Abstract
The vertebrate lens is a tissue composed of terminally differentiated fiber cells and anterior lens epithelial cells. The abundant, preferential expression of the soluble proteins called crystallins creates a transparent, refractive index gradient in the lens. Several transcription factors such as Pax6, Sox1, and L-Maf have been shown to regulate lens development. Here we show that mice lacking the transcription factor c-Maf are microphthalmic secondary to defective lens formation, specifically from the failure of posterior lens fiber elongation. The marked impairment of crystallin gene expression observed is likely explained by the ability of c-Maf to transactivate the crystallin gene promoter. Thus, c-Maf is required for the differentiation of the vertebrate lens.
The vertebrate lens presents a system in which tissue-specific transcription factors control a differentiative program. The growing list of transcription factors essential for eye morphogenesis demonstrates the exquisite complexity of this system, in which determination, embryonic induction, cellular differentiation, cross-regulation, and regeneration are all required (1). A family of proteins called the crystallins is responsible for the transparent and refractive properties of the lens.
Crystallins constitute 90% of the soluble proteins in the lens (2). There are three major crystallin classes in the lens, the α-, β-, and γ-crystallins, as well as several taxon-specific crystallins (3). In the mouse and rat, α-crystallins are the first to be expressed in the embryonic lens; they appear in both epithelial cells and fiber cells. The β- and γ-crystallins appear at a later stage (4); their expression is restricted to the fiber cells. Recent studies have shown that Pax6, Sox1, and L-Maf are important proteins in regulating lens development and crystallin gene expression in the lens (5–8).
The v-maf oncogene is the earliest described member of the maf family of genes, which encode transcription factors containing a basic region/leucine zipper domain (9). Large Maf subfamily members contain a putative activation domain at the N terminus, whereas small Maf subfamily group members lack a distinct activation domain (10). Maf family members share structural similarity both within and outside of the basic leucine zipper region (10), and they bind a common recognition element, 12-o-tetradecanoylphorbol 13-acetate (TPA) Maf response element (T-MARE) or cyclic AMP response element (CRE) Maf response element (C-MARE) (11). Expression pattern studies of the large Maf, c-Maf (12–14), have demonstrated that it is expressed in the eye lens of the embryonic rat (15, 16) in a highly regulated temperospatial fashion. By immunocytochemistry, c-Maf is detected in the posterior lens fiber cells but is completely absent in the anterior lens epithelial cells at embryonic day 13 (E13).
A putative role for c-Maf in lens development is suggested both by its expression in posterior lens fibers at E13 (15, 16) and by the presence of c-Maf MAREs in crystallin gene promoters (8, 17, 18). We deleted the c-maf gene by gene targeting. Mice homozygous for the mutation have small eyes or microphthalmia. In the mutant eyes, the elongation of the posterior lens fiber cells is defective and crystallin gene expression is severely impaired. We also show that the c-Maf protein can transactivate the γF-crystallin promoter, whose MARE has been shown to be critical for its activity (8). Thus, c-Maf is required for the differentiation of the vertebrate lens as a result of its action on crystallin gene expression.
MATERIALS AND METHODS
Targeting Vector and Gene Disruption.
c-maf genomic clones were isolated from a 129-genomic phage library and mapped with the use of several restriction enzymes. The targeting vector was linearized at the NotI site and electroporated into embryonic stem cells maintained in cell-conditioned medium in the presence of leukemia inhibitory factor. After selection in G418, correct replacement events were identified by Southern blot hybridization with the use of an external 5′ probe and an external 3′ probe. Homologous recombination events had occurred in 2 of 170 G418-resistant embryonic stem cell lines. Both of these lines were injected in C57BL/6 blastocyst embryos. Only one line resulted in germ-line transmission. Heterozygous offspring were crossed to BALB/c mice.
Nuclear Extract Preparation and Immunoblotting.
Freshly isolated kidney was homogenized manually with a Dounce homogenizer. Cells were washed once with cold PBS/0.1% BSA. Cells were resuspended in 0.9 ml of solution A (10 mM Tris, pH 7.5/10 mM NaCl/3 mM MgCl2) with protease inhibitors (Complete tablets, Boehringer Mannheim). Nonidet P-40 (5% at 100 μl) was added and mixed by inverting several times. After 5 min on ice, cells were spun at 1800 × g at 4°C. Supernatant was removed, and pellet was washed gently with 1 ml of solution A and repelleted. After complete removal of supernatant, the appropriate volume of solution C (20 mM Hepes/0.42 M NaCl/1.5 mM MgCl2/0.2 mM EDTA/25% glycerol/0.01% NaN3, with protease inhibitors) was added—twice the volume of the pellet. Pellet was resuspended by pipetting and left on ice for 30–40 min, mixing once or twice during incubation. The mixture was pelleted by spinning 10 min at top speed in a Microfuge at 4°C. Supernatant was removed and transferred to a fresh, prechilled tube. An equal volume of solution D (20 mM Hepes/50 mM KCl/0.2 mM EDTA/20% glycerol/0.01% NaN3, with protease inhibitors) was added and mixed well. Extracts were stored at −70°C.
Extracts were separated on SDS/9% polyacrylamide gels and transferred onto Optitran nitrocellulose membranes (Schleicher & Schuell). Immunoblots were incubated 1 h at room temperature in blocking solution (Tris-buffered saline with 5% milk and 0.05% Tween-20) followed by the primary antibody diluted (1:1000) in 1% blocking solution for 1–2 h; the primary antibody was a rabbit anti-mouse antiserum (prepared by J. Zhang, Medical University of South Carolina, Charleston). Primary incubation was followed by incubation with horseradish peroxidase-conjugated goat anti-rabbit-IgG antibody (Santa Cruz Biotechnology) for 1 h at room temperature and developed by enhanced chemiluminescence (Amersham).
Histological Analysis of Mutant Mice.
For light microscopy, mouse embryos at different stages of development were fixed in 1.25% glutaraldehyde/2% formaldehyde in phosphate-buffered saline for at least 24 h. Postnatal mouse eyes were enucleated and then fixed as above. Tissues were postfixed in 1% osmium tetroxide, dehydrated through a graded series of ethanol, and embedded in Epon. Transverse 1-μm sections were taken.
Reverse Transcription (RT)–PCR Amplification.
Eyes were isolated from adult mutant mice and wild-type littermates. For RT-PCR, RNA was extracted with Trizol (GIBCO/BRL) and reverse transcribed into cDNA with hexamer primers. Crystallin amplification, including primer sequence and conditions, was performed as described (7). In brief, we used cDNA generated from a 5-week-old mouse eye. Dilution was determined to obtain linear growth amplification. PCR was performed by use of Taq DNA polymerase (Boehringer Mannheim). To control for genomic contamination, all sets of primers for RT-PCR bracket an intron or introns. Primers for γ-crystallins are based on a previous study (4). Amplification products were separated on a 1.2% agarose gel.
Crystallin Gene Transactivation.
Genomic DNA was isolated from splenocytes from a DBA2 mouse by using DNAzol (GIBCO/BRL). From the published promoter sequence (26), primers were designed (5′ primer GCCCCACCTGCAACAAACAAC and 3′ primer GGCAGGTCAGATGGGATGGTG) to amplify a 1.4-kb region of the promoter. This promoter was subcloned into the pBS/luciferase plasmid (gift of Kenneth Murphy, Washington University, St. Louis) after initial ligation into Topo-TA (Invitrogen).
Transfections were performed as described (41). In brief, 106 cells were plated in a 25-cm2 flask and grown overnight at 37°C in a 5%CO2/95% air atmosphere. Plasmid DNA was mixed with 0.25 M CaCl2. BES-buffered saline (0.5 ml of 2× BBS was added to DNA and incubated 10–20 min at room temperature. Meanwhile, medium was removed from cells and 9 ml of fresh DMEM/10% fetal calf serum was added. Ca-phosphate-DNA solution was added dropwise to cells while swirling the flask, and cells were incubated 15–24 h at 37°C under 5% CO2. After that time, medium was removed, and cells were washed twice with medium, refed, and incubated for 24 h. At 24 h, luciferase was assayed with a Luciferase Assay kit (Promega).
RESULTS AND DISCUSSION
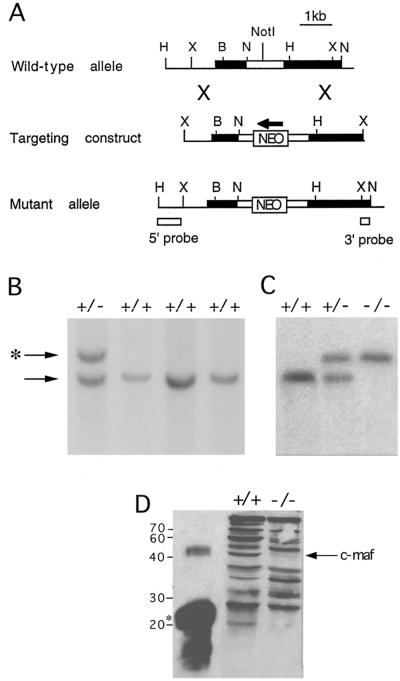
To establish definitively the function of c-Maf in vivo, a mutation of the c-maf gene was introduced into the germ line of mice by inserting a neomycin-resistance gene (neo) at the codon for amino acid 170 (Fig. 1A). One embryonic stem clone (Fig. 1B) transmitted the disrupted allele to offspring, and c-maf+/− mice were intercrossed to generate c-maf−/− mice (Fig. 1C). Western blot analysis of kidney extracts revealed an absence of immunoreactive c-Maf protein in c-maf−/− animals. (Fig. 1D).
Figure 1.
Targeted disruption of the murine c-maf gene. (A) Intron–exon structure of genomic DNA including the c-maf gene isolated from a 129/sv genomic library (Top). The thick black line represents the transcribed part of the c-maf locus, which is a single exon. The open box represents the ORF. Insertion of neo at the NotI site (amino acid codon 170) disrupts the gene 150 bp 3′ of the acidic transactivation of the c-Maf protein (Middle). The resultant mutant allele with the position of the probes used in the diagnostic Southern blot analysis is shown (Bottom). B, BamHI; H, HindIII; N, NcoI; and X, XhoI. (B) Southern blot analysis of embryonic stem cell DNA. As predicted from the restriction map of the wild-type locus, digestion of DNA with NcoI generates a 3-kb fragment, which is replaced by a 4-kb fragment when hybridized with the 3′ probe. (C) Southern blot analysis of tail DNA from mice resulting from the matings of c-maf+/− mice. (D) Western blot of kidney nuclear extracts from +/+ or −/− mice using a polyclonal anti-c-Maf antiserum. Control lane is a truncated recombinant c-Maf protein indicated by an asterisk.
Disruption of the c-maf gene affected both intrauterine and postnatal survival. Of 176 pups born, only 8 were homozygous null (−/−) as compared with 68 wild-type (+/+), 12% of the expected Mendelian ratio. Heterozygote (+/−) mice were born at the expected Mendelian frequency and appeared completely normal. Serial timed matings revealed that intrauterine death of c-maf−/− embryos was occurring at E17.5–E18.5 for reasons not yet understood. At birth, c-maf−/− mice showed no gross abnormalities, but from postnatal day 15 onward, mutant animals could easily be identified by their runted size and the presence of microphthalmia. Approximately one-third of affected animals did not survive past weaning.
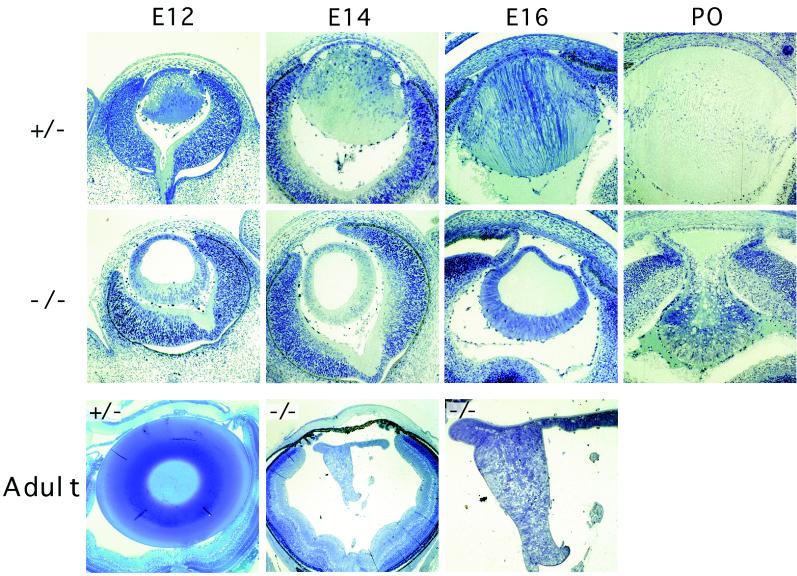
To determine the cause for the severe microphthalmia in the c-maf−/− mice, histological analysis of the eyes at different stages of development was performed. Examination of the eye revealed a selective defect in the formation of the lens in the mutant animals (Fig. 2). Normally after formation of the lens vesicle, cells of the posterior region cease dividing, elongate toward the anterior epithelial wall, and differentiate into lens fiber cells, thereby filling the cavity of the lens vesicle. In the c-maf mutant lens, this elongation fails to occur, resulting in a small and hollow lens cavity; this accounts for the presence of microphthalmia. The defect became apparent by E12, consistent with both the timing and location of c-Maf expression (15, 16, 19) and the onset of primary lens fiber differentiation during eye development (20). Human primary aphakia (absence of the lens with microphthalmia) is accompanied by major deformities of the anterior and posterior segments of the eye. The c-maf mutant mouse represents a model of secondary aphakia in which the defect is limited to the lens and the anterior and posterior segments of the eye are well formed. In the c-maf−/− eye, the cornea is well separated from the lens and the formation of the neural retina is grossly normal (Fig. 2).
Figure 2.
Histological analysis of heterozygote (+/−) and c-maf mutant (−/−) adult, neonatal, and embryonic lens. Richardson staining of c-maf−/− lens and heterozygote+/− lens at E12, E14, E16, postnatal day zero (P0), and 5-week-old adult. Loss of c-maf results in failure of posterior fiber elongation, apparent by E12, with subsequent absence of lens formation. Sections (1 μm) of Epon plastic embedded material were stained with Richardson stain.
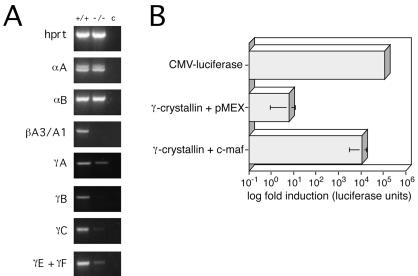
Crystallins are the most abundant soluble protein in lens fiber cells and are critical for proper development of the lens (4, 7, 21). For example, mutations in β- and γ-crystallin genes have been demonstrated to be the cause of lens fiber cell differentiation defects and cataracts in naturally occurring mutant mice (22–25). The stage at which the c-maf−/− lens defect becomes apparent (E12) correlates with the initiation of γ-crystallin gene expression (4). Although MAREs are present in the promoter regions of some crystallin genes (18, 26), a role for c-Maf in regulating these genes in the mammalian lens has not been demonstrated. We therefore examined the expression of multiple crystallin genes in the adult lens by semiquantitative RT-PCR (Fig. 3A). A dramatic reduction in the expression of members of the γ-crystallin gene cluster, which consists of the highly homologous genes γA, γB, γC, γD, γE, and γF, and the βA3/A1 gene was found. The expression of the α-class genes, αA and αB, was normal. These data fit well with the earlier activation of the α-crystallins during development, and the activation of the γ-crystallin gene cluster at E12, coincident with the expression of c-Maf. The normal expression of αA and αB demonstrates also that the down-regulation or absence of the γ- and β-crystallins is not simply the result of the smaller or degenerating adult c-maf−/− lens.
Figure 3.
c-Maf controls the expression of multiple crystallin genes. (A) Semiquantitative RT-PCR analysis of αA-, αB-, γA-, B-, C-, E/F-crystallins in the adult wild-type (+/+) and c-maf mutant (−/−) lens. For each gene, amplification of hypoxanthine phosphoribosyltransferase (hprt) cDNA was used as an internal control. Lane c indicates PCR performed in absence of reverse transcriptase. (B) c-Maf-dependent transactivation of the γF-crystallin promoter. The Psi2 fibroblast cell line was cotransfected with a c-Maf expression plasmid, pMex/maf, or control pMex vector alone, and a γF-crystallin promoter-luciferase reporter plasmid containing 1.4 kb of upstream sequence. One representative experiment of four is shown. The fold transactivation of the γF-crystallin promoter by c-Maf ranged from 100- to 1000-fold.
c-Maf has not previously been shown to control the transcription of γ-crystallin genes. We tested the ability of c-Maf to transactivate the γF-crystallin promoter by transient transfection assays in a fibroblast cell line. Ectopic expression of c-Maf increased reporter activity 100- to 1000-fold in four independent experiments when compared with the parental expression vector, pMEX (Fig. 3B); one representative experiment is shown. This is consistent with the recent demonstration that mutation of the MARE in the γF-crystallin promoter abolishes its function (8). We conclude that c-Maf is required for the induction of the γ-crystallins and that the failure of lens fiber differentiation in the absence of c-Maf is secondary to the absence of these critical lens-specific proteins.
A number of transcription factors have been shown to control eye development, in some instances through direct regulation of lens-specific genes. Mutations of the Pax6 transcription factor are responsible for the Small eye (complete absence of nose and eyes) phenotype in mice and aniridia phenotype in humans, whereas supernumerary eyes are induced by ectopic expression of eyeless, the Pax6 homologue, in Drosophila (27–31). Disruption of the Sox1 gene, shown to regulate the γ-crystallin gene cluster, results in a similar arrest of lens development (7) although the phenotype is not as severe as that observed in c-maf−/− mice. The difference in severity of the phenotype may be because of absence of β-crystallin as well as γ-crystallins in c-maf-deficient but not in Sox1-deficient mice. Other transcription factors such as Six3, Rx, Lhx2, and RAR/RXR (32–36) as well as secreted factors BMP7 and SPARC/osteonectin (37–39) are also involved in eye development and/or cataract formation but, unlike c-Maf, are expressed at sites in the eye in addition to the lens. The members of the large Maf subfamily, c-Maf, MafB, and NRL, are expressed in the eye of the mouse, rat, chicken, Xenopus, and zebrafish (8, 15, 16). Recently, L-Maf, a new member of the avian Maf family, was shown to be lens-specific; its ectopic expression converts chicken embryonic ectoderm into lens fibers, suggesting that L-Maf may participate in both lens induction and lens fiber differentiation (8). A mammalian orthologue of L-Maf has not been identified, although its sequence most closely resembles the mouse MafB gene, implicated in development of the erythroid lineage in the chicken (40). The expression patterns of c-Maf and MafB in the rat lens differ both spatially and temporally (15, 16). MafB expression is restricted to the epithelial cells of the lens, whereas c-Maf is distributed throughout lens fiber cells with highest expression in lens equator cells, a site where differentiation into lens fiber from lens epithelium is initiated (15, 16, 19). Taken together, these data suggest distinct but critical roles for both c-Maf and L-Maf in lens development and differentiation. c-Maf mutant mice provide an important tool for investigating the role of crystallins in lens development and serve as a model of secondary human aphakia. Systematic genetic analyses of patients with this rare disease may uncover mutations of c-Maf itself or of downstream targets of c-Maf such as the crystallins.
Acknowledgments
We thank Drs. Susanne J. Szabo, Rebecca Lieberson, Andrea Wurster, and Mohamed Oukka for a careful review of the manuscript. This work was supported by National Institutes of Health Grant AI/AG 37833 (L.H.G.), a gift from The G. Harold and Leila Y. Mathers Charitable Foundation (L.H.G.), a Leukemia Society of America Scholar Award (M.J.G.), an Arthritis Foundation Investigator Award (I.-C.H.), the Foundation Fighting Blindness (T.L.), and a career development award from Research to Prevent Blindness (T.L.).
ABBREVIATIONS
- En
embryonic day n
- MARE
Maf response element
- RT
reverse transcription
- TPA
12-o-tetradecanoylphorbol 13-acetate
- CRE
cyclic AMP response element
References
- 1.Cvekl A, Piatigorsky J. BioEssays. 1996;18:621–630. doi: 10.1002/bies.950180805. [DOI] [PubMed] [Google Scholar]
- 2.de Jong W W. In: Molecular and Cellular Biology of the Eye Lens. Blomendal H, editor. New York: Wiley; 1981. pp. 221–278. [Google Scholar]
- 3.Wistow G J, Piatigorsky J. Annu Rev Biochem. 1988;57:479–504. doi: 10.1146/annurev.bi.57.070188.002403. [DOI] [PubMed] [Google Scholar]
- 4.Goring D R, Breitman M L, Tsui L C. Exp Eye Res. 1992;54:785–795. doi: 10.1016/0014-4835(92)90034-p. [DOI] [PubMed] [Google Scholar]
- 5.Kamachi Y, Sockanathan S, Liu Q, Breitman M L, Lovell-Badge R, Kondoh H. EMBO J. 1995;14:3510–3519. doi: 10.1002/j.1460-2075.1995.tb07357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duncan M K, Haynes J I, Cvekl A, Piatigorsky J. Mol Cell Biol. 1998;18:5579–5586. doi: 10.1128/mcb.18.9.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishiguchi S, Wood H, Kondoh H, Lovell-Badge R, Episkopou V. Genes Dev. 1998;12:776–781. doi: 10.1101/gad.12.6.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogino H, Yasuda K. Science. 1998;280:115–118. doi: 10.1126/science.280.5360.115. [DOI] [PubMed] [Google Scholar]
- 9.Nishizawa M, Kataoka K, Goto N, Fujiwara K T, Kawai S. Proc Natl Acad Sci USA. 1989;86:7711–7715. doi: 10.1073/pnas.86.20.7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blank V, Andrews N C. Trends Biochem Sci. 1997;22:437–441. doi: 10.1016/s0968-0004(97)01105-5. [DOI] [PubMed] [Google Scholar]
- 11.Kerppola T K, Curran T. Oncogene. 1994;9:3149–3158. [PubMed] [Google Scholar]
- 12.Kataoka K, Nishizawa M, Kawai S. J Virol. 1993;67:2133–2141. doi: 10.1128/jvi.67.4.2133-2141.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurschner C, Morgan J I. Mol Cell Biol. 1995;15:246–254. doi: 10.1128/mcb.15.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho I C, Hodge M R, Rooney J W, Glimcher L H. Cell. 1996;85:973–983. doi: 10.1016/s0092-8674(00)81299-4. [DOI] [PubMed] [Google Scholar]
- 15.Sakai M, Imaki J, Yoshida K, Ogata A, Matsushima-Hibaya Y, Kuboki Y, Nishizawa M, Nishi S. Oncogene. 1997;14:745–750. doi: 10.1038/sj.onc.1200869. [DOI] [PubMed] [Google Scholar]
- 16.Yoshida K, Imaki J, Koyama Y, Harada T, Shinmei Y, Oishi C, Matsushima-Hibiya Y, Matsuda A, Nishi S, Matsuda H, Sakai M. Inv Ophthalmol Visual Sci. 1997;38:2679–2683. [PubMed] [Google Scholar]
- 17.Matsuo I, Yasuda K. Nucleic Acids Res. 1992;20:3701–3712. doi: 10.1093/nar/20.14.3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharon-Friling R, Richardson J, Sperbeck S, Lee D, Rauchman M, Maas R, Swaroop A, Wistow G. Mol Cell Biol. 1998;18:2067–2076. doi: 10.1128/mcb.18.4.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakayama H, Yamasaki H, Nishizawa M, Goto N. Int J Dev Biol. 1995;39:957–964. [PubMed] [Google Scholar]
- 20.Pei Y F, Rhodin J A G. Anat Rec. 1970;168:105–126. doi: 10.1002/ar.1091680109. [DOI] [PubMed] [Google Scholar]
- 21.Brady J P, Garland D, Duglas-Tabor Y, Robison W G, Jr, Groome A, Wawrousek E F. Proc Natl Acad Sci USA. 1997;94:884–889. doi: 10.1073/pnas.94.3.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cartier M, Breitman M L, Tsui L C. Nat Genet. 1992;2:42–45. doi: 10.1038/ng0992-42. [DOI] [PubMed] [Google Scholar]
- 23.Graw J. Biol Chem. 1997;378:1331–1348. [PubMed] [Google Scholar]
- 24.Kador P F, Fukui H N, Fukushi S, Jernigan H M, Jr, Kinoshita J H. Exp Eye Res. 1980;30:59–68. doi: 10.1016/0014-4835(80)90124-4. [DOI] [PubMed] [Google Scholar]
- 25.Chambers C, Russell P. J Biol Chem. 1991;266:6742–6746. [PubMed] [Google Scholar]
- 26.Lok S, Breitman M L, Chepelinsky A B, Piatigorsky J, Gold R J, Tsui L C. Mol Cell Biol. 1985;5:2221–2230. doi: 10.1128/mcb.5.9.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hill R E, Favor J, Hogan B L, Ton C C, Saunders G F, Hanson I M, Prosser J, Jordan T, Hastie N D, van Heyningen V. Nature (London) 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- 28.Ton C C, Hirvonen H, Miwa H, Weil M M, Monaghan P, Jordan T, van Heyningen V, Hastie N D, Meijers-Heijboer H, Drechsler M, et al. Cell. 1991;67:1059–1074. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- 29.Walther C, Gruss P. Development (Cambridge, UK) 1991;113:1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- 30.Quiring R, Walldorf U, Kloter U, Gehring W J. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- 31.Halder G, Callaerts P, Gehring W J. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- 32.Tini M, Otulakowski G, Breitman M L, Tsui L C, Giguere V. Genes Dev. 1993;7:295–307. doi: 10.1101/gad.7.2.295. [DOI] [PubMed] [Google Scholar]
- 33.Oliver G, Mailhos A, Wehr R, Copeland N G, Jenkins N A, Gruss P. Development (Cambridge, UK) 1995;121:4045–4055. doi: 10.1242/dev.121.12.4045. [DOI] [PubMed] [Google Scholar]
- 34.Oliver G, Loosli F, Koster R, Wittbrodt J, Gruss P. Mech Dev. 1996;60:233–239. doi: 10.1016/s0925-4773(96)00632-6. [DOI] [PubMed] [Google Scholar]
- 35.Porter F D, Drago J, Xu Y, Cheema S S, Wassif C, Huang S P, Lee E, Grinberg A, Massalas J S, Bodine D, Alt F, Westphal H. Development (Cambridge, UK) 1997;124:2935–2944. doi: 10.1242/dev.124.15.2935. [DOI] [PubMed] [Google Scholar]
- 36.Mathers P H, Grinberg A, Mahon K A, Jamrich M. Nature (London) 1997;387:603–607. doi: 10.1038/42475. [DOI] [PubMed] [Google Scholar]
- 37.Dudley A T, Lyons K M, Robertson E J. Genes Dev. 1995;9:2795–2807. doi: 10.1101/gad.9.22.2795. [DOI] [PubMed] [Google Scholar]
- 38.Luo G, Hofmann C, Bronckers A L, Sohocki M, Bradley A, Karsenty G. Genes Dev. 1995;9:2808–2820. doi: 10.1101/gad.9.22.2808. [DOI] [PubMed] [Google Scholar]
- 39.Gilmour D T, Lyon G J, Carlton M B, Sanes J R, Cunningham J M, Anderson J R, Hogan B L, Evans M J, Colledge W H. EMBO J. 1998;17:1860–1870. doi: 10.1093/emboj/17.7.1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sieweke M H, Tekotte H, Frampton J, Graf T. Cell. 1996;85:49–60. doi: 10.1016/s0092-8674(00)81081-8. [DOI] [PubMed] [Google Scholar]
- 41.Chen C, Okayama H. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]