Abstract
Human rhinoviruses (HRVs) are important contributors to respiratory disease, but their healthcare burden remains unclear, primarily because of the lack of sensitive, accurate, and convenient means of determining their causal role. To address this, we developed and clinically validated the sensitivity and specificity of a real-time reverse transcription-PCR (RT-PCR) assay targeting the viral 5′ noncoding region defined by sequences obtained from all 100 currently recognized HRV prototype strains and 85 recently circulating field isolates. The assay successfully amplified all HRVs tested and could reproducibly detect 50 HRV RNA transcript copies, with a dynamic range of over 7 logs. In contrast, a quantified RNA transcript of human enterovirus 68 (HEV68) that showed the greatest sequence homology to the HRV primers and probe set was not detected below a concentration of 5 × 105 copies per reaction. Nucleic acid extracts of 111 coded respiratory specimens that were culture positive for HRV or HEV were tested with the HRV real-time RT-PCR assay and by two independent laboratories that used different in-house HRV/HEV RT-PCR assays. Eighty-seven HRV-culture-positive specimens were correctly identified by the real-time RT-PCR assay, and 4 of the 24 HEV-positive samples were positive for HRV. HRV-specific sequences subsequently were identified in these four specimens, suggesting HRV/HEV coinfection in these patients. The assay was successfully applied in an investigation of a coincidental outbreak of HRV respiratory illness among laboratory staff.
Human rhinovirus (HRV) infections are among the most frequent causes of the common cold (42), and more recent studies have linked HRVs to more severe lower respiratory illnesses in otherwise healthy young children (34, 35), the elderly (16, 36, 48), and the immunocompromised (13, 19). Persons with underlying respiratory diseases, like asthma, chronic bronchitis, and cystic fibrosis (12, 26, 46), also may have an increased risk of severe HRV-associated complications.
Together with the human enteroviruses (HEVs), HRVs are classified within the family Picornaviridae (27). There are currently 100 distinct HRV serotypes assigned to two species, A and B (2), and new genetic variants of HRV have been reported recently (29, 33); one former HRV serotype, HRV87, recently was shown by sequence analysis to be a strain of HEV68 (6, 43). The clinical presentation of HRV infection is of little diagnostic value, and laboratory diagnosis has been complicated by the failure of some strains to grow in cell culture and by their extreme antigenic variability, precluding the routine use of antigen detection methods or serology. Moreover, distinguishing HRVs from HEVs, which also may be present in respiratory specimens, by acid liability testing has been shown to be ineffective for many strains (5, 17, 18). Reverse transcription-PCR (RT-PCR) assays therefore have become the method of choice for the sensitive detection and differentiation of HRVs and have greatly enhanced our appreciation of the role of these viruses in human disease.
Numerous molecular assays have been described for the HRVs/HEVs. These assays typically target the 5′ noncoding region (5′NCR) of the viral genome that contains highly conserved sequences suitable for molecular assay development. However, most of these assays require postamplification processing of the amplicon by gel electrophoresis, probe hybridization, sequencing, or restriction analysis to confirm and differentiate HRVs from HEVs (1, 3, 4, 5, 14, 24, 31, 34, 41). More recently, real-time RT-PCR assays have been described for HRVs/HEVs (8, 9, 25, 38, 44) that offer potentially rapid, sensitive, and quantitative results and that are less prone to amplicon contamination. However, because of the more extensive genetic variability of the HRVs and the lack of available sequence data in the public domain, few real-time RT-PCR assays have been described specifically for the HRVs (8, 9, 44, 51), and none to our knowledge has been shown to successfully detect all recognized HRV prototype strains. In this study, we sequenced a portion of the 5′NCR of all HRV prototype strains as well as multiple recently circulating field isolates and used these data to develop and evaluate an HRV real-time RT-PCR assay.
(Data from this report were presented in part at the 23rd Clinical Virology Symposium, Clearwater, FL, 29 April to 2 May 2007.)
MATERIALS AND METHODS
Virus strains and clinical specimens.
One hundred HRV prototype strains (HRV1A, HRV1B, HRV2 to HRV86, and HRV88 to HRV100) kindly provided by ViroPharma Inc. (30) and 85 HRV field isolates obtained from several sources between 1999 and 2007 were available for study. HRV isolates were sequenced directly or after one additional passage in HeLa Ohio cells. Infected cells were incubated at 35°C in 5% CO2 with gentle rocking until reaching full cytopathic effect. Isolates then were freeze-thawed twice and clarified by low-speed centrifugation, and supernatants were collected and stored at −70°C. Forty-eight HEV laboratory strains grown in primary monkey kidney or human RD cells and prepared as described above included echoviruses 1 to 6, 8, 9, 11 to 25, and 29 to 31; coxsackievirus types A2, A4 to A6, A8 to A10, A16, A21, A24, and B1 to B6; enterovirus types 68, 70, and 71; and poliovirus types 1, 2, and 3. Other respiratory viruses available for specificity testing included respiratory syncytial virus, human metapneumovirus, human parainfluenza viruses 1 to 4, adenovirus, coronaviruses 229E and OC43, influenza viruses A and B, and human bocavirus (32). Coded respiratory specimens culture positive for HRV or HEV were provided by the California Department of Health Services, the University of Washington, the Vanderbilt Medical Center, and the University of Rochester Medical Center for clinical validation studies. Nasal and throat swab specimens were self obtained by symptomatic laboratory staff and were expressed in 2 ml of chilled viral transport medium (Hank's buffered salt solution with 0.5% gelatin) and frozen at −70°C prior to testing.
Sample extraction.
Total nucleic acid extracts were prepared from 100 μl of infected cell culture lysate or 200 μl of clinical specimen using the NucliSens easyMAG extraction system by following the manufacturer's instructions (bioMérieux, Durham, NC).
Partial 5′NCR sequencing.
Extracted viral RNA was reverse transcribed using random hexamer primers (Promega, Madison, WI) at 52°C for 60 min with Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA) by following the manufacturer's instructions. Five microliters of the cDNA then was amplified in two separate PCRs using HRV species A- and B-specific primer sets (Table 1) with the HotStarTaq master mix kit (Qiagen, Chatsworth, CA). PCR cycling conditions were the following: an initial activation step at 95°C for 15 min, followed by 35 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min, with a final extension of 72°C for 5 min on a GeneAmp PCR system 9700 (Applied Biosystems). Amplified products were purified with the QIAquick PCR purification kit (Qiagen), and sequencing was performed in both directions using the amplification primers and the ABI Prism BigDye Terminator cycle sequencing ready reaction kit, version 3.1, on an ABI 3100 DNA sequencer (Applied Biosystems). Sequencher software (version 3.1.1; Gene Codes, Ann Arbor, MI) was used for sequence assembly and editing.
TABLE 1.
HRV/HEV primers and probes
| Primer and/or probea | Sequenceb (5′-3′) | Position |
|---|---|---|
| Real-time RT-PCR | ||
| Primer, forward | CPXGCCZGCGTGGC | 356-369c |
| Primer, reverse | GAAACACGGACACCCAAAGTA | 563-543c |
| Probe | TCCTCCGGCCCCTGAATGYGGC | 444-465c |
| HRVA 5′NCR sequencing | ||
| Primer, forward | GTACTCTGTTATTCCGGTAACTTTGYAYGCCA | 49-80c |
| Primer, reverse | CCAACATTCTGTCTAGATACYTGDGCVCCCAT | 655-623c |
| HRVB 5′NCR sequencing | ||
| Primer, forward | ACTCTGGTACTATGTACCTTTGTACGCCTGTT | 48-80d |
| Primer, reverse | CCACTCTTCTGTGTAGACACYTGDGCDCCCAT | 661-629d |
| HRV14 RNA transcript | ||
| Primer, forward, T7 | TAATACGACTCACTATAGGGCAAGCACTTCTGTTT | 179-193d |
| Primer, reverse, SP6 | ATTTAGGTGACACTATAGAAGCATCTGGTAATTTCC | 1089-1074d |
| HEV68 RNA transcript | ||
| Primer, forward, T7 | TAATACGACTCACTATAGGGTCTTATGAGCAAGCACT | 52-68e |
| Primer, reverse, SP6 | ATTTAGGTGACACTATAGAAATTACTTCAAAATAACTCAG | 573-554e |
Probes were 5′-end labeled with 6-carboxyfluorescein and 3′-end labeled with Black Hole Quencher 1. HRVA, HRV species A; HRVB, HRV species B.
Y = dC or dT; D = dA, dT, or dG; V = dA, dC, or dG; X = LNA-dA; Z = LNA-dT (Glen Research Corporation, Sterling, VA). P is a pyrimidine derivative, a degenerate base mimicking a C/T mix (Glen Research Corporation). Underlined sequences are T7 and SP6 promoter sites.
The nucleotide numbering is based on that of the HRV1B sequence (accession no. D00239).
The nucleotide numbering is based on that of the HRV14 sequence (accession no. K02121).
The nucleotide numbering is based on that of the HEV68 sequence (formerly HRV87) (accession no. AY062273).
Primers and probes.
Conserved regions of the 5′NCR were identified from alignments of nucleotide sequences available in the GenBank database (HEV sequence accession numbers: AF108187, AF169670, AF303040, AF405316, AF405321, AF412341 to AF412376, AF412383, AJ007342, AY028214 to AY028218, AY062275, L76395, L76409, L76411, L76413, L76411, L76412, X89534, X89535, X89539, and z78133; HRV sequence accession numbers: AF108149 to AF108186, AF542419 to AF542421, AF542424 to AF542448, D00239, DQ473485 to DQ473512, DQ473509, E01069, EF186077, L24917, M16248, NC_001490, and X02316) or were obtained during this study (HRV sequence accession numbers EU095987 to EU096086). Primers and probes were selected with the aid of Primer Express software, version 2.0.0 (Applied Biosystems, Foster City, CA), and Netprimer (Premier Biosoft International). Primers and probes that showed no major nonspecific homologies on BLAST analysis were synthesized by the Centers for Disease Control and Prevention (CDC) Biotechnology Core Facility. TaqMan probes were labeled at the 5′ end with the reporter molecule 6-carboxyfluorescein and at the 3′ end with the quencher Black Hole Quencher 1 (Biosearch Technologies, Inc., Novato, CA). Locked nucleic acid (Exiqon A/S, Vedbaek, Denmark) analogues (LNA) that increase thermodynamic stability (37) were introduced during oligonucleotide synthesis to achieve a balanced melting temperature between the forward and reverse primers. Optimal primer and probe concentrations were determined by checkerboard titrations against synthetic RNA transcripts (see below). Primer and probe sets that gave the highest amplification efficiencies at optimized conditions and with no identifiable cross-reactions were chosen for further study (Table 1).
Real-time RT-PCR assay.
The real-time RT-PCR assay was performed using the iScript one-step RT-PCR kit for probes (Bio-Rad, Hercules, CA). Each 25-μl reaction mixture contained 1 μM forward and reverse primers, 0.1 μM probe, and 5 μl of nucleic acid extract. The amplification was performed on an iCycler iQ real-time detection system (Bio-Rad) using the following thermocycling conditions: 10 min at 48°C for RT, 3 min at 95°C for polymerase activation, and then 45 cycles of 15 s at 95°C and 1 min at 60°C. Each run included template and nontemplate controls. Specimen extracts also were tested for the human RNase P gene to monitor specimen quality as previously described (10).
RNA transcript synthesis.
RNA transcripts were prepared from the 5′NCR of representative HRV and HEV strains for analytical sensitivity and specificity studies. PCR amplicons were prepared from HRV14 and HEV68 using primers that bracketed the real-time RT-PCR signature and that contained 5′-end T7 and Sp6 promoter sequences to serve as templates for RNA polymerase (Table 1). The expected sequences were confirmed by amplicon sequencing. The transcripts were synthesized and purified using MegaScript and MegaClear kits (Ambion, Inc., Austin, TX), respectively, and were quantified using the NanoDrop 1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). Positive-sense transcripts for HRV14 and HEV68 were 951 and 545 nucleotides (nt) in length, respectively, with RNA yields of 2.03 × 1011 copies/μl for HRV14 and 3.10 × 1012 copies/μl for HEV68. To generate standard curves for quantitative determinations, replicate serial 10-fold dilutions of the transcripts were prepared in 10 mM Tris-EDTA buffer containing yeast tRNA (50 ng/ml) (Ambion) and were stored at −70°C until use.
Nucleotide sequence accession numbers.
The HRV sequences determined in the course of this work were deposited in GenBank under accession numbers EU095987 to EU096086.
RESULTS
Assay development and optimization.
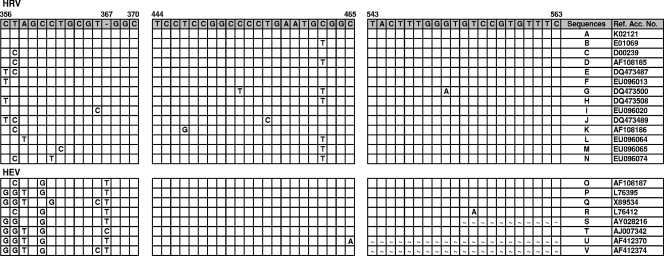
The 5′NCR of all 100 HRV prototype strains and 85 recently circulating field isolates were sequenced and aligned with those of representative HRV/HEV strains available from GenBank (NIH). A conserved region identified between nt 356 and 563 (HRV1B accession no. D00239) (Fig. 1) was selected to develop primer and probe sets for evaluation. The sets that gave the best initial performance with several representative HRV strains were chosen for subsequent studies. The reverse primer and probe were highly conserved among all available HRV/HEV sequences. In contrast, the forward primer was located in a variable region that contained a signature T indel at nt 367 that distinguished all HRVs from HEVs and that could be exploited for differential amplification. To compensate for the necessarily shorter length of the forward primer, LNA were introduced to achieve a balanced midpoint temperature with the reverse primer. The performance of the assay was optimized using the iScript one-step RT-PCR kit for probes (Bio-Rad). Limited examinations of other commercial real-time RT-PCR reagent kits found that the QuantiTect probe PCR kit (Qiagen) and Ag-Path-ID one-step RT-PCR kit (Applied Biosystems) performed comparably to the iScript kit, whereas amplification was less efficient with the TaqMan one-step RT-PCR master mix (Applied Biosystems) or failed entirely with the SuperScript III platinum one-step quantitative RT-PCR kit (Invitrogen) on repeat evaluations (data not shown).
FIG. 1.
Alignment of partial 5′NCR sequences of 100 HRV and 52 HEV serotypes in regions corresponding to primers and probes used for the HRV real-time RT-PCR assay. Consensus sequences and nucleotide variations from the consensus are shown for each alignment. A blank square indicates that that base is identical to the consensus sequence base. The dash indicates the presence of an indel at position 367. ∼, Not determined. The nucleotide position numbering is based on the sequence of HRV1B (accession no. D00239). Letters in the Sequences column indicate the following HRV strains: A, 3, 4, 6, 13, 14, 17 to 19, 27, 37, 41, 48, 49, 53, 61, 73, 79, 82 to 84, 90, 92, 93, 96, and 97; B, 2, 8 to 11, 15, 16, 20, 21, 23 to 25, 29, 30, 32, 34, 38, 40, 44, 46, 50, 54 to 57, 60, 62, 66 to 68, 74, 76, 80, 81, 85, 95, 98, and 100; C, 1A, 1B, 22, 43, 51, 64, 71, 75, 86, and 94; D, 7, 12, 31, 36, 39, 45, 47, 58, and 89; E, 5, 35, 42, 52, 65, 69, and 91; F, 26 and 99; G, 59 and 63; H, 28; I, 33; J, 70; K, 72; L, 77; M, 78; and N, 88. Letters in the Sequences column indicate the following HEV types: O, coxsackievirus types A1, A2, A11, A13, A15, A17 to A22, and A24, HEV68 and HEV70, and poliovirus types 1 and 3; P, coxsackievirus types A3 to A7, A10, A12, and A14, coxsackievirus types B1, B3, and B5, echoviruses 1, 6 to 8, 13, 16 to 21, 27, 29, 31, and 32, and enterovirus types 69 and 71; Q, echovirus types 4, 14, and 15; R, poliovirus type 2; S, coxsackievirus type A8; T, echovirus type 11; U, echovirus type 26; and V, echovirus type 33. Ref. Acc. No., reference accession number.
Assay evaluation with HRV/HEV isolates.
Viral culture lysates of all 100 HRV prototype strains, 85 HRV field isolates, and 48 HEV laboratory strains were tested by the real-time RT-PCR assay. Undiluted RNA extracts of all HRV prototype strains and field isolates gave strongly positive reactions (median cycle threshold [CT] value, 13.7; range, 9.3 to 25.3). In contrast, 34 HEVs were nonreactive, and 14 (echoviruses 1, 3, 5, 6, 13, and 21; poliovirus types 1 and 2; enterovirus types 68 and 71; coxsackievirus types A4, A6, and A24; and coxsackievirus type B1) gave weakly positive reactions (median CT value, 34; range, 33 to 34.8); positive reactions with HEVs appeared to be related to the virus titer and not to any particular virus type. Tenfold serial dilutions of total nucleic acid extracts of HRV2, HRV5, HRV33, HRV88, echovirus 1 and coxsackievirus type B1 were tested in parallel by the real-time RT-PCR assay and a conventional RT-PCR assay that amplifies an ∼115-bp region of the 5′NCR of both HRVs and HEVs (45). Whereas the relative sensitivities of the two assays for the HRVs were equivalent, the real-time RT-PCR assay was ≥105-fold less sensitive than the conventional assay with HEVs echovirus 1 and coxsackievirus type B1.
Assay analytical sensitivity and specificity.
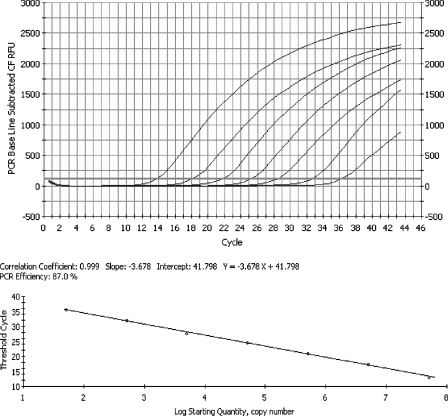
Serial 10-fold dilutions of HRV14 RNA transcripts that showed 100% sequence identity with the real-time RT-PCR primers and probe set were tested to assess the amplification efficiency. A linear amplification was achieved over a 7-log dynamic range from 5 × 101 to 5 × 107 copies per reaction (Fig. 2). The assay's detection limit was determined with 24 replicates of 50, 5, and 1 transcript copy per reaction. At 50 copies, 100% of the replicates were positive; at 5 copies, 9 (37.5%) of the replicates were positive; and at 1 copy, 2 (8.3%) of the replicates were positive. In contrast, the HEV68 transcript was not detected below a concentration of approximately 5 × 105 copies per reaction. Nucleic acid extracts of other respiratory viruses, including human respiratory syncytial virus, human metapneumovirus, parainfluenza viruses 1 to 4, adenovirus, coronaviruses 229E and OC43, influenza viruses A and B, and human bocavirus were negative by the real-time RT-PCR assay.
FIG. 2.
Representative real-time RT-PCR amplification plot obtained with serial 10-fold dilutions (5 × 101 to 5 × 107 copies per reaction) of HRV14 RNA transcript. The top panel shows a baseline subtractive curve fit (CF) view of the data, with relative fluorescence units (RFU) plotted against cycle numbers. The default setting of 10 times the standard deviation of the RFU measured in all wells over the baseline cycles was used to calculate the CT for a positive reaction (horizontal line). The bottom panel shows a standard curve analysis of the DNA amplification plots with CT values plotted proportionately against the logarithm of the input copy number. The dynamic range of assay spans 7 logs, with an R2 value of 0.999.
Assay reproducibility.
To assess intra- and interassay reproducibility, 10-fold serial dilutions of the HRV14 RNA transcripts, from 5 × 101 to 5 × 107 copies per reaction, were tested in triplicate on three succeeding days. Over the linear range of the assay, the coefficient of variation of the mean CT values ranged from 0.24 to 0.94% within runs and from 0.91 to 2.68% between runs.
Assay evaluation with clinical specimens.
To assess the performance of the real-time RT-PCR assay with clinical samples, extracts of 111 coded respiratory specimens previously determined to be culture positive for HRV or HEV were prepared and tested simultaneously with the HRV real-time RT-PCR assay and by two independent laboratories (laboratories A and B) that used different in-house HRV/HEV RT-PCR assays. Of 87 HRV culture-positive specimens tested, all were identified as HRV by the real-time RT-PCR assay (median CT value, 26.3; range, 14.9 to 38.5) (Table 2); HRV also was identified in all 87 specimens by one or both of the reference in-house RT-PCR assays. Of 24 HEV culture-positive specimens, 4 were positive for HRV by the real-time RT-PCR assay (median CT value, 28.8; range, 26.2 to 32.1); one of these four also was identified as HRV by laboratory B. HEV isolates available from three of the four specimens were not amplified by the real-time RT-PCR, whereas amplicon sequences obtained from all four clinical specimens were found to be HRV, suggesting that these specimens contained both HRV and HEV.
TABLE 2.
Comparison of HRV real-time RT-PCR assay to reference assays for detection of HRV and HEV in clinical specimensc
| Virus detected | No. of samples | Results by the CDC HRV real-time RT-PCR
|
Results from laboratory:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Aa
|
Bb
|
||||||||||
| HRV real-time RT-PCR
|
HEV real-time RT-PCR
|
Gel (HRV)
|
HEV real-time RT-PCR
|
||||||||
| Pos | Neg | Pos | Neg | Pos | Neg | Pos | Neg | Pos | Neg | ||
| HRV | 87 | 87 | 0 | 54 | 33 | 1 | 86 | 87 | 0 | 0 | 87 |
| HEV | 24 | 4 | 20 | 0 | 24 | 22 | 2 | 2 | 1 | 21 | 3 |
The in-house assays used by laboratory A were hydrolysis probe real-time RT-PCR assays for HRV and HEV.
The in-house assay used by laboratory B for HEV was a hydrolysis probe real-time RT-PCR assay. HEV-negative samples that subsequently yielded an appropriately sized band following gel electrophoresis were considered positive for HRV.
Pos, positive; neg, negative.
Outbreak investigation.
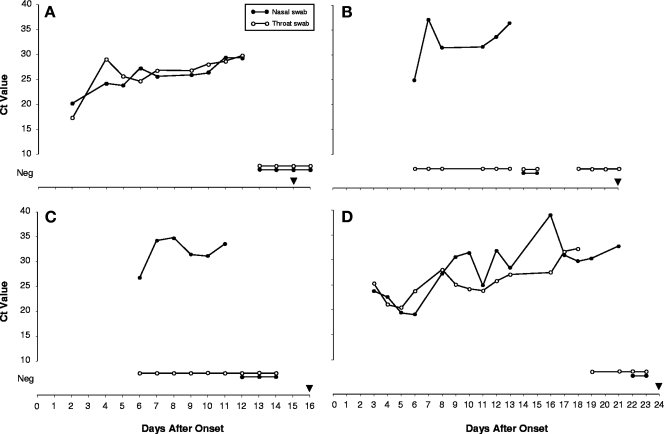
Between 10 and 22 September 2006, six CDC laboratory staff members developed respiratory illnesses characterized by one or more of the following symptoms: cough, congestion, myalgia, chills, or fever, and five were absent from work for 1 or more days. Testing samples from these individuals with a comprehensive panel of respiratory virus PCR assays identified five cases of HRV infection. Serial throat and nasal swab specimens were self collected from four of the HRV-positive individuals beginning 2 and 6 days after the onset of symptoms and continued until at least two consecutive specimens tested negative (Fig. 3). The duration of detectable HRV ranged from 11 to 21 days (median, 12.5 days). With the exception of case A, in which HRV was detected at comparable levels from both throat and nasal swabs, throat swabs either were consistently negative for HRV (cases B and C) or became negative earlier than did nasal swabs (case D). The duration of symptoms for the five HRV-positive cases ranged from 12 to 24 days (median, 16 days); one individual (case D) had a prolonged paroxysmal cough that persisted for 24 days. The duration of reported symptoms exceeded the duration of detectable HRV by real-time RT-PCR for all cases. The sequencing of a partial region of the HRV VP1 gene from the specimens obtained from the five cases identified two genetically distinct HRV strains that showed the closest sequence identities to HRV86 (amino acid identity score, 83.5%) and HRV69 (amino acid identity score, 84.6%), respectively.
FIG. 3.
HRV detected by real-time RT-PCR in serial nasal and throat swab specimens from four laboratory staff members (A, B, C, and D) with acute respiratory illness. CT values are plotted against the number of days after the onset of illness. Neg, HRV not detected; black inverted triangle, the end of reported respiratory symptoms.
DISCUSSION
The 5′NCR has long been the preferred site for designing molecular diagnostic assays for HRVs/HEVs due to the availability of highly conserved sequences that support the complex secondary structures of the HRV/HEV internal ribosome entry site (50). Whereas the locations of these conserved sequences offer considerable flexibility for designing targeted primers/probes for HEV real-time RT-PCR assays (25, 38, 47), the development of comparable assays for HRVs has been hampered by their greater genetic variability and the paucity of published HRV sequence data from the 5′NCR. The few real-time assays that have been described for HRVs were not evaluated against or failed to detect all known HRV serotypes (8, 9, 44, 51), or they used Sybr green instead of probe-based fluorescence detection (8); the results of Sybr green testing can be difficult to interpret (49). A comprehensive analysis of HRV 5′NCR sequences obtained in this study identified a region suitable for the development of an HRV-specific probe-based real-time RT-PCR assay that was demonstrated to detect all HRV prototype stains, multiple recently circulating field isolates, including newly identified genetic variants (33), and HRV culture-positive clinical specimens. The assay was further shown to discriminate between HRVs and HEVs in clinical specimens and successfully identified HRV coinfections in specimens that were culture positive only for HEV.
During the course of this study, an outbreak of HRV respiratory illness occurred among laboratory workers that was investigated to assess the viral shedding patterns in four of those affected. HRV was detected by real-time RT-PCR for up to 3 weeks after the onset of symptoms in one case; however, most cases showed progressively decreasing virus loads, becoming RT-PCR negative within 2 weeks of the onset of symptoms, and all cases continued to be symptomatic after the cessation of HRV shedding. Studies of children have found HRV for up to 5 to 6 weeks by RT-PCR (20); however, results of epidemiological and human volunteer studies of shedding patterns in healthy adults are more consistent with our findings (15). In most cases, nasal swabs were better than throat swabs for virus recovery, reflecting the preferred site of HRV replication in the anterior nasal mucosa (21). The ability of the real-time RT-PCR assay to detect two genetically novel HRV strains responsible for this outbreak and its primer/probe sequence compatibility with the newly recognized group of HRVs represented by strain HRV-QPM (33) confirm its robust diagnostic capabilities.
Despite these advantages, our real-time RT-PCR assay has several potential limitations. Although all HRV strains evaluated were successfully detected, the full genetic diversity of HRVs may not be fully represented in our study. Unrecognized genetic heterogeneity in the primer/probe region could compromise assay performance. Moreover, despite our efforts to design an HRV-specific assay, some HEVs present at high titers in the respiratory specimen could be misidentified as HRV. In addition to these technical issues, the potential for long-term shedding of HRVs in some populations (20, 23, 28, 51) and the high rates of asymptomatic HRV infection found in some studies (11, 39, 51) make it more difficult to establish an etiologic link to disease. By restricting the testing to HRVs alone, other members of the Picornaviridae that have been implicated in acute respiratory disease would not be identified. Respiratory illnesses indistinguishable from those caused by HRVs occur in up to 21% of non-poliovirus HEV infections (40), and picornaviruses of the more recently identified Parechovirus genus also have been linked to respiratory disease (22). Therefore, a more comprehensive diagnostic strategy would combine the HRV real-time RT-PCR assay with real-time assays for the other respiratory picornavirus pathogens (7).
Our real-time RT-PCR assay permits rapid, sensitive, and specific detection of HRVs in a format more convenient for diagnostic laboratories. Expanding routine testing for these viruses will help better define the epidemiology of HRV infection and the spectrum and burden of HRV disease.
Acknowledgments
We thank David Schnurr of the Viral and Rickettsial Disease Laboratory, California Department of Health Services, and Kenneth Schnabel of the University of Rochester School of Medicine and Dentistry for providing clinical specimens for this study.
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the views of the funding agency.
Footnotes
Published ahead of print on 5 December 2007.
REFERENCES
- 1.Andeweg, A. C., T. M. Bestebroer, M. Huybreghs, T. G. Kimman, and J. C. de Jong. 1999. Improved detection of rhinoviruses in clinical samples by using a newly developed nested reverse transcription-PCR assay. J. Clin. Microbiol. 37524-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andries, K., B. Dewindt, J. Snoeks, L. Wouters, H. Moereels, P. J. Lewi, and P. A. Janssen. 1990. Two groups of rhinoviruses revealed by a panel of antiviral compounds present sequence divergence and differential pathogenicity. J. Virol. 641117-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atmar, R. L., and P. R. Georghiou. 1993. Classification of respiratory tract picornavirus isolates as enteroviruses or rhinoviruses by using reverse transcription-polymerase chain reaction. J. Clin. Microbiol. 312544-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billaud, G., S. Peny, V. Legay, B. Lina, and M. Valette. 2003. Detection of rhinovirus and enterovirus in upper respiratory tract samples using a multiplex nested PCR. J. Virol. Methods 108223-228. [DOI] [PubMed] [Google Scholar]
- 5.Blomqvist, S., A. Skytta, M. Roivainen, and T. Hovi. 1999. Rapid detection of human rhinoviruses in nasopharyngeal aspirates by a microwell reverse transcription-PCR-hybridization assay. J. Clin. Microbiol. 372813-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blomqvist, S., C. Savolainen, L. Raman, M. Roivainen, and T. Hovi. 2002. Human rhinovirus 87 and enterovirus 68 represent a unique serotype with rhinovirus and enterovirus features. J. Clin. Microbiol. 404218-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corless, C. E., M. Guiver, R. Borrow, V. Edwards-Jones, A. J. Fox, E. B. Kaczmarski, and K. J. Mutton. 2002. Development and evaluation of a real-time RT-PCR for the detection of enterovirus and parechovirus RNA in CSF and throat swab samples. J. Med. Virol. 67555-562. [DOI] [PubMed] [Google Scholar]
- 8.Dagher, H., H. Donninger, P. Hutchinson, R. Ghildyal, and P. Bardin. 2004. Rhinovirus detection: comparison of real-time and conventional PCR. J. Virol. Methods 117113-121. [DOI] [PubMed] [Google Scholar]
- 9.Deffernez, C., W. Wunderli, Y. Thomas, S. Yerly, L. Perrin, and L. Kaiser. 2004. Amplicon sequencing and improved detection of human rhinovirus in respiratory samples. J. Clin. Microbiol. 423212-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emery, S. L., D. D. Erdman, M. D. Bowen, B. R. Newton, J. M. Winchell, R. F. Meyer, S. Tong, B. T. Cook, B. P. Holloway, K. A. McCaustland, P. A. Rota, B. Bankamp, L. E. Lowe, T. G. Ksiazek, W. J. Bellini, and L. J. Anderson. 2004. Real-time reverse transcription-polymerase chain reaction assay for SARS-associated coronavirus. Emerg. Infect. Dis. 10311-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox, J. P., M. K. Cooney, C. E. Hall, and H. M. Foy. 1985. Rhinoviruses in Seattle families, 1975-1979. Am. J. Epidemiol. 122830-846. [DOI] [PubMed] [Google Scholar]
- 12.Friedlander, S. L., and W. W. Busse. 2005. The role of rhinovirus in asthma exacerbations. J. Allergy Clin. Immunol. 116267-273. [DOI] [PubMed] [Google Scholar]
- 13.Ghosh, S., R. Champlin, R. Couch, J. England, I. Raad, S. Malik, M. Luna, and E. Whimbey. 1999. Rhinovirus infections in myelosuppressed adult blood and marrow transplant recipients. Clin. Infect. Dis. 29528-532. [DOI] [PubMed] [Google Scholar]
- 14.Halonen, P., E. Rocha, J. Hierholzer, B. Holloway, T. Hyypia, P. Hurskainen, and M. Pallansch. 1995. Detection of enteroviruses and rhinoviruses in clinical specimens by PCR and liquid-phase hybridization. J. Clin. Microbiol. 33648-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hendley, J. O., and J. M. Gwaltney. 1988. Mechanisms of transmission of rhinovirus infections. Epidemiol. Rev. 10243-258. [PubMed] [Google Scholar]
- 16.Hicks, L. A., C. W. Shepard, P. H. Britz, D. D. Erdman, M. Fischer, B. L. Flannery, A. J. Peck, X. Lu, W. L. Thacker, R. F. Benson, M. L. Tondella, M. E. Moll, C. G. Whitney, L. J. Anderson, and D. R. Feikin. 2006. Two outbreaks of severe respiratory disease in nursing homes associated with rhinovirus. J. Am. Geriatr. Soc. 54284-289. [DOI] [PubMed] [Google Scholar]
- 17.Hyypiä, T., T. Puhakka, O. Ruuskanen, M. Mäkelä, A. Arola, and P. Arstila. 1998. Molecular diagnosis of human rhinovirus infections: comparison with virus isolation. J. Clin. Microbiol. 362081-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ireland, D. C., J. Kent, and K. G. Nicholson. 1993. Improved detection of rhinoviruses in nasal and throat swabs by seminested RT-PCR. J. Med. Virol. 4096-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ison, M. G., F. G. Hayden, L. Kaiser, L. Corey, and M. Boeckh. 2003. Rhinovirus infections in hematopoietic stem cell transplant recipients with pneumonia. Clin. Infect. Dis. 361139-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jartti, T., P. Lehtinen, T. Vuorinen, M. Koskenvuo, and O. Ruuskanen. 2004. Persistence of rhinovirus and enterovirus RNA after acute respiratory illness in children. J. Med. Virol. 72695-699. [DOI] [PubMed] [Google Scholar]
- 21.Johnston, S. L., and D. A. J. Tyrreil. 1995. Rhinoviruses, p. 553-563. In E. H. Lennette, D. A. Lennette, and E. T. Lennette (ed.), Diagnostic procedures for viral, rickettsial, and chlamydial infections, 7th ed. American Public Health Association, Washington, DC.
- 22.Joki-Korpela, P., and T. Hyypia. 2001. Parechoviruses, a novel group of human picornaviruses. Ann. Med. 33466-471. [DOI] [PubMed] [Google Scholar]
- 23.Kaiser, L., J. D. Aubert, J. C. Pache, C. Deffernez, T. Rochat, J. Garbino, W. Wunderli, P. Meylan, S. Yerly, L. Perrin, I. Letovanec, L. Nicod, C. Tapparel, and P. M. Soccal. 2006. Chronic rhinoviral infection in lung transplant recipients. Am. J. Respir. Crit. Care Med. 1741392-1399. [DOI] [PubMed] [Google Scholar]
- 24.Kämmerer, U., B. Kunkel, and K. Korn. 1994. Nested PCR for specific detection and rapid identification of human picornaviruses. J. Clin. Microbiol. 32285-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kares, S., M. Lonnrot, P. Vuorinen, S. Oikarinen, S. Taurianen, and H. Hyoty. 2004. Real-time PCR for rapid diagnosis of entero- and rhinovirus infections using LightCycler. J. Clin. Virol. 2999-104. [DOI] [PubMed] [Google Scholar]
- 26.Khetsuriani, N., N. N. Kazerouni, D. D. Erdman, X. Lu, S. C. Redd, L. J. Anderson, and W. G. Teague. 2007. Prevalence of viral respiratory tract infections in children with asthma. J. Allergy Clin. Immunol. 119314-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King, A. M. Q., F. Brown, P. Christian, T. Hovi, T. Hyypiä, N. J. Knowles, S. M. Lemon, P. D. Minor, A. C. Palmenberg, T. Skern, and G. Stanway. 2000. Picornaviridae, p. 657-678. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh report of the International Committee for the Taxonomy of Viruses. Academic Press, San Diego, CA.
- 28.Kling, S., H. Donninger, Z. Williams, J. Vermeulen, E. Weinberg, K. Latiff, R. Ghildyal, and P. Bardin. 2005. Persistence of rhinovirus RNA after asthma exacerbation in children. Clin. Exp. Allergy 35672-678. [DOI] [PubMed] [Google Scholar]
- 29.Lamson, D., N. Renwick, V. Kapoor, Z. Liu, G. Palacios, J. Ju, A. Dean, K. St. George, T. Briese, and W. I. Lipkin. 2006. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype that caused influenza-like illness in New York State during 2004-2005. J. Infect. Dis. 1941398-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ledford, R. M., N. R. Patel, T. M. Demenczuk, A. Watanyar, T. Herbertz, M. S. Collett, and D. C. Pevear. 2004. VP1 sequencing of all human rhinovirus serotypes: insights into genus phylogeny and susceptibility to antiviral capsid-binding compounds. J. Virol. 783663-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loens, K., H. Goossens, C. de Laat, H. Foolen, P. Oudshoorn, S. Pattyn, P. Sillekens, and M. Ieven. 2006. Detection of rhinoviruses by tissue culture and two independent amplification techniques, nucleic acid sequence-based amplification and reverse transcription-PCR, in children with acute respiratory infections during a winter season. J. Clin. Microbiol. 44166-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu, X., M. Chittaganpitch, S. J. Olsen, I. M. Mackay, T. P. Sloots, A. M. Fry, and D. D. Erdman. 2006. Real-time PCR assays for detection of bocavirus in human specimens. J. Clin. Microbiol. 443231-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McErlean, P., L. A. Shackelton, S. B. Lambert, M. D. Nissen, T. P. Sloots, and I. M. Mackay. 2007. Characterisation of a newly identified human rhinovirus, HRV-QPM, discovered in infants with bronchiolitis. J. Clin. Virol. 3967-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, E. K., X. Lu, D. D. Erdman, K. A. Poehling, Y. Zhu, M. R. Griffin, T. V. Hartert, L. J. Anderson, G. A. Weinberg, C. B. Hall, M. K. Iwane, K. M. Edwards, and the New Vaccine Surveillance Network. 2007. Rhinovirus-associated hospitalizations in young children. J. Infect. Dis. 195773-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monto, A. S., A. M. Fendrick, and M. W. Sarnes. 2001. Respiratory illness caused by picornavirus infection: a review of clinical outcomes. Clin. Ther. 231615-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicholson, K. G., J. Kent, V. Hammersley, and E. Cancio. 1996. Risk factors for lower respiratory complications of rhinovirus infections in elderly people living in the community: prospective cohort study. Br. Med. J. 3131119-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nielsen, C. B., S. K. Singh, J. Wengel, and J. P. Jacobsen. 1999. The solution structure of a locked nucleic acid (LNA) hybridized to DNA. J. Biomol. Struct. Dyn. 17175-191. [DOI] [PubMed] [Google Scholar]
- 38.Nijhuis, M., N. van Maarseveen, R. Schuurman, S. Verkuijlen, M. de Vos, K. Hendriksen, and A. M. van Loon. 2002. Rapid and sensitive routine detection of all members of the genus enterovirus in different clinical specimens by real-time PCR. J. Clin. Microbiol. 403666-3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nokso-Koivisto, J., T. J. Kinnari, P. Lindahl, T. Hovi, and A. Pitkäranta. 2002. Human picornavirus and coronavirus RNA in nasopharynx of children without concurrent respiratory symptoms. J. Med. Virol. 66417-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pallansch, M. A., and R. P. Roos. 2001. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enterovirus, p. 723-775. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 41.Papadopoulos, N. G., J. Hunter, G. Sanderson, J. Meyer, and S. L. Johnston. 1999. Rhinovirus identification by BglI digestion of picornavirus RT-PCR amplicons. J. Virol. Methods 80179-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pitkäranta, A., and F. G. Hayden. 1998. Rhinoviruses: important respiratory pathogens. Ann. Med. 30529-537. [DOI] [PubMed] [Google Scholar]
- 43.Savolainen, C., S. Blomqvist, M. N. Mulders, and T. Hovi. 2002. Genetic clustering of all 102 human rhinovirus prototype strains: serotype 87 is close to human enterovirus 70. J. Gen. Virol. 83333-340. [DOI] [PubMed] [Google Scholar]
- 44.Scheltinga, S. A., K. E. Templeton, M. F. Beersma, and E. C. Claas. 2005. Diagnosis of human metapneumovirus and rhinovirus in patients with respiratory tract infections by an internally controlled multiplex real-time RNA PCR. J. Clin. Virol. 33306-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schrag, S. J., J. T. Brooks, C. Van Beneden, U. D. Parashar, P. M. Griffin, L. J. Anderson, W. J. Bellini, R. F. Benson, D. D. Erdman, A. Klimov, T. G. Ksiazek, T. C. Peret, D. F. Talkington, W. L. Thacker, M. L. Tondella, J. S. Sampson, A. W. Hightower, D. F. Nordenberg, B. D. Plikaytis, A. S. Khan, N. E. Rosenstein, T. A. Treadwell, C. G. Whitney, A. E. Fiore, T. M. Durant, J. F. Perz, A. Wasley, D. Feikin, J. L. Herndon, W. A. Bower, B. W. Klibourn, D. A. Levy, V. G. Coronado, J. Buffington, C. A. Dykewicz, R. F. Khabbaz, and M. E. Chamberland. 2004. SARS surveillance during emergency public health response, United States, March-July 2003. Emerg. Infect. Dis. 10185-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smyth, A. R., R. L. Smyth, C. Y. Tong, C. A. Hart, and D. P. Heaf. 1995. Effect of respiratory virus infections including rhinovirus on clinical status in cystic fibrosis. Arch. Dis. Child. 73117-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verstrepen, W. A., S. Kuhn, M. M. Kockx, M. E. Van De Vyvere, and A. H. Mertens. 2001. Rapid detection of enterovirus RNA in cerebrospinal fluid specimens with a novel single-tube real-time reverse transcription-PCR assay. J. Clin. Microbiol. 394093-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wald, T., P. Shult, P. Krause, B. Miller, P. Drinka, and S. Gravenstein. 1995. A rhinovirus outbreak among residents of a long-term care facility. Ann. Intern. Med. 123588-593. [DOI] [PubMed] [Google Scholar]
- 49.Wittwer, C. T., M. G. Herrmann, A. A. Moss, and R. P. Rasmussen. 1997. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques 22130-138. [DOI] [PubMed] [Google Scholar]
- 50.Witwer, C., S. Rauscher, I. L. Hofacker, and P. F. Stadler. 2001. Conserved RNA secondary structures in Picornaviridae genomes. Nucleic. Acids Res. 295079-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wright, P. F., A. M. Deatly, R. A. Karron, R. B. Belshe, J. R. Shi, W. C. Gruber, Y. Zhu, and V. B. Randolph. 2007. Comparison of results of detection of rhinovirus by PCR and viral culture in human nasal wash specimens from subjects with and without clinical symptoms of respiratory illness. J. Clin. Microbiol. 452126-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]