Abstract
The genus Aspergillus is ideally suited for the investigation of RNA silencing evolution because it includes species that have experienced a variety of RNA silencing gene changes. Our work on this subject begins here with the model species Aspergillus nidulans. Filamentous ascomycete fungi generally each encode two of the core RNA silencing proteins, Dicer and Argonaute, but A. nidulans appears to have lost one of each to gene truncation events. Although a role in growth, development, or RNA silencing was not detected for the truncated genes, they do produce spliced and poly(A)-tailed transcripts, suggesting that they may have an undetermined biological function. Population analysis demonstrates that the truncated genes are fixed at the species level and that their full-length orthologs in a closely related species are also unstable. With these gene truncation events, A. nidulans encodes only a single intact Dicer and Argonaute. Their deletion results in morphologically and reproductively normal strains that are incapable of experimental RNA silencing. Thus, our results suggest that the remaining A. nidulans RNA silencing genes have a “nonhousekeeping” function, such as defense against viruses and transposons.
RNA silencing proteins, including Dicers, Argonautes, and RNA-dependent RNA polymerases (RdRPs), are found in all eukaryotic lineages thus far investigated, making it likely that RNA silencing evolved before divergence of the last common eukaryotic ancestor (13). Since then, RNA silencing proteins have retained or developed a number of important biological roles, including genome defense, chromatin modification, and gene regulation (for reviews, see references 31, 44, and 70). Despite the importance of RNA silencing, it is not uniformly conserved in all eukaryotes, with current evidence indicating that four of six eukaryotic supergroups have members that do not encode Dicers, Argonautes, and RdRPs (13). However, while these types of genes have been lost by some species, they have been expanded in others. This is perhaps most apparent in basidiomycete fungi, where a combination of RNA silencing gene gain and loss has resulted in species that encode between zero and eight Dicers, zero and three Argonautes, and zero and nine RdRPs (48). It is thus apparent that RNA silencing gene changes are common in eukaryotic evolution; however, the forces contributing to these changes, as well as the methods by which species adapt to them, are unclear.
Dicers are characterized by a number of domains, including an N-terminal helicase domain and two C-terminal RNase III domains. These proteins cleave long double-stranded RNA (dsRNA) into 21- to 26-bp fragments (4, 22). The resulting small RNA is typically classified based on the dsRNA source from which it originates and includes types such as repeat-associated small interfering RNA (rasiRNA), microRNA (miRNA), and small interfering RNA (siRNA) (46). Endogenous sources of dsRNA are repetitive DNA, noncoding regulatory genes, and foreign genetic elements such as viruses and transposons. Some of these sources require an RdRP to produce the double-stranded Dicer substrate (3, 15, 17, 20, 43, 64, 65, 67). Exogenous dsRNA can also be introduced into cells or whole organisms experimentally. One method involves transforming organisms with inverted repeat transgenes (IRTs) so that IRT transcription will produce hairpin RNA molecules to serve as Dicer substrates (41, 55, 73).
Dicer processed small RNA is incorporated into an Argonaute-containing ribonucleoprotein effector complex (9, 14, 46). Examples include RITS, which contains a rasiRNA and directs heterochromatin assembly (71); miRNP, which contains a miRNA and inhibits the translation of mRNA (47, 52, 60), and RISC, which contains a siRNA or miRNA and directs RNA cleavage (29, 33, 45). The various small RNA likely guide their effector complexes to specific targets by complementary base pairing (22, 45). Argonautes typically contain an N-terminal PAZ domain and a C-terminal Piwi domain. Within the RISC complex, the PAZ domain is thought to bind the siRNA (42, 57, 66, 75), and the Piwi domain is thought to cleave the target mRNA (2, 42, 57, 66).
With regard to filamentous fungi, the Neurospora crassa RNA silencing machinery is the most thoroughly characterized. Two N. crassa RNA silencing phenomena are meiotic silencing (40, 62, 63) and quelling (16, 18, 19, 58), both of which appear to be efficient genome defense mechanisms. Meiotic silencing is activated by unpaired DNA during the sexual cycle, while quelling occurs during the vegetative cycle and depends on high numbers of tandemly arranged transgenes. A two-pathway hypothesis has been proposed to explain the evolutionary origin of N. crassa meiotic silencing and quelling. This hypothesis suggests that a single group of ancestral RNA silencing genes duplicated in an early ancestor of the filamentous ascomycetes, leading to two paralogous groups of RNA silencing genes with evolutionarily divergent functions (5, 23). The hypothesis is supported by phylogenetic evidence from both Aspergillus fumigatus and N. crassa (5, 23) and genetic evidence from studies with N. crassa. This genetic evidence links meiotic silencing to the Dicer DCL-1 (1), the Argonaute SMS-2 (40), and the RdRP SAD-1 (63) and quelling to Argonaute QDE-2 (11) and RdRP QDE-1 (17). However, either of the N. crassa Dicers (DCL-1 and DCL-2) is sufficient for quelling (11), demonstrating that there is not always a clear division of labor between the two pathways.
RNA silencing is crucial for normal growth and developmental processes in higher eukaryotes (26, 27, 32, 36, 37, 53, 56, 74), but it is unclear how important RNA silencing is for growth and development in fungi. Although at least some N. crassa RNA silencing mutants are sterile in homozygous crosses (1, 40, 63), other morphological changes have not been reported for N. crassa single or double Dicer mutants (12), or Dicer mutants of the tree pathogen, Cryphonectria parasitica (59). In Magnaporthe oryzae and Mucor circinelloides, other filamentous fungi whose RNA silencing genes have been partially characterized, Dicer mutants appear to have slight morphological abnormalities (35, 49). Finally, in the fission yeast Schizosaccharomyces pombe, RNA silencing mutations disrupt normal cell cycle regulation (10) and cause chromosome segregation defects (72). These phenotypes have not yet been associated with RNA silencing defects in other fungi.
A group of closely related species that have followed different evolutionary paths in regard to RNA silencing should benefit studies of RNA silencing gene evolution. Herein, a survey of the Dicers and Argonaute genes in seven Aspergillus species demonstrates that Aspergillus fungi are well suited for such studies because they include species that have experienced RNA silencing gene gain or loss. The present study thoroughly characterizes the loss of RNA silencing genes—a Dicer and an Argonaute—to gene truncation events in the model species Aspergillus nidulans. In addition, the remaining A. nidulans RNA silencing genes (a Dicer, an Argonaute, and two RdRPs) are characterized with respect to experimental RNA silencing, growth, and development. A companion study investigates the role of RNA silencing in the defense against mycoviruses (28a).
MATERIALS AND METHODS
Phylogenetic trees, domain identification, and nucleotide polymorphism analysis.
Accession numbers for protein sequences used in the present study are provided in the supplemental material. Ascomycete Dicers and Argonautes were identified by searching (blastp) the GenBank (National Center for Biotechnology Information) and fungus-specific databases with the consensus sequences of RNase III (cd00593.1) and Piwi (pfam02171.11) domains, as well as the sequence of known fungal Dicer and Argonaute proteins. Species-specific databases included the following: www.cadre.man.ac.uk (A. fumigatus), www.broad.mit.edu (A. nidulans, Gibberella zeae, M. oryzae, and N. crassa), www.bio.nite.go.jp (Aspergillus oryzae), and www.sanger.ac.uk (Schizosaccharomyces pombe). Non-annotated Aspergillus databases (www.ncbi.nlm.nih.gov; Aspergillus clavatus NRRL1, Aspergillus flavus NRRL3357, Aspergillus terreus NIH2624, and Neosartorya fischeri NRRL181) were searched (tblastn) for contigs containing putative Dicer and Argonaute orthologs (see the supplemental material). Putative Dicer and Argonaute gene sequences were retrieved and aligned to the annotated orthologs found in the A. fumigatus or A. oryzae genome databases to predict exon-intron boundaries. The putative protein sequences were aligned by using MUSCLE (21) (European Bioinformatics Institute [www.ebi.ac.uk/muscle]). Alignments were imported onto the San Diego Supercomputer Center's Biology Workbench (workbench.sdsc.edu) to produce unrooted, noniterated trees using the DrawTree function.
For nucleotide polymorphism analysis, PCR-amplified fragments were generally gel purified and cloned into pCR2.1-TOPO (Invitrogen) before sequencing. Only one to two clones were sequenced for each locus; thus, some polymorphic residues may be a result of PCR and/or sequencing error. Sequences were imported into Bio Edit (28) and MEGA (39) for processing and analysis and aligned by using CLUSTAL W (68) (see also the supplementary material).
Strains and culture conditions.
The genotypes of the A. nidulans FGSC A4 (referred to here as simply A4) derivatives used in the present study are listed in Table 1. The four wild isolates of A. nidulans (G143, H109, HcB, and HcE) were collected from various locations around the United Kingdom and represent four distinct heterokaryon compatibility groups (25). The isolates were provided along with two wild Aspergillus rugulosus isolates (strains 203 and 211) by David Geiser (Penn State University). An industrial A. rugulosus isolate, SRRC 1173 (38), was also used in the present study. To confirm that the various isolates were of independent lineages, Southern analysis of genomic DNA from the A. nidulans and A. rugulosus isolates was performed with a probe for A. nidulans transposon relic An5242.3. Each wild isolate produced a unique pattern, while all A4 derivatives produced an identical pattern (see the supplementary material). For physiological assays and crossing assays, a series of crosses was first performed to place all RNA silencing gene deletion alleles and RdRP deletion alleles in the same genetic background (see the supplemental material). The combined gene deletions include ΔdclB, ΔrsdA, ΔrrpB, and ΔrrpC. When all deletion alleles are in the same genetic background the strain is referred to as ΔRNAi.
TABLE 1.
Strains used in this study
| Category and straina | Genotype |
|---|---|
| Reference strains | |
| RTMH13.B3* | ΔstcE::argB; veA1 |
| RTMH13.B1* | ΔstcE::argB; aflR(IRT)::trpC veA1 |
| RTMH13.F2 | pyrG89; metG1, ΔstcE::argB; veA1 |
| RTMH13.F3* | pyrG89; metG1 ΔstcE::argB; aflR(IRT)::trpC veA1 |
| Transformants | |
| TEAB65.C1‡ | pyrG89; metG1 ΔstcE::argB; ΔrsdA::pyrG; aflR(IRT)::trpC veA1 |
| TJW64.17, 46 | pyrG89; wA3; pyroA4 ΔstcE::argB; ΔdclA::pyroA, trpC801 veA1 |
| TBRG3.6 | pyrG89; ΔppdB::metG; metG1 ΔstcE::argB; aflR(IRT)::trpC veA1 |
| TTMH158.1 | pyrG89; metG1 ΔstcE::argB; ΔdclB::pyrG; aflR(IRT)::trpC veA1 |
| TTMH160.3 | pyrG89; wA3; gpdA(p)::dclB::pyroA ΔstcE::argB; veA1 |
| Analysis of dclA function in IRT-RNA silencing (Fig. 3) | |
| RTMH192.1 | pyrG89; wA3; ΔstcE::argB; veA1 |
| RTMH192.2, 5, 10 | pyrG89; ΔstcE::argB; aflR(IRT)::trpC ΔdclA::pyroA veA1 |
| RTMH192.3, 6, 8 | pyrG89; ΔstcE::argB; aflR(IRT)::trpC veA1 |
| RTMH192.4 | pyrG89; ΔstcE::argB; ΔdclA::pyroA, veA1 |
| RTMH192.7 | pyrG89; wA3; ΔstcE::argB; aflR(IRT)::trpC, ΔdclA::pyroA veA1 |
| RTMH192.9 | pyrG89; wA3; ΔstcE::argB; aflR(IRT)::trpC, veA1 |
| Analysis of ppdB and rsdA function in IRT-RNA silencing (Fig. 4) | |
| RTMH193.1‡ | pyrG89; ΔppdB::metG; metG1 ΔstcE::argB; ΔrsdA::pyrG; aflR(IRT)::trpC veA1 |
| RTMH193.4 | pyrG89; metG1 ΔstcE::argB; ΔrsdA::pyrG; aflR(IRT)::trpC veA1 |
| RTMH193.10 | pyrG89; ΔppdB::metG; ΔstcE::argB; aflR(IRT)::trpC veA1 |
| Analysis of dclB function in IRT-RNA silencing (Fig. 7) | |
| RTMH215.6, 7 | pyrG89; ΔstcE::argB; ΔdclB::pyrG; veA1 |
| RTMH215.12, 13 | pyrG89; ΔstcE::argB; ΔdclB::pyrG; aflR(IRT)::trpC veA1 |
| RTMH215.4, 17 | pyrG89; gpdA(p)::dclB::pyroA ΔstcE::argB; ΔdclB::pyrG; veA1 |
| RTMH215.20, 29 | pyrG89; gpdA(p)::dclB::pyroA ΔstcE::argB; ΔdclB::pyrG; aflR(IRT)::trpC veA1 |
| Analysis of radial growth and spore production (Table 3) | |
| RDIT9.32† | WT |
| RTMH211.14 | WT |
| RTMH229.24 | WT |
| RTMH218.7, 39 | ΔrrpB::pyrG; ΔrrpC::metG ΔdclB::pyrG; ΔrsdA::pyrG |
| RTMH211.6, 11 | ΔppdB::metG |
| RTMH212.42, 65 | ΔdclA::pyroA |
| Analysis of crossing (Table 4) | |
| RTMH227.29, 47 | yA2; ΔrrpB::pyrG; ΔrrpC::metG ΔdclB::pyrG; ΔrsdA::pyrG |
| RTMH227.67, 68 | wA3; ΔrrpB::pyrG; ΔrrpC::metG ΔdclB::pyrG; ΔrsdA::pyrG |
| RTMH229.25, 37 | yA2 |
| RTMH229.19 | wA3 |
| RTMH233.C | wA3 |
Commas are used between the suffixes of strain names to indicate independently isolated transformants or recombinants of the same genotype. The aflR IRT transgene used in this study is the aflR(IRT1300) transgene referenced earlier (30). Strain sources: *, Hammond and Keller (30), †, Tsitsigiannis et al. (69). ‡, Strains that carry an ectopic copy of the rsdA deletion vector; this does not appear to have influenced any of the results (data not shown). Additional strains used in this study, such as the transformation hosts and crossing intermediates, are described in the supplemental material.
Glucose minimal medium (GMM) with appropriate supplements (61) was used for all experiments unless otherwise indicated. For genomic DNA isolation 1 g of yeast extract was added to 1 liter of liquid GMM. Yeast-glucose-trace element medium (YGT) as described previously (8) was used for crossing assays. A. nidulans conidia were inoculated into 25 ml of liquid GMM (2.5 × 107 conidia for RNA isolation) and incubated under nonstringent dark conditions at 37°C unless otherwise indicated.
Gene deletions, transformations, and crosses.
The sequences for all oligonucleotide PCR primers used in the present study are provided in the supplemental material. Genes were deleted by double-homologous recombination. Details of the deletion and complementation plasmids used in the present study are provided there as well. Transformations were performed essentially as described by Yu and Adams (76). Changes to the protocol included the use of 3 to 4 mg of Sigma lysing enzyme (L1412-10G; Sigma) per ml of OM-A protoplasting buffer or the use of the glucanase-driselase-lyticase lysing enzyme mix suggested by Jung et al. (34). Because selecting for outcrossed cleistothecia from a cross between prototrophic A. nidulans strains can be difficult, the ΔrsdA transformants described previously (30) were not used, and a new ΔrsdA methionine-auxotroph was created instead (described in the supplemental material).
Because A. nidulans is capable of both selfing and crossing, an assay (inspired by Bruggeman et al. [6]) involving spore color mutants was used to determine whether A. nidulans RNA silencing mutants were deficient in crossing ability. Yellow (yA2) and white (wA3) spore color gene mutations segregate independently during meiosis, so that crosses between strains that are yellow (wA yA2) and white (wA3 yA) result in progeny that are yellow (wA yA2), green (wA yA), or white (wA3 yA2 or wA3 yA). Scoring the conidial color of ascospore-derived colonies from a single cleistothecium thus allows one to distinguish between selfed and crossed cleistothecia. For the assay, 5 × 105 conidia of each crossing partner were mixed in 5.0 ml of molten YGT (0.7% agar, ∼48°C) and spread onto the surface of 30.0 ml of solid YGT. Plates were wrapped with parafilm and cultured in the dark for 10 days. For scoring, cleistothecia were harvested and cleaned of fungal tissue by rolling on 3% water agar. During harvesting there was an intentional bias toward the larger (than average) cleistothecia, since this was assumed to be an indicator of maturity. Cleaned cleistothecia were burst open in 500 μl of sterile water, and 5 to 10 μl of each ascospore suspension was spread onto solid GMM. After 3 days at 37°C, the colony conidial color was recorded and used to determine whether each cleistothecium was the result of selfing (yellow or white colonies) or crossing (white, yellow, and green colonies). Plates were stored at 4°C during cleistothecium scoring, which was performed over several weeks.
Southern and Northern blotting.
All transformants and recombinants were single spore purified, and genotypes were confirmed by Southern blotting using Hybond-XL nylon membranes (Amersham Biosciences) with the alkali transfer protocol as described by the manufacturer. Southern blotting-based identification of all deletion strains was performed with probes for the deleted sequences (see Results) and at least one flanking region of the deleted gene (see the supplemental material). For Northern analysis, total RNA was harvested from lyophilized tissues by using TRIzol reagent (Invitrogen) essentially as suggested by the manufacturer. RNA was separated by formaldehyde-denaturing gel electrophoresis and blotted onto Hybond-XL nylon membranes by capillary transfer in 10× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). A picture of the ethidium bromide-stained rRNA is included with all Northern blots to indicate the relative levels of RNA in each lane.
Determining RNA silencing function via NOR production.
Previous analysis of A. nidulans RNA silencing demonstrated that an IRT containing A. nidulans aflR sequences can be used to silence aflR expression through RNA silencing (IRT-RNA silencing [30]). Because A. nidulans strains carrying a ΔstcE allele produce the compound norsolorinic acid (NOR) in an aflR-dependent manner (7), it is possible to determine whether IRT-RNA silencing is functional by measuring NOR production in strains carrying both an aflR IRT and a ΔstcE allele (30). In essence, the lack of NOR in the presence of ΔstcE and the aflR IRT indicates that IRT-RNA silencing is functional. To measure NOR production, conidia were qualitatively transferred to the center of a 30.0-ml plate of GMM (plus supplements if required), and cultures were grown for 5 to 6 days. A 1.4-cm-diameter core was then taken from the center of the colony and assayed by thin-layer chromatography (TLC) as previously described (30).
GenBank accession numbers.
The GenBank accession numbers for the strains discussed in this study were as follows: (i) for A. nidulans RTMH13.C5 (an A4 derivative), internal transcribed spacer (ITS), EU287942; (ii) for A. nidulans G143, dclA (target 1), EU289898; and ppdB (target 3), EU289899; (iii) for A. rugulosus 211, dclA (targets 1 and 2), EU289900; ppdB (target 4), EU289903; rrpA relic (partial), EU289905; actin (partial), EU289911; ITS, EU289912; dclB (partial), EU289913; and ppdA (partial), EU289914; (iv) for A. rugulosus 1173, dclA (target 2), EU289901; ppdB (target 4), EU289904; rrpA relic (partial), EU289906; actin (partial), EU289915; ITS, EU289916; dclB (partial), EU289917; and ppdA (partial), EU289918; and (v) for A. rugulosus 203, ppdB (target 4), EU289902; actin (partial), EU289907; ITS, EU289908; dclB (partial), EU289909; and ppdA (partial), EU289910.
RESULTS
Dicers and Argonautes in the filamentous ascomycetes.
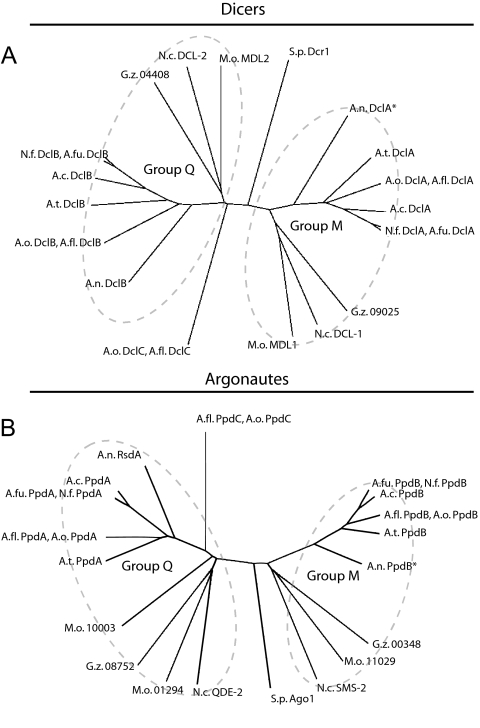
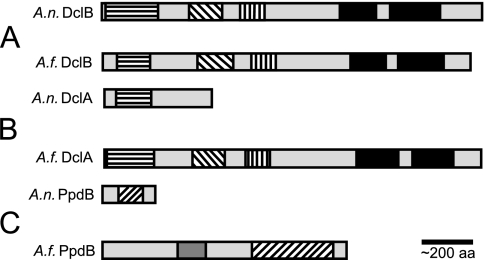
As originally noted for the N. crassa and A. fumigatus Dicers and Argonautes (5, 23), two paralogous groups of RNA silencing proteins are typical of filamentous ascomycetes (Fig. 1). Although quelling and meiotic silencing have only been described in N. crassa, we refer to the paralogous groups as “Q” and “M” in the present study for reference purposes. Most analyzed fungi encode a single member for groups Q and M, although a few deviate from this pattern. Aspergillus oryzae and Aspergillus flavus each encode three Dicers and three Argonautes, apparently due to group Q duplication. Similarly, Magnaporthe oryzae encodes three Argonautes, apparently due to duplication of its group Q Argonaute (Fig. 1). The opposite phenomenon of RNA silencing gene loss appears to have occurred in A. nidulans. Although there is evidence for both A. nidulans group M and Q proteins (Fig. 1), only the A. nidulans group Q proteins appear to be full length (Fig. 2). For example, while the group Q Dicer (DclB) is predicted to have all of the domains of a typical Dicer, the group M Dicer (DclA) is predicted to have only a Dead Box helicase domain (Fig. 2A and B). Similarly, while the group Q Argonaute (RsdA) is predicted to have both domains of a typical Argonaute, the group M Argonaute (PpdB) is predicted to have only a Piwi, or partial Piwi, domain (Fig. 2C) (30). Thus, in contrast to N. crassa, M. oryzae, G. zeae, and other aspergilli, A. nidulans has lost full-length group M RNA silencing proteins.
FIG. 1.
Ascomycete Dicers and Argonautes. The predicted sequences for putative Dicers (A) and Argonautes (B) were used to assemble unrooted, noniterated phylogenetic trees (see Materials and Methods). Proteins were arbitrarily divided into two general groups (Q or M). Names for Aspergillus proteins, except A. nidulans RsdA, were derived from the names of the corresponding A. fumigatus orthologs proposed by Galagan et al. (23). Uncharacterized proteins from non-Aspergillus species were not given names. Abbreviations: A.c., A. clavatus; A.fl., A. flavus; A.fu., A. fumigatus; A.n., A. nidulans; A.o., A. oryzae; A.t., A. terreus, G.z., G. zeae; M.o., M. oryzae; N.c., N. crassa; N.f., N. fischeri; S.p., S. pombe.
FIG. 2.
Predicted domains of A. nidulans and A. fumigatus Dicer and Argonaute proteins. The predicted sequences of A. nidulans and A. fumigatus DclB, DclA and PpdB were used to search the National Center for Biotechnology Information conserved domain database for domain identification. The truncated nature of A. nidulans dclA and ppdB is revealed by comparison to their A. fumigatus orthologs. The A. nidulans group Q Argonaute, RsdA, was characterized in a previous study (30). An MPH1 domain overlaps the DEAD and Hel C domains in both A. fumigatus Dicer proteins and A. nidulans DclB (not shown). Dicer domains: DEAD-like helicase (pfam00270, smart00487), horizontal bars; Helicase C (cd00079), diagonal bars; Duf283 (pfam03368), vertical bars; RNase III (cd00593), black boxes; MPH1 (cog1111.1). Argonaute domains: PAZ (cd02846), dark gray boxes; Piwi (pfam02171), diagonal bars.
Truncation events have occurred at two A. nidulans RNA silencing gene loci.
The missing domains of A. nidulans dclA and ppdB are not due to an annotation mistake. An extensive analysis of sequences flanking the truncated dclA and ppdB genes did not locate “missing” RNase III or PAZ domains (data not shown), suggesting that the annotation prediction of gene truncation is correct for both genes. Thus, the dclA and ppdB loci have lost their respective 3′ and 5′ ends through major truncation events. This hypothesis is further supported below.
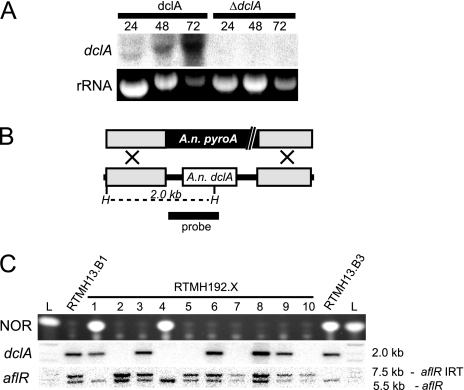
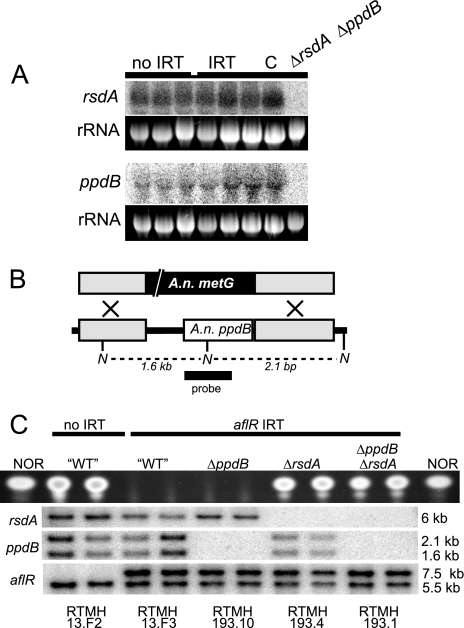
Despite their truncated nature, transcripts are detected from both the dclA and the ppdB loci (Fig. 3A and 4A). In addition, transcript mapping by partial cDNA analysis and/or 3′ RACE (rapid amplification of cDNA ends) demonstrates that intron splicing and poly(A) tailing occur on dclA and ppdB transcripts (data not shown). This suggests that the truncated genes may encode truncated proteins with biological function. However, protein tagging experiments failed to detect a translated protein from either transcript (data not shown), and gene deletion affected neither experimental RNA silencing nor physiological characteristics such as growth or development (Fig. 3 and 4 and see also Table 3 and the supplemental material).
FIG. 3.
The truncated A. nidulans dclA locus is transcribed but not required for IRT-RNA silencing. (A) Northern blotting identifies dclA transcripts in total RNA from 24-, 48-, and 72-h cultures, an increase in transcription was observed at late time points. The dclA transcript migrates slightly below the 18S rRNA band, as expected by its truncated nature. Strains: dclA strain, RTMH192.3; ΔdclA mutant, RTMH192.2. (B) Schematic representation of dclA replacement with A. nidulans pyroA. Gray boxes represent the dclA-flanking sequences used in the deletion vector. The white box represents the predicted A. nidulans dclA open reading frame (ORF). H, HindIII sites. (C) TLC analysis of NOR production and Southern blotting (HindIII digest) results for control strains and recombinants from a cross between a ΔdclA transformant (TJW64.17) and an aflR IRT-carrying strain (RTMH13.F3). In the Southern blots, the absence of a band for dclA is indicative of the deletion genotype (see probe template in panel B) and the presence of two bands for aflR is indicative of the aflR IRT genotype. Note that the loss of NOR production, and thus RNA silencing of aflR, is normal regardless of dclA presence. Strain names are listed above each lane. L, 1-kb ladder or NOR standard.
FIG. 4.
The truncated A. nidulans ppdB locus is transcribed but not required for IRT-RNA silencing. (A) Northern blotting identifies ppdB and rsdA transcripts in total RNA from 48-h cultures. The rsdA transcript migrates slightly above the 26S rRNA band, while the ppdB transcript migrates slightly below the 18S rRNA band, as expected by its truncated nature. Strains: no IRT, RTMH13.B3; IRT, RTMH 13.B1; C, RTMH192.3; ΔrsdA ΔppdB, RTMH193.1. (B) Schematic representation of ppdB replacement with A. nidulans metG. Gray boxes represent the ppdB-flanking sequences used in the deletion vector. The white box represents the predicted ppdB ORF. N, NcoI sites. (C) TLC analysis of NOR production and Southern blotting (NcoI digest for ppdB; HindIII digest for rsdA and aflR) results for control strains and recombinants from a cross between a ΔppdB transformant (TBRG3.6) and an ΔrsdA transformant (TEAB65.C1). In the Southern blots, the absence of bands for ppdB and rsdA (30) is indicative of the deletion genotypes (see probe templates in panel B and also the supplemental material), and the presence of two bands for aflR is indicative of the aflR IRT genotype. Note that RNA silencing of aflR is dependent on rsdA but not on ppdB. Strain names are listed below each lane.
TABLE 3.
Growth and spore production
| Analysis, expt, and straina | Genotype | Mean diam (mm) ± SD | Mean no. (105/mm2) ± SD
|
|
|---|---|---|---|---|
| Conidia | Ascospores | |||
| Radial growth | ||||
| Expt 1 | ||||
| RTMH211.14 | WT | 58.5 ± 1.1 | ||
| RTMH229.24 | WT | 59.3 ± 0.6 | ||
| RTMH218.7 | ΔdclB ΔrsdA ΔrrpB/C | 59.0 ± 1.0 | ||
| RTMH218.39 | ΔdclB ΔrsdA ΔrrpB/C | 59.1 ± 0.6 | ||
| Expt 2 | ||||
| RTMH211-14 | WT | 57.0 ± 1.4 | ||
| RTMH229-24 | WT | 57.3 ± 0.5 | ||
| RTMH212-42 | ΔdclA | 57.9 ± 0.8 | ||
| RTMH212-65 | ΔdclA | 56.4 ± 1.1 | ||
| RTMH211-6 | ΔppdB | 58.6 ± 1.6 | ||
| RTMH211-11 | ΔppdB | 57.8 ± 1.9 | ||
| Spore production | ||||
| Expt 3 | ||||
| RTMH211.14 | WT | 2.17 ± 0.25 | 1.04 ± 0.15 | |
| RTMH218.39 | ΔdclB ΔrsdA ΔrrpB/C | 1.86 ± 0.29 | 0.92 ± 0.19 | |
| RTMH218.7 | ΔdclB ΔrsdA ΔrrpB/C | 2.06 ± 0.14 | 0.83 ± 0.18 | |
| RDIT9.32 | WT | 2.19 ± 0.22 | 0.70 ± 0.15 | |
| Expt 4 | ||||
| RTMH211-14 | WT | 4.90 ± 0.32 | 0.11 ± 0.05 | |
| RTMH211-6 | ΔppdB | 5.12 ± 0.35 | 0.10 ± 0.04 | |
| RTMH212-65 | ΔdclA | 5.78 ± 0.49 | 0.05 ± 0.01 | |
| Expt 5 | ||||
| RTMH229-24 | WT | 4.33 ± 0.52 | 0.23 ± 0.08 | |
| RTMH212-65 | ΔdclA | 4.26 ± 0.49 | 0.21 ± 0.04 | |
Radial growth rate was determined by placing 2 μl of a conidial suspension (100 conidia per μl) in the center of a solid plate of 25.0 ml of GMM, followed by incubation for 5 days at 37°C. Experiment 1, n = 9 to 10; experiment 2, n = 6. Spore production was determined in 6-day-old cultures (37°C, 12 h light). Replicates were prepared by mixing 106 conidia in 5.0 ml of liquid molten GMM agar (0.7%, ∼48°C), followed by plating onto 30.0 ml (experiment 3) or 25.0 ml (experiments 4 and 5) solid GMM. Three 1.4-cm-diameter cores were harvested from each plate and ground in 0.01% Tween 80. Dilutions were then made for hemacytometer-based spore counting. Experiment 3, n = 3; experiment 4, n = 4 to 5; experiment 5, n = 5. The only difference detected in these experiments was in experiment 4, where ΔdclA produced more conidia and fewer ascospores than the wild-type (WT) and ΔppdB mutant strains. However, these results were not consistent with a subsequent experiment (experiment 5), suggesting that experimental error or an unknown genetic difference was a factor in the results for experiment 4.
The A. nidulans dclA and ppdB truncations are common to wild A. nidulans isolates.
Because all other analyzed filamentous ascomycetes were found to encode full-length orthologs of dclA and ppdB, it seemed possible that the dclA and ppdB truncations might be specific to the common A. nidulans laboratory strain A4 and its derivatives. Therefore, the dclA and ppdB loci of wild A. nidulans isolates were investigated. Analysis of genomic fragments spanning the 3′ dclA intergenic region, 5′ ppdB intergenic region and parts of the dclA and ppdB coding regions indicated that the four wild A. nidulans isolates carry the same dclA and ppdB truncations as A4 (Fig. 5 and 6). The fact that longer PCR products are amplified from the 5′ ppdB intergenic region of G143 and HcB than from any of the other isolates (Fig. 6B) is due to the presence of a 5.5-kb insertion with significant identity to a transposon relic found at locus AN8648.3 (and others) of the A. nidulans genome (blastx, E = 0).
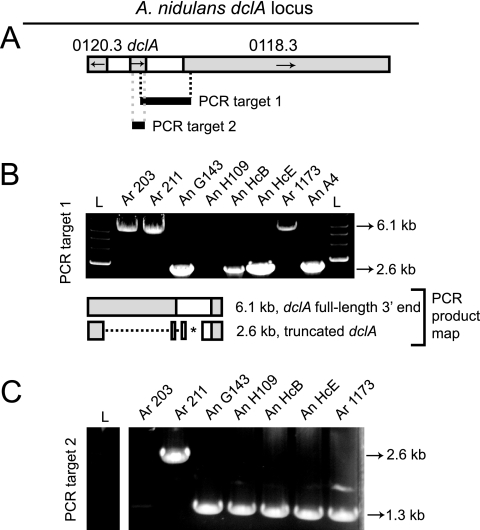
FIG. 5.
Comparison of A. nidulans and A. rugulosus dclA. (A) A schematic of the predicted A. nidulans dclA locus is shown with its immediate flanking genes. Genes are gray and intergenic regions are white. PCR was used to amplify two DNA fragments (target 1 and target 2) from wild A. nidulans isolates (An) and A. rugulosus isolates (Ar). (B) Target 1 is 6.1 kb long in all A. rugulosus isolates and 2.6 kb in all A. nidulans isolates, suggesting that the 3′ end of A. rugulosus dclA is full length while the A. nidulans dclA truncation is fixed at the species level. Although a product is not amplified for A. nidulans HcB, a different primer set amplifies a band similar to one from A4 for the same approximate location (data not shown). A PCR product map depicts the A. nidulans truncation. It comprises three major deletions (dashed line) and a single insertion (*). (C) Target 2 is 2.6 kb in A. rugulosus 211 and 1.3 kb in all other isolates. Sequencing indicates that this is due to a transposon insertion. The A. rugulosus 203 1.3-kp product is present but, for undetermined reasons, is faint relative to the PCR products amplified from the other isolates. L, 1-kb ladder; An A4, strain RTMH13.C5.
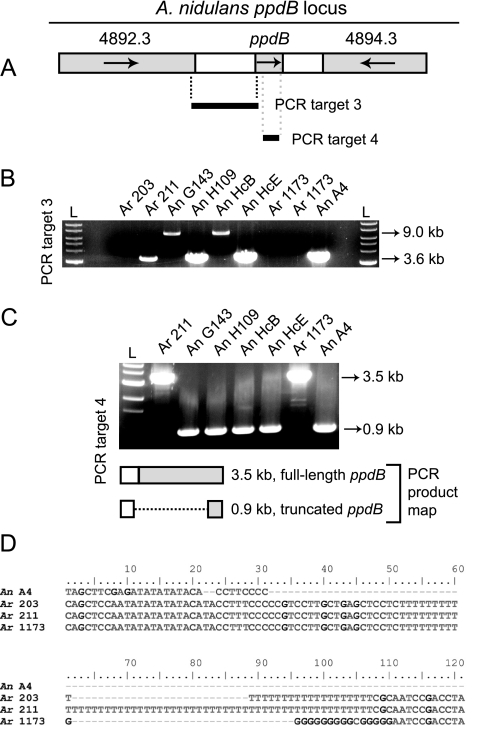
FIG. 6.
Comparison of the A. nidulans and A. rugulosus ppdB loci. (A) A schematic of the predicted A. nidulans ppdB locus is shown with its immediate flanking genes. Genes are gray, and intergenic regions are white. PCR was used to amplify two DNA fragments (target 3 and target 4) from wild A. nidulans isolates (An) and A. rugulosus isolates (Ar). (B) Target 3 is 3.6 kb for isolates from both species, except for A. nidulans G143 and HcB. Sequencing indicates that this is due to a transposon insertion in the 5′ ppdB-intergenic region of A. nidulans G143 (HcB was not sequenced). Although bands were not amplified for A. rugulosus 203 and A. rugulosus SRRC 1173, a different primer set amplified a band of similar size to one from A4 for the same approximate location (data not shown). (C) Target 4 is 3.5 kb for all A. rugulosus isolates and 0.9 kb for all A. nidulans isolates. A 3.5-kb PCR product was also amplified from A. rugulosus 203 with the same primer set (product not shown; the sequence is provided in the supplemental material). A PCR product map depicts how the 3.5-kb band is characteristic of a full-length ppdB and the A. nidulans ppdB truncation (dashed line) extends to the 5′ intergenic region. Differences in ORF location (gray shading in panels A and C) are due to differences in predicted ppdB start codon locations for the A. nidulans truncated ppdB and A. rugulosus ppdB. For panels B and C: L, 1-kb ladder; An A4, strain RTMH13.C5. (D) A variable polynucleotide repeat exists next to the A. nidulans ppdB truncation point in the three A. rugulosus isolates.
A. rugulosus encodes full-length versions of dclA and ppdB.
To determine whether the gene truncations also exist in the closely related species A. rugulosus (54), dclA and ppdB loci in this species were investigated. The close relationship between A. rugulosus and A. nidulans is illustrated by the fact that there is essentially no nucleotide polymorphism between their ITS sequences (Table 2) . PCR amplification of genomic fragments spanning the 3′ dclA-intergenic region and the ppdB coding region resulted in products that are longer than those observed in A. nidulans (Fig. 5B and 6C), suggesting that both A. rugulosus genes are full length. For example, a 6.1-kp product is amplified from the A. rugulosus 3′ dclA intergenic region instead of the 2.6-kp product amplified from the orthologous locus in A. nidulans (Fig. 5B), and a 3.5-kp product is amplified from the A. rugulosus ppdB coding region instead of the 0.9-kp product amplified from the orthologous locus in A. nidulans (Fig. 6C). Sequencing of the A. rugulosus products demonstrated that this species does indeed carry full-length versions of dclA and ppdB (see the supplemental material).
TABLE 2.
Nucleotide polymorphism analysis
| Locus | Alignment length (no. of nucleotides) | % Nucleotide polymorphisma in A. rugulosus strain:
|
||
|---|---|---|---|---|
| 203 | 211 | 1173 | ||
| ITS | 604 | 0.0 | 0.3 | 0 |
| Actin | 711 | 1.8 | 1.7 | 1.8 |
| ppdA/rsdA (Piwi) | 973 | 5.0 | 5.0 | 4.3 |
| ppdB (Piwi)* | 415 | 5.3 | 5.3 | 5.3 |
| dclB (helicase) | 1,010 | 6.4 | 6.4 | 6.6 |
| dclA (helicase) | 1,124 | ND | 10.8c | 11.4 |
| Intronsb | 509 | ND | 10.6 | 11.2 |
| Degenerate rrpA | 520 | ND | 15.0 | 15.6 |
| ppdB intergenic† | 421 | 17.1 | 17.6 | 16.6 |
The percent nucleotide polymorphism was calculated as the number of variable nucleotides and indels (between aligned sequences from A. nidulans and the specified A. rugulosus isolate) divided by the length of the alignment. The alignments are provided in the supplemental material. Predicted introns were removed from the actin, Dicer, and Argonaute fragments before the analysis. The A. nidulans ppdB PCR product from Fig. 6 (target 4) contains both coding sequences (*) and intergenic sequences (†); therefore, it was divided accordingly before alignment and analysis. ND, not determined.
A concatenated intron containing predicted intron sequences from the actin, Dicer, and Argonaute fragments was analyzed. The GT/AG consensus splice sites were removed before alignment.
The transposon insertion located in the A. rugulosus 211 dclA helicase domain was excluded from the alignment.
Comparative analysis between the A. rugulosus and A. nidulans sequences indicates that the A. nidulans dclA locus is missing three DNA fragments (approximately 3, 0.7, and 0.3 kb) and encodes an insertion of 0.6 kb in its the 3′ intergenic region (Fig. 5B). The insertion contains two short elements (50 to 100 bp) that are repeated approximately 15 times throughout the A. nidulans genome (see the supplemental material). The A. nidulans ppdB locus is characterized by a single major deletion of 2.6 kb that appears to have originated approximately 20 nucleotides upstream of a polynucleotide repeat of variable length and sequence in the three A. rugulosus isolates (Fig. 6D).
Although the A. rugulosus dclA and ppdB loci encode full-length genes; whether or not they produce functional proteins has not been determined. A. rugulosus 211 (dclA), however, is not likely to be functional because sequencing of a DNA fragment spanning its DEAD-like helicase domain (Fig. 5C, note longer-than-normal PCR product) identified an insertion similar to a transposase found in the A. oryzae genome (blastx, e−121, GI:92019814) (see the supplemental material). Thus, as in A. nidulans, the group M RNA silencing genes of A. rugulosus appears to be unusually susceptible to mutation.
Nucleotide polymorphism between A. nidulans and A. rugulosus loci.
Nucleotide polymorphism levels between several loci from A. nidulans and A. rugulosus isolates are compared in Table 2. The ITS sequences are essentially identical between all A. nidulans and A. rugulosus isolates, with only one to three variable nucleotides or indels found in an ITS clone from one A. nidulans isolate (HcE) and one A. rugulosus isolate (isolate 211) (see the supplemental material). A fragment of actin also has very little polymorphism between the species (1.7 to 1.8%). An alignment of six concatenated introns was found to have much more variability than the ITS region or the actin sequences, with a nucleotide polymorphism range of 10.6 to 11.2%. A fragment from the 5′ ppdB intergenic region also has a relatively high level of polymorphism at 16.6 to 17.6%. Similar to what has been reported for A. nidulans (30), the A. rugulosus rrpA locus encodes a degenerate rrpA (see the supplemental material) and, accordingly, a high level of polymorphism exists at the rrpA locus (15 to 15.6%).
With respect to the group M RNA silencing proteins, a fragment of dclA spanning the Dead Box helicase domain (Fig. 5C, target 2) has 10.8 to 11.4% nucleotide variability between A. nidulans and A. rugulosus species, whereas a fragment of ppdB has less polymorphism between the species (5.3%). A similar analysis for the group Q RNA silencing genes finds a polymorphism level of 4.3 to 5.0% between the Argonautes (rsdA/ppdA) and 6.4 to 6.6% between the Dicers (dclB). The range in nucleotide polymorphisms and how they may relate to dcl and ppd evolution are discussed below.
A. nidulans dclB is required for experimental RNA silencing.
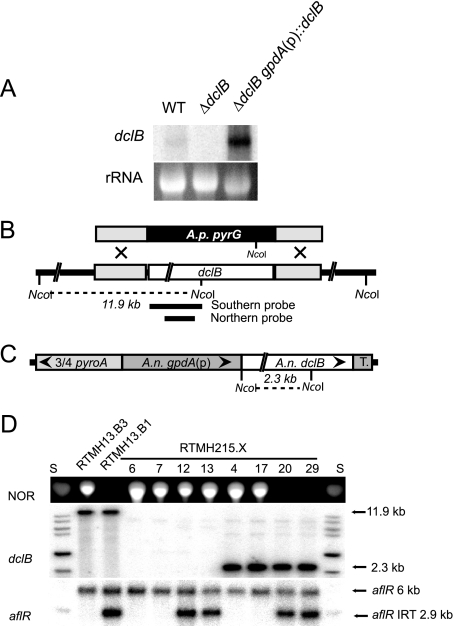
With the truncation of DclA, DclB is the only full-length Dicer found in the A. nidulans genome. Its deletion indicates that it is essential for IRT-RNA silencing, which is restored when the deletion allele is complemented by an ectopically expressed gpdA(p)::dclB transgene (Fig. 7D). Northern blotting revealed a single dclB transcript that is absent from the ΔdclB genetic background and transcribed at higher-than-normal levels when expressed from the ectopic gpdA(p)::dclB transgene (Fig. 7A).
FIG. 7.
A. nidulans dclB is required for IRT-RNA silencing. (A) Northern blotting identifies dclB transcript in total RNA from 48-h cultures. The ethidium bromide-stained 26S rRNA band is shown. Strains: WT, RTMH13.B3; ΔdclB, RTMH215.6; ΔdclB gpdA(p)::dclB, RTMH215.4. (B) Schematic representation of dclB replacement with pyrG. Gray boxes represent the dclB flanking sequences used in the deletion vector. The white box represents the predicted A. nidulans dclB ORF. (C) Schematic representation of dclB complementation by targeting a gpdA::dclB construct to the pyroA locus. (D) NOR analysis and Southern blotting (HindIII digest for dclB, NcoI-HindIII digests for aflR) results for control strains and recombinants from a cross between a ΔdclB transformant (TTMH158.1) and a transformant carrying the gpdA(p)::dclB transgene (TTMH160.3). Note that the absence of a band for dclB indicates the deletion genotype (see the probe template in panel B) and that the presence of two bands for aflR indicates the IRT genotype. Because normal levels of NOR are detected for the ΔdclB aflR(IRT) genotype, the data indicate that RNA silencing is dependent on dclB. S, NOR standard or 1-kb ladder.
A. nidulans lacking RNA silencing genes and RdRPs are phenotypically normal.
RdRPs have important roles in RNA silencing processes (see above). A. nidulans encodes two RdRPs, RrpB and RrpC, which is one less than is typically encoded by a filamentous ascomycete (30). To efficiently identify a role for any of the A. nidulans RNA silencing proteins in a physiological process, the deletion alleles for dclB, rsdA, rrpB (30), and rrpC (30) were combined into the same genetic background (referred to as ΔRNAi). Measurements of radial growth and spore production revealed no significant differences between A. nidulans wild-type strains and strains lacking the group Q RNA silencing genes and the RdRPs (Table 3 and the supplemental material). This demonstrates that RNA silencing genes are not essential for normal growth, conidiation, or ascosporogenesis.
At least some N. crassa RNA silencing genes are required for the sexual cycle (1, 40, 63), but the fact that A. nidulans RNA silencing mutants produce wild-type levels of ascospores (Table 3) indicates that RNA silencing genes are not required for A. nidulans meiosis. Furthermore, an experiment specifically designed to identify mating deficiencies revealed no differences in homozygous wild-type or ΔRNAi crosses, although a statistical favoring of ΔRNAi selfing was seen in the heterozygous wild-type-to-ΔRNAi crosses (Table 4). Overall, these data demonstrate that RNA silencing genes are not a fundamental requirement of mating or meiosis in ascomycete fungi.
TABLE 4.
Crossing assaya
| Cross | Genotypes | % Selfed cleistothecia ± SD | % Crossed cleistothecia ± SD |
|---|---|---|---|
| Expt 1 | |||
| A | ΔRNAi(y) × ΔRNAi(w) | 52.8 ± 9.7 (y), 13.8 ± 6.3 (w), 66.7 ± 8.3 (t) | 33.3 ± 8.3 |
| B | ΔRNAi(y) × WT(w) | 67.5 ± 3.5 (y), 25.8 ± 1.1 (w), 93.3 ± 4.7 (t) | 6.7 ± 4.7 |
| C | WT(y) × ΔRNAi(w) | 26.6 ± 7.0 (y), 67.5 ± 8.2 (w), 94.2 ± 1.2 (t) | 5.9 ± 1.2 |
| D | WT(y) × WT(w) | 33.9 ± 3.5 (y), 31.7 ± 6.7 (w), 65.6 ± 3.5 (t) | 34.4 ± 3.4 |
| Expt 2 | |||
| E | ΔRNAi(y) × ΔRNAi(w) | 18.8 ± 2.5 (y), 46.1 ± 3.5 (w), 65.0 ± 3.3 (t) | 35.0 ± 3.3 |
| F | ΔRNAi(y) × WT(w) | 72.5 ± 1.2 (y), 22.5 ± 3.5 (w), 95.0 ± 2.4 (t) | 4.2 ± 3.5 |
| G | WT(y) × ΔRNAi(w) | 35.0 ± 4.7 (y), 60.0 ± 9.4 (w), 95.0 ± 4.7 (t) | 5.0 ± 4.7 |
| H | WT(y) × WT(w) | 32.8 ± 6.3 (y), 30.6 ± 1.0 (w), 63.3 ± 7.3 (t) | 36.7 ± 7.3 |
Wild-type (WT) strains are wild type at all loci except yA or wA. ΔRNAi strains are quadruple knockouts (ΔdclB, ΔrsdA, Δrrp, and ΔrrpC). “(y)” refers to yellow-conidia-progeny-producing cleistothecia, “(w)” refers to white-conidia-progeny-producing cleistothecia, and “(t)” refers to the total number of selfed cleistothecia. Sixty cleistothecia were scored for each replica plate, and three replica plates were analyzed for crosses A, D, E, and H, while two replica plates were analyzed for crosses B, C, F, and G. Experiment 1 strains: ΔRNAi (y), RTMH227.29; ΔRNAi (w), RTMH227.67; WT (y), RTMH229.37; WT (w), RTMH229-19. Experiment 2 strains: ΔRNAi (y), RTMH227.47; ΔRNAi (w), RTMH227.68; WT (y), RTMH229-25; WT (w), RTMH233.C.
DISCUSSION
The evolutionary forces driving eukaryotic RNA silencing gene change are unclear. The data presented here indicate that Aspergillus fungi are well suited for elucidating these forces. The model fungus A. nidulans is especially useful since it has recently experienced RNA silencing gene truncation in two widely conserved genes: dclA, a Dicer encoding gene, and ppdB, an Argonaute encoding gene. These two genes are predicted to be part of the same paralogous RNA silencing gene group (M) commonly found in filamentous ascomycetes. Although the truncated genes are transcribed, they are not required for experimental RNA silencing, growth, or developmental processes (at least under standard culture conditions). Thus, the affect of dclA and ppdB truncation on A. nidulans biology is unknown. However, their truncation did leave A. nidulans with only a single Dicer and a single Argonaute to mediate all RNA silencing based processes. These are dclB and rsdA, members of the other paralogous RNA silencing gene group (Q) commonly found in filamentous ascomycetes. These proteins are required for experimental RNA silencing but are not required for normal growth and development (at least under standard culture conditions). This latter finding contrasts significantly with a requirement for functional RNA silencing in growth and/or development in higher eukaryotes and hints that an elaborate miRNA-like gene-regulation mechanism does not exist in A. nidulans. This hypothesis is consistent with the current state of fungal RNA silencing research, that is, miRNA has not yet been discovered in the fungal kingdom.
The population analysis performed in the present study suggests that the A. nidulans dclA and ppdB truncations are fixed at the species level. A caveat to this finding is that all of the analyzed A. nidulans isolates originated in the United Kingdom. Thus, the status of dclA and ppdB in A. nidulans isolates from other parts of the world is unknown. Regardless, it is still surprising that the dclA and ppdB truncations were selected for in the United Kingdom isolates, given that dclA and ppdB orthologs are conserved in all other filamentous-ascomycete genomes investigated. One of several simple hypotheses to account for this phenomenon is that the gene truncation events somehow correlate—or previously correlated—with an increase in fitness. The data presented here suggest that such a hypothetical fitness gain does not influence RNA silencing, growth, or development. Another hypothesis is that group M RNA silencing has lost relevance in A. nidulans, and thus the truncations represent the natural degeneration of defunct genes.
Our data indicate that the A. nidulans dclA and ppdB truncations must have occurred sometime after divergence from an ancestor shared with its close relative A. rugulosus. Although a theoretical divergence date for these species has not been determined, it appears to be recent since there is a high level of nucleotide identity shared between some A. nidulans and A. rugulosus loci. For example, their ITS sequences are nearly identical (Table 2), suggesting that they could even be classified as the same species. The fact that A. nidulans and A. rugulosus both encode a degenerate rrpA locus indicates that this locus can be used to estimate the amount of polymorphism that occurs in loci experiencing little or no selection. A relatively high level of polymorphism is found at this locus (15.0 to 15.6%) and at another probable low-selection locus, the 5′ ppdB-intergenic region (16.6 to 17.6%). In comparison, dclA and ppdB loci have lower levels of polymorphism, with ppdB having less than dclA. A similar analysis for the group Q RNA silencing proteins indicates that Dicers are not necessarily more likely than Argonautes to acquire polymorphic residues; thus, the lower polymorphism level of ppdB may be an important clue toward understanding the order of the truncation events. A simple explanation is that A. nidulans dclA was truncated before ppdB and has thus had more time to accumulate mutations. However, this would suggest that selection to maintain ppdB was not lost with dclA truncation, possibly because its product had a function that was independent of DclA.
The consequences of group M Dicer and Argonaute gene truncation on A. nidulans biology are unknown. If Aspergillus dclA and ppdB control repetitive DNA, one might predict that the A. nidulans dclA and ppdB truncations would result in its increase. However, recent estimates indicate that levels of repetitive DNA are not very different between A. nidulans and A. fumigatus, with each species predicted to contain 3.0 and 2.9%, respectively (24). Despite this similarity, it seems possible that the dclA and ppdB truncations could have removed some restraints on repetitive DNA in A. nidulans, perhaps leading to increased mobility and a higher turnover rate, if not a detectable increase in overall amount.
It is perhaps pertinent to note the correlation between group M RNA silencing gene truncations/disruptions and repetitive DNA. We have found such elements 3′ of dclA (Fig. 5B), 5′ of ppdB (Fig. 6B), and in the middle of the A. rugulosus 211 dclA helicase domain (Fig. 5C). In addition, a polynucleotide repeat appears to have contributed to A. nidulans ppdB truncation (inferred from the position of this repeat in A. rugulosus, Fig. 6D). It is unclear whether these findings are simply the result of the natural degeneration of defunct genes, or whether they are related to the antagonistic relationship between RNA silencing and selfish genetic elements. If the former is correct, it is at least ironic that the elements which RNA silencing genes are thought to control are contributing to RNA silencing gene demise in A. nidulans.
With respect to its intact RNA silencing machinery, A. nidulans does not require dclB, rsdA, rrpB, or rrpC for growth and developmental processes under standard culture conditions (nonstandard conditions have not been thoroughly investigated). This is somewhat consistent with the slight nature of the phenotypic abnormalities reported for RNA silencing mutants in other filamentous fungi (M. oryzae and M. circinelloides) (35, 49) but contrasts significantly with the requirement for some RNA silencing genes for sexual reproduction in N. crassa (1, 40, 63).
A peculiar finding concerning A. nidulans development came from the A. nidulans crossing assays. In heterozygous crosses between wild-type and ΔRNAi, the level of crossed cleistothecia was significantly lower than in homozygous crosses (Table 4). The fact that crossed-cleistothecium levels dropped only in the heterozygous crosses suggests that absence of RNA silencing in one parent is not the basis of the phenomenon. A possibility that has not yet been investigated is that the pyrG (2 to 3) and metG (0 to 1) selectable markers in the ΔRNAi genotype (due to the gene replacements) are a contributing factor. Most importantly, however, the results of these experiments clearly demonstrate that RNA silencing genes are not required for meiosis or crossing in A. nidulans.
The lack of a detected role for the A. nidulans group Q Dicer and Argonaute and the A. nidulans RdRPs in growth and development suggests that they may not be involved in “housekeeping” functions (as defined by reference 51) but may be used for an auxiliary process such as defense against viruses. Accordingly, a companion study reports that Aspergillus viruses are targets and suppressors of A. nidulans RNA silencing (28a). Other possible roles for the intact A. nidulans RNA silencing machinery include transposon control (as has been demonstrated for N. crassa [50]) and chromatin regulation (as has been demonstrated for S. pombe [for a review, see reference 44]). Future experiments will examine these specific possibilities.
For fungi in general, RNA silencing gene evolution appears to be more complex than in any other type of eukaryote, and the continued study of Aspergillus species should help elucidate its driving forces. While the A. nidulans dclA and ppdB truncations were characterized here, the same genes have been conserved or duplicated by other aspergilli. These species are thus prime candidates for comparative studies that should shed light on the evolutionary significance of RNA silencing gene gain and loss in eukaryotes.
Supplementary Material
Acknowledgments
We gratefully acknowledge David Geiser (Penn State University) for the wild Aspergillus isolates.
This project was supported by a College of Agricultural and Life Sciences, Wisconsin Distinguished Graduate Fellowship to T.M.H.; by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service (2005-35201-15350); and by an NSF grant (MCB-0236393) to N.P.K. The work at Orsay was supported by the Université Paris-Sud, the CNRS, and the Institut Universitaire de France. Y.R.-D. was supported by a predoctoral fellowship of the Ministère de l'Education Supérieure et la Recherche (France) and CONACYT (Mexico).
Footnotes
Published ahead of print on 7 December 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Alexander, W. G., N. B. Raju, H. Xiao, T. M. Hammond, T. D. Perdue, R. L. Metzenberg, P. J. Pukkila, and P. K. T. Shiu. 2007. DCL-1 colocalizes with other components of the MSUD machinery and is required for silencing. Fungal Genet. Biol. doi: 10.1016/j.fgb. 2007.10.006. [DOI] [PubMed]
- 2.Baumberger, N., and D. C. Baulcombe. 2005. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 10211928-11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beclin, C., S. Boutet, P. Waterhouse, and H. Vaucheret. 2002. A branched pathway for transgene-induced RNA silencing in plants. Curr. Biol. 12684-688. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409363-366. [DOI] [PubMed] [Google Scholar]
- 5.Borkovich, K. A., L. A. Alex, O. Yarden, M. Freitag, G. E. Turner, N. D. Read, S. Seiler, D. Bell-Pedersen, J. Paietta, N. Plesofsky, M. Plamann, M. Goodrich-Tanrikulu, U. Schulte, G. Mannhaupt, F. E. Nargang, A. Radford, C. Selitrennikoff, J. E. Galagan, J. C. Dunlap, J. J. Loros, D. Catcheside, H. Inoue, R. Aramayo, M. Polymenis, E. U. Selker, M. S. Sachs, G. A. Marzluf, I. Paulsen, R. Davis, D. J. Ebbole, A. Zelter, E. R. Kalkman, R. O'Rourke, F. Bowring, J. Yeadon, C. Ishii, K. Suzuki, W. Sakai, and R. Pratt. 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 681-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruggeman, J., A. J. Debets, P. J. Wijngaarden, J. A. deVisser, and R. F. Hoekstra. 2003. Sex slows down the accumulation of deleterious mutations in the homothallic fungus Aspergillus nidulans. Genetics 164479-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butchko, R. A., T. H. Adams, and N. P. Keller. 1999. Aspergillus nidulans mutants defective in stc gene cluster regulation. Genetics 153715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calvo, A. M., J. Bok, W. Brooks, and N. P. Keller. 2004. veA is required for toxin and sclerotial production in Aspergillus parasiticus. Appl. Environ. Microbiol. 704733-4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carmell, M. A., Z. Xuan, M. Q. Zhang, and G. J. Hannon. 2002. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 162733-2742. [DOI] [PubMed] [Google Scholar]
- 10.Carmichael, J. B., P. Provost, K. Ekwall, and T. C. Hobman. 2004. ago1 and dcr1, two core components of the RNA interference pathway, functionally diverge from rdp1 in regulating cell cycle events in Schizosaccharomyces pombe. Mol. Biol. Cell 151425-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Catalanotto, C., G. Azzalin, G. Macino, and C. Cogoni. 2002. Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes. Dev. 16790-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Catalanotto, C., M. Pallotta, P. ReFalo, M. S. Sachs, L. Vayssie, G. Macino, and C. Cogoni. 2004. Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol. Cell. Biol. 242536-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerutti, H., and J. A. Casas-Mollano. 2006. On the origin and functions of RNA-mediated silencing: from protists to man. Curr. Genet. 5081-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerutti, L., N. Mian, and A. Bateman. 2000. Domains in gene silencing and cell differentiation proteins: the novel PAZ domain and redefinition of the Piwi domain. Trends Biochem. Sci. 25481-482. [DOI] [PubMed] [Google Scholar]
- 15.Chicas, A., C. Cogoni, and G. Macino. 2004. RNAi-dependent and RNAi-independent mechanisms contribute to the silencing of RIPed sequences in Neurospora crassa. Nucleic Acids Res. 324237-4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cogoni, C., J. T. Irelan, M. Schumacher, T. J. Schmidhauser, E. U. Selker, and G. Macino. 1996. Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA-DNA interactions or DNA methylation. EMBO J. 153153-3163. [PMC free article] [PubMed] [Google Scholar]
- 17.Cogoni, C., and G. Macino. 1999. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399166-169. [DOI] [PubMed] [Google Scholar]
- 18.Cogoni, C., and G. Macino. 1997. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc. Natl. Acad. Sci. USA 9410233-10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cogoni, C., N. Romano, and G. Macino. 1994. Suppression of gene expression by homologous transgenes. Antonie van Leeuwenhoek 65205-209. [DOI] [PubMed] [Google Scholar]
- 20.Dalmay, T., A. Hamilton, S. Rudd, S. Angell, and D. C. Baulcombe. 2000. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101543-553. [DOI] [PubMed] [Google Scholar]
- 21.Edgar, R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elbashir, S. M., W. Lendeckel, and T. Tuschl. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galagan, J. E., S. E. Calvo, K. A. Borkovich, E. U. Selker, N. D. Read, D. Jaffe, W. FitzHugh, L. J. Ma, S. Smirnov, S. Purcell, B. Rehman, T. Elkins, R. Engels, S. Wang, C. B. Nielsen, J. Butler, M. Endrizzi, D. Qui, P. Ianakiev, D. Bell-Pedersen, M. A. Nelson, M. Werner-Washburne, C. P. Selitrennikoff, J. A. Kinsey, E. L. Braun, A. Zelter, U. Schulte, G. O. Kothe, G. Jedd, W. Mewes, C. Staben, E. Marcotte, D. Greenberg, A. Roy, K. Foley, J. Naylor, N. Stange-Thomann, R. Barrett, S. Gnerre, M. Kamal, M. Kamvysselis, E. Mauceli, C. Bielke, S. Rudd, D. Frishman, S. Krystofova, C. Rasmussen, R. L. Metzenberg, D. D. Perkins, S. Kroken, C. Cogoni, G. Macino, D. Catcheside, W. Li, R. J. Pratt, S. A. Osmani, C. P. DeSouza, L. Glass, M. J. Orbach, J. A. Berglund, R. Voelker, O. Yarden, M. Plamann, S. Seiler, J. Dunlap, A. Radford, R. Aramayo, D. O. Natvig, L. A. Alex, G. Mannhaupt, D. J. Ebbole, M. Freitag, I. Paulsen, M. S. Sachs, E. S. Lander, C. Nusbaum, and B. Birren. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422859-868. [DOI] [PubMed] [Google Scholar]
- 24.Galagan, J. E., S. E. Calvo, C. Cuomo, L. J. Ma, J. R. Wortman, S. Batzoglou, S. I. Lee, M. Basturkmen, C. C. Spevak, J. Clutterbuck, V. Kapitonov, J. Jurka, C. Scazzocchio, M. Farman, J. Butler, S. Purcell, S. Harris, G. H. Braus, O. Draht, S. Busch, C. D'Enfert, C. Bouchier, G. H. Goldman, D. Bell-Pedersen, S. Griffiths-Jones, J. H. Doonan, J. Yu, K. Vienken, A. Pain, M. Freitag, E. U. Selker, D. B. Archer, M. A. Penalva, B. R. Oakley, M. Momany, T. Tanaka, T. Kumagai, K. Asai, M. Machida, W. C. Nierman, D. W. Denning, M. Caddick, M. Hynes, M. Paoletti, R. Fischer, B. Miller, P. Dyer, M. S. Sachs, S. A. Osmani, and B. W. Birren. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 4381105-1115. [DOI] [PubMed] [Google Scholar]
- 25.Geiser, D. M., M. L. Arnold, and W. E. Timberlake. 1994. Sexual origins of British Aspergillus nidulans isolates. Proc. Natl. Acad. Sci. USA 912349-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giraldez, A. J., R. M. Cinalli, M. E. Glasner, A. J. Enright, J. M. Thomson, S. Baskerville, S. M. Hammond, D. P. Bartel, and A. F. Schier. 2005. MicroRNAs regulate brain morphogenesis in zebra fish. Science 308833-838. [DOI] [PubMed] [Google Scholar]
- 27.Grishok, A., A. E. Pasquinelli, D. Conte, N. Li, S. Parrish, I. Ha, D. L. Baillie, A. Fire, G. Ruvkun, and C. C. Mello. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control Caenorhabditis elegans developmental timing. Cell 10623-34. [DOI] [PubMed] [Google Scholar]
- 28.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 4195-98. [Google Scholar]
- 28a.Hammond, T. M., M. D. Andrewski, M. J. Roossinck, and N. P. Keller. 2008. Aspergillus mycoviruses are targets and suppressors of RNA silencing. Eukaryot. Cell 7350-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammond, S. M., E. Bernstein, D. Beach, and G. J. Hannon. 2000. An RNA-directed nuclease mediates posttranscriptional gene silencing in Drosophila cells. Nature 404293-296. [DOI] [PubMed] [Google Scholar]
- 30.Hammond, T. M., and N. P. Keller. 2005. RNA silencing in Aspergillus nidulans is independent of RNA-dependent RNA polymerases. Genetics 169607-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He, L., and G. J. Hannon. 2004. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5522-531. [DOI] [PubMed] [Google Scholar]
- 32.Hutvagner, G., J. McLachlan, A. E. Pasquinelli, E. Balint, T. Tuschl, and P. D. Zamore. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293834-838. [DOI] [PubMed] [Google Scholar]
- 33.Hutvagner, G., and P. D. Zamore. 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 2972056-2060. [DOI] [PubMed] [Google Scholar]
- 34.Jung, M. K., Y. Ovechkina, N. Prigozhina, C. E. Oakley, and B. R. Oakley. 2000. The use of β-d-glucanase as a substitute for Novozyme 234 in immunofluorescence and protoplasting. Fungal Gent. Newsl. 4765-66. [Google Scholar]
- 35.Kadotani, N., H. Nakayashiki, Y. Tosa, and S. Mayama. 2004. One of the two Dicer-like proteins in the filamentous fungi Magnaporthe oryzae genome is responsible for hairpin RNA-triggered RNA silencing and related small interfering RNA accumulation. J. Biol. Chem. 27944467-44474. [DOI] [PubMed] [Google Scholar]
- 36.Kanellopoulou, C., S. A. Muljo, A. L. Kung, S. Ganesan, R. Drapkin, T. Jenuwein, D. M. Livingston, and K. Rajewsky. 2005. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 19489-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ketting, R. F., S. E. Fischer, E. Bernstein, T. Sijen, G. J. Hannon, and R. H. Plasterk. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in Caenorhabditis elegans. Genes Dev. 152654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klich, M., C. Mendoza, E. Mullaney, N. Keller, and J. W. Bennett. 2001. A new sterigmatocystin-producing Emericella variant from agricultural desert soils. Syst. Appl. Microbiol. 24131-138. [DOI] [PubMed] [Google Scholar]
- 39.Kumar, S., K. Tamura, and M. Nei. 1994. MEGA: molecular evolutionary genetics analysis software for microcomputers. Comput. Appl. Biosci. 10189-191. [DOI] [PubMed] [Google Scholar]
- 40.Lee, D. W., R. J. Pratt, M. McLaughlin, and R. Aramayo. 2003. An Argonaute-like protein is required for meiotic silencing. Genetics 164821-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, H., T. R. Cottrell, L. M. Pierini, W. E. Goldman, and T. L. Doering. 2002. RNA interference in the pathogenic fungus Cryptococcus neoformans. Genetics 160463-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu, J., M. A. Carmell, F. V. Rivas, C. G. Marsden, J. M. Thomson, J. J. Song, S. M. Hammond, L. Joshua-Tor, and G. J. Hannon. 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 3051437-1441. [DOI] [PubMed] [Google Scholar]
- 43.Martens, H., J. Novotny, J. Oberstrass, T. L. Steck, P. Postlethwait, and W. Nellen. 2002. RNAi in Dictyostelium: the role of RNA-directed RNA polymerases and double-stranded RNase. Mol. Biol. Cell 13445-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martienssen, R. A., M. Zaratiegui, and D. B. Goto. 2005. RNA interference and heterochromatin in the fission yeast Schizosaccharomyces pombe. Trends Genet. 21450-456. [DOI] [PubMed] [Google Scholar]
- 45.Martinez, J., A. Patkaniowska, H. Urlaub, R. Luhrmann, and T. Tuschl. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110563-574. [DOI] [PubMed] [Google Scholar]
- 46.Meister, G., and T. Tuschl. 2004. Mechanisms of gene silencing by double-stranded RNA. Nature 431343-349. [DOI] [PubMed] [Google Scholar]
- 47.Mourelatos, Z., J. Dostie, S. Paushkin, A. Sharma, B. Charroux, L. Abel, J. Rappsilber, M. Mann, and G. Dreyfuss. 2002. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 16720-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakayashiki, H., N. Kadotani, and S. Mayama. 2006. Evolution and diversification of RNA silencing proteins in fungi. J. Mol. Evol. 63127-135. [DOI] [PubMed] [Google Scholar]
- 49.Nicolas, F. E., J. P. de Haro, S. Torres-Martinez, and R. M. Ruiz-Vazquez. 2007. Mutants defective in a Mucor circinelloides dicer-like gene are not compromised in siRNA silencing but display developmental defects. Fungal Genet. Biol. 44504-516. [DOI] [PubMed] [Google Scholar]
- 50.Nolan, T., L. Braccini, G. Azzalin, A. De Toni, G. Macino, and C. Cogoni. 2005. The posttranscriptional gene silencing machinery functions independently of DNA methylation to repress a LINE1-like retrotransposon in Neurospora crassa. Nucleic Acids Res. 331564-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Obbard, D. J., F. M. Jiggins, D. L. Halligan, and T. J. Little. 2006. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr. Biol. 16580-585. [DOI] [PubMed] [Google Scholar]
- 52.Olsen, P. H., and V. Ambros. 1999. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216671-680. [DOI] [PubMed] [Google Scholar]
- 53.Park, W., J. Li, R. Song, J. Messing, and X. Chen. 2002. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 121484-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peterson, S. W. 2000. Phylogenetic relationships in Aspergillus based on rDNA sequence analysis, p. 323-356. In R. A. Samson and J. I. Pitt (ed.), Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Harwood Academic Publishers, New York, NY.
- 55.Piccin, A., A. Salameh, C. Benna, F. Sandrelli, G. Mazzotta, M. Zordan, E. Rosato, C. P. Kyriacou, and R. Costa. 2001. Efficient and heritable functional knock-out of an adult phenotype in Drosophila using a GAL4-driven hairpin RNA incorporating a heterologous spacer. Nucleic Acids Res. 29E55-E56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reinhart, B. J., E. G. Weinstein, M. W. Rhoades, B. Bartel, and D. P. Bartel. 2002. MicroRNAs in plants. Genes Dev. 161616-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rivas, F. V., N. H. Tolia, J. J. Song, J. P. Aragon, J. Liu, G. J. Hannon, and L. Joshua-Tor. 2005. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 12340-349. [DOI] [PubMed] [Google Scholar]
- 58.Romano, N., and G. Macino. 1992. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 63343-3353. [DOI] [PubMed] [Google Scholar]
- 59.Segers, G. C., X. Zhang, F. Deng, Q. Sun, and D. L. Nuss. 2007. Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc. Natl. Acad. Sci. USA 10412902-12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seggerson, K., L. Tang, and E. G. Moss. 2002. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev. Biol. 243215-225. [DOI] [PubMed] [Google Scholar]
- 61.Shimizu, K., and N. P. Keller. 2001. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shiu, P. K., and R. L. Metzenberg. 2002. Meiotic silencing by unpaired DNA: properties, regulation and suppression. Genetics 1611483-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shiu, P. K., N. B. Raju, D. Zickler, and R. L. Metzenberg. 2001. Meiotic silencing by unpaired DNA. Cell 107905-916. [DOI] [PubMed] [Google Scholar]
- 64.Sijen, T., J. Fleenor, F. Simmer, K. L. Thijssen, S. Parrish, L. Timmons, R. H. Plasterk, and A. Fire. 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107465-476. [DOI] [PubMed] [Google Scholar]
- 65.Smardon, A., J. M. Spoerke, S. C. Stacey, M. E. Klein, N. Mackin, and E. M. Maine. 2000. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 10169-178. [DOI] [PubMed] [Google Scholar]
- 66.Song, J. J., S. K. Smith, G. J. Hannon, and L. Joshua-Tor. 2004. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 3051434-1437. [DOI] [PubMed] [Google Scholar]
- 67.Sugiyama, T., H. Cam, A. Verdel, D. Moazed, and S. I. Grewal. 2005. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc. Natl. Acad. Sci. USA 102152-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsitsigiannis, D. I., R. Zarnowski, and N. P. Keller. 2004. The lipid body protein, PpoA, coordinates sexual and asexual sporulation in Aspergillus nidulans. J. Biol. Chem. 27911344-11353. [DOI] [PubMed] [Google Scholar]
- 70.Vaucheret, H. 2006. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 20759-771. [DOI] [PubMed] [Google Scholar]
- 71.Verdel, A., S. Jia, S. Gerber, T. Sugiyama, S. Gygi, S. I. Grewal, and D. Moazed. 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303672-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Volpe, T., V. Schramke, G. L. Hamilton, S. A. White, G. Teng, R. A. Martienssen, and R. C. Allshire. 2003. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 11137-146. [DOI] [PubMed] [Google Scholar]
- 73.Wesley, S. V., C. A. Helliwell, N. A. Smith, M. B. Wang, D. T. Rouse, Q. Liu, P. S. Gooding, S. P. Singh, D. Abbott, P. A. Stoutjesdijk, S. P. Robinson, A. P. Gleave, A. G. Green, and P. M. Waterhouse. 2001. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27581-590. [DOI] [PubMed] [Google Scholar]
- 74.Wienholds, E., M. J. Koudijs, F. J. van Eeden, E. Cuppen, and R. H. Plasterk. 2003. The microRNA-producing enzyme Dicer1 is essential for zebra fish development. Nat. Genet. 35217-218. [DOI] [PubMed] [Google Scholar]
- 75.Yan, K. S., S. Yan, A. Farooq, A. Han, L. Zeng, and M. M. Zhou. 2003. Structure and conserved RNA binding of the PAZ domain. Nature 426468-474. [DOI] [PubMed] [Google Scholar]
- 76.Yu, J. H., and T. H. Adams. 1999. Filamentous fungi, p. 417-434. In A. L. Demain and J. E. Davies (ed.), Manual of industrial microbiology and biotechnology. ASM Press, Washington, DC.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.