Dobson CM (2004) Principles of protein folding, misfolding and aggregation. Semin Cell Dev Biol 15: 3–16 [DOI] [PubMed] [Google Scholar]
- Doyle SM, Shorter J, Zolkiewski M, Hoskins JR, Lindquist S, Wickner S (2007a) Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat Struct Mol Biol 14: 114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SM, Hoskins JR, Wickner S (2007b) Collaboration between the ClpB AAA+ remodeling protein and the DnaK chaperone system. Proc Natl Acad Sci USA 104: 11138–11144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RJ (1997) Do molecular chaperones have to be proteins? Biochem Biophys Res Commun 238: 687–692 [DOI] [PubMed] [Google Scholar]
- Franzmann TM, Wühr M, Richter K, Walter S, Buchner J (2005) The activation mechanism of Hsp26 does not require dissociation of the oligomer. J Mol Biol 350: 1083–1093 [DOI] [PubMed] [Google Scholar]
- Gamer J, Bujard H, Bukau B (1992) Physical interaction between heat shock proteins DnaK, DnaJ, and GrpE and the bacterial heat shock transcription factor sigma 32. Cell 69: 833–842 [DOI] [PubMed] [Google Scholar]
- Germaniuk A, Liberek K, Marszalek J (2002) A bi-chaperone (Hsp70–Hsp78) system restores mitochondrial DNA synthesis following thermal inactivation of Mip1p polymerase. J Biol Chem 277: 27801–27808 [DOI] [PubMed] [Google Scholar]
- Giese KC, Vierling E (2002) Changes in oligomerization are essential for the chaperone activity of a small heat shock protein in vivo and in vitro. J Biol Chem 277: 46310–46318 [DOI] [PubMed] [Google Scholar]
- Glover JR, Lindquist S (1998) Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94: 73–82 [DOI] [PubMed] [Google Scholar]
- Goldfless SJ, Morag AS, Belisle KA, Sutera VA, Lovett ST (2006) DNA repeat rearrangements mediated by DnaK-dependent replication fork repair. Mol Cell 21: 595–604 [DOI] [PubMed] [Google Scholar]
- Goloubinoff P, Mogk A, Zvi AP, Tomayasu T, Bukau B (1999) Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc Natl Acad Sci USA 96: 13732–13737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimshaw J, Jelesarov I, Siegenthaler RH, Christen P (2003) Thermosensor action of GrpE. The DnaK chaperone system at heat shock temperatures. J Biol Chem 278: 19048–19053 [DOI] [PubMed] [Google Scholar]
- Groemping Y, Reinstein J (2001) Folding properties of the nucleotide exchange factor GrpE from Thermus thermophilus: GrpE is a thermosensor that mediates heat shock response. J Mol Biol 314: 167–178 [DOI] [PubMed] [Google Scholar]
- Haslbeck M, Walke S, Stromer T, Ehrnsperger M, White HE, Chen S, Saibil H, Buchner J (1999) Hsp26: a temperature-regulated chaperone. EMBO J 18: 6744–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M (2002) sHsps and their role in the chaperone network. Cell Mol Life Sci 59: 1649–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslbeck M, Franzmann T, Weinfurtner D, Buchner J (2005) Some like it hot: the structure and function of small heat shock proteins. Nat Struct Mol Biol 12: 842–846 [DOI] [PubMed] [Google Scholar]
- Haslberger T, Weibezahn H, Zahn R, Lee S, Tsai FTF, Bukau B, Mogk A (2007) M domains couple the ClpB threading motor with the DnaK chaperone activity. Mol Cell 25: 247–260 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295: 1852–1858 [DOI] [PubMed] [Google Scholar]
- Hinnerwisch J, Fenton WA, Furtak KA, Farr GW, Horwich AL (2005) Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell 121: 1029–1041 [DOI] [PubMed] [Google Scholar]
- Jahn TR, Radford SE (2008) Folding versus aggregation: Polypeptide conformations on competing pathways. Arch Biochem Biophys 469: 100–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob U, Gaestel M, Engel K, Buchner J (1993) Small heat shock proteins are molecular chaperones. J Biol Chem 268: 1517–1520 [PubMed] [Google Scholar]
- Kitagawa M, Wada C, Yoshioka S, Yura T (1991) Expression of ClpB, an analog of the ATP-dependent protease regulatory subunit in Escherichia coli, is controlled by a heat shock sigma factor (sigma 32). J Bacteriol 173: 4247–4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Matsumura Y, Tsuchido T (2000) Small heat shock proteins, IbpA and IbpB, are involved in resistances to heat and superoxide stresses in Escherichia coli. FEMS Microbiol Lett 184: 165–171 [DOI] [PubMed] [Google Scholar]
- Krzewska J, Langer T, Liberek K (2001) Mitochondrial Hsp78, a member of the Clp/Hsp100 family in Saccharomyces cerevisiae, cooperates with Hsp70 in protein refolding. FEBS Lett 489: 92–96 [DOI] [PubMed] [Google Scholar]
- Laskowska E, Kuczynska-Wiœnik D, Skorko-Glonek J, Taylor A (1996a) Degradation of proteases Lon, Clp and HtrA, of Escherichia coli proteins aggregated in vivo by heat shock; HtrA protease action in vivo and in vitro. Mol Microbiol 22: 555–571 [DOI] [PubMed] [Google Scholar]
- Laskowska E, Wawrzynów A, Taylor A (1996b) IbpA and IbpB, the new heat-shock proteins, bind to endogenous Escherichia coli proteins aggregated intracellularly by heat shock. Biochimie 78: 117–122 [DOI] [PubMed] [Google Scholar]
- Lee GJ, Vierling E (2000) A small heat shock protein cooperates with heat shock protein 70 systems to reactivate a heat-denatured protein. Plant Physiol 122: 189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Sowa ME, Watanabe YH, Sigler PB, Chiu W, Yoshida M, Tsai FTF (2003) The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell 115: 229–240 [DOI] [PubMed] [Google Scholar]
- Lee S, Choi JM, Tsai FT (2007) Visualizing the ATPase cycle in a protein disaggregating machine: structural basis for substrate binding by ClpB. Mol Cell 25: 261–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowska A, Gierszewska M, Marszałek J, Liberek K (2006) Hsp78 chaperone functions in restoration of mitochondrial network following heat stress. Biochim Biophys Acta 1763: 141–151 [DOI] [PubMed] [Google Scholar]
- Lewandowska A, Matuszewska M, Liberek K (2007) Conformational properties of aggregated polypeptides determine ClpB-dependence in the disaggregation process. J Mol Biol 371: 800–811 [DOI] [PubMed] [Google Scholar]
- Li GC, Werb Z (1982) Correlation between synthesis of heat shock proteins and development of thermotolerance in Chinese hamster fibroblasts. Proc Natl Acad Sci USA 79: 3218–3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberek K, Georgopoulos C, Zylicz M (1988) Role of the Escherichia coli DnaK and DnaJ heat shock proteins in the initiation of bacteriophage lambda DNA replication. Proc Natl Acad Sci USA 85: 6632–6636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M (1991) Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase of DnaK. Proc Natl Acad Sci USA 88: 2874–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberek K, Georgopoulos C (1993) Autoregulation of the Escherichia coli heat shock response by the DnaK and DnaJ heat shock proteins. Proc Natl Acad Sci USA 90: 11019–11023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum R, Tkach JM, Vierling E, Glover JM (2004) Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J Biol Chem 279: 29139–29146 [DOI] [PubMed] [Google Scholar]
- Matuszewska M, Kuczynska-Wisnik D, Laskowska E, Liberek K (2005) The small heat shock protein IbpA of Escherichia coli cooperates with IbpB in stabilization of thermally aggregated proteins in a disaggregation competent state. J Biol Chem 280: 12292–12298 [DOI] [PubMed] [Google Scholar]
- Mayer MP, Bukau B (2005) Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci 62: 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A, Tomoyasu T, Goloubinoff P, Rüdiger S, Röder D, Langen H, Bukau B (1999) Identification of thermolabile Escherichia coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J 18: 6934–6949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A, Deuerling E, Vorderwulbecke S, Vierling E, Bukau B (2003) Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol Microbiol 50: 585–595 [DOI] [PubMed] [Google Scholar]
- Motohashi K, Watanabe Y, Yohda M, Yoshida M (1999) Heat-inactivated proteins are rescued by the DnaK. J-GrpE set and ClpB chaperones. Proc Natl Acad Sci USA 96: 7184–7189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto H, Suzuki N, Roy SK (2000) Constitutive expression of a small heat-shock protein confers cellular thermotolerance and thermal protection to the photosynthetic apparatus in cyanobacteria. FEBS Lett 483: 169–174 [DOI] [PubMed] [Google Scholar]
- Park SH, Bolender N, Eisele F, Kostova Z, Takeuchi J, Coffino P, Wolf DH (2007) The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin–proteasome system. Mol Biol Cell 18: 153–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsell DA, Kowal AS, Singer MA, Lindquist S (1994) Protein disaggregation mediated by heat-shock protein Hsp104. Nature 373: 475–478 [DOI] [PubMed] [Google Scholar]
- Pelham HR (1986) Speculations on the functions of the major heat shock and glucose-regulated proteins. Cell 46: 959–961 [DOI] [PubMed] [Google Scholar]
- Plesofsky-Vig N, Brambl R (1995) Disruption of the gene for hsp30, an alpha-crystallin-related heat shock protein of Neurospora crassa, causes defects in thermotolerance. Proc Natl Acad Sci USA 92: 5032–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB (1998) Prions. Proc Natl Acad Sci USA 95: 13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Hong SW, Vierling E, Lindquist S (2000) Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12: 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger TR, Morimoto RI, Hatzimanikatis V (2006) Bistability explains threshold phenomena in protein aggregation both in vivo and in vitro. Biophys J 90: 886–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Lindquist S (1990) HSP104 required for induced thermotolerance. Science 248: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Schaupp A, Marcinowski M, Grimminger V, Bösl B, Walter S (2007) Processing of proteins by the molecular chaperone Hsp104. J Mol Biol 370: 674–686 [DOI] [PubMed] [Google Scholar]
- Schirmer EC, Glover JR, Singer MA, Lindquist S (1996) HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci 21: 289–296 [PubMed] [Google Scholar]
- Schmitt M, Neupert W, Langer T (1996) The molecular chaperone Hsp78 confers compartment-specific thermotolerance to mitochondria. J Cell Biol 134: 1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires CL, Pendersen S, Ross B, Squires C (1991) ClpB is the Escherichia coli heat shock protein F84.1. J Bacteriol 173: 4254–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlieker C, Weibezahn J, Patzelt H, Tessarz P, Strub C, Zeth K, Erbse A, Schneider-Mergener J, Chin JW, Schultz PG, Bukau B, Mogk A (2004) Substrate recognition by the AAA+ chaperone ClpB. Nat Struct Mol Biol 11: 607–615 [DOI] [PubMed] [Google Scholar]
- Skowyra D, Georgopoulos C, Zylicz M (1990) The E. coli dnaK gene product, the hsp70 homolog, can reactivate heat-inactivated RNA polymerase in an ATP hydrolysis-dependent manner. Cell 62: 939–944 [DOI] [PubMed] [Google Scholar]
- Stefani M (2004) Protein misfolding and aggregation: new examples in medicine and biology of the dark side of the protein world. Biochim Biophys Acta 1739: 5–25 [DOI] [PubMed] [Google Scholar]
- Studer S, Narberhaus F (2000) Chaperone activity and homo- and hetero-oligomer formation of bacterial small heat shock proteins. J Biol Chem 275: 37212–37218 [DOI] [PubMed] [Google Scholar]
- Tartaglia GG, Pechmann S, Dobson CM, Vendruscolo M (2007) Life on the edge: a link between gene expression levels and aggregation rates of human proteins. Trends Biochem Sci 32: 204–206 [DOI] [PubMed] [Google Scholar]
- Teter SA, Houry WA, Ang D, Tradler T, Rockabrand D, Fischer G, Blum P, Georgopoulos C, Hartl FU (1999) Polypeptide flux through bacterial Hsp70: DnaK cooperates with trigger factor in chaperoning nascent chains. Cell 97: 755–765 [DOI] [PubMed] [Google Scholar]
- Vale RD (2000) AAA proteins. Lords of the ring. J Cell Biol 150: F13–F19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel M, Bukau B, Mayer MP (2006) Allosteric regulation of Hsp70 chaperones by a proline switch. Mol Cell 21: 359–367 [DOI] [PubMed] [Google Scholar]
- Wawrzynow A, Wojtkowiak D, Marszalek J, Banecki B, Jonsen M, Graves B, Georgopoulos C, Zylicz M (1995) The ClpX heat-shock protein of Escherichia coli, the ATP-dependent substrate specificity component of the ClpP–ClpX protease, is a novel molecular chaperone. EMBO J 14: 1867–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, Zentgraf H, Weber-Ban EU, Dougan DA, Tsai FTF, Mogk A, Bukau B (2004) Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 119: 653–665 [DOI] [PubMed] [Google Scholar]
- Wojtkowiak D, Georgopoulos C, Zylicz M (1993) Isolation and characterization of ClpX, a new ATP-dependent specificity component of the Clp protease of Escherichia coli. J Biol Chem 268: 22609–22617 [PubMed] [Google Scholar]
- Wickner S, Gottesman S, Skowyra D, Hoskins J, McKenney K, Maurizi MR (1994) A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc Natl Acad Sci USA 91: 12218–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavodszky P, Kardos J, Svingor A, Petsko GA (1998) Adjustment of conformational flexibility is a key event in the thermal adaptation of proteins. Proc Natl Acad Sci USA 95: 7406–7411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemienowicz A, Skowyra D, Zeilstra-Ryalls J, Fayet O, Georgopoulos C, Zylicz M (1993) Both the Escherichia coli chaperone systems, GroEL/GroES and DnaK/DnaJ/GrpE, can reactivate heat-treated RNA polymerase. Different mechanisms for the same activity. J Biol Chem 268: 25425–25431 [PubMed] [Google Scholar]
- Ziętkiewicz S, Krzewska J, Liberek K (2004) Successive & synergistic action of the Hsp70 and Hsp100 chaperones in protein disaggregation. J Biol Chem 279: 44376–44383 [DOI] [PubMed] [Google Scholar]
- Ziętkiewicz S, Lewandowska A, Stocki P, Liberek K (2006) Hsp70 chaperone machine remodels protein aggregates at the initial step of Hsp70–Hsp100-dependent disaggregation. J Biol Chem 281: 7022–7029 [DOI] [PubMed] [Google Scholar]
- Zolkiewski M (2006) A camel passes through the eye of a needle: protein unfolding activity of Clp ATPases. Mol Microbiol 61: 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolkiewski M (1999) ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. A novel multi-chaperone system from Escherichia coli. J Biol Chem 274: 28083–28086 [DOI] [PubMed] [Google Scholar]
- Zzaman S, Reddy JM, Bastia D (2004) The DnaK–DnaJ–GrpE chaperone system activates inert wild type pi initiator protein of R6K into a form active in replication initiation. J Biol Chem 279: 50886–50894 [DOI] [PubMed] [Google Scholar]