Abstract
The addition of the poly(A) tail to the ends of eukaryotic mRNAs is catalyzed by poly(A) polymerase (PAP). PAP activity is known to be highly regulated, for example, by alternative splicing and phosphorylation. In this study we show that the small ubiquitin-like modifier (SUMO) plays multiple roles in regulating PAP function. Our discovery of SUMO-conjugated PAP began with the observation of a striking pattern of abundant higher-molecular-weight forms of PAP in certain mouse tissues and cell lines. PAP constitutes an unusual SUMO substrate in that, despite the absence of any consensus sumoylation sites, PAP interacts very strongly with the SUMO E2 enzyme ubc9 and can be extensively sumoylated both in vitro and in vivo. Six sites of sumoylation in PAP were identified, with two overlapping one of two nuclear localization signals (NLS). Strikingly, mutation of the two lysines at the NLS to arginines, or coexpression of a SUMO protease with wild-type PAP, caused PAP to be localized to the cytoplasm, demonstrating that sumoylation is required to facilitate PAP nuclear localization. Sumoylation also contributes to PAP stability, as down-regulation of sumoylation led to decreases in PAP levels. Finally, the activity of purified PAP was shown to be inhibited by in vitro sumoylation. Our study thus shows that SUMO regulates PAP in numerous distinct ways and is integral to normal PAP function.
[Keywords: PAP, sumoylation, polyadenylation, ubc9, localization]
Formation of nearly all eukaryotic mRNA 3′ ends involves endonucleolytic cleavage followed by the addition of a poly(A) tail. The poly(A) tail is important for mRNA stability, transport into the cytoplasm, and efficient translation initiation (Huang and Carmichael 1996; Preiss et al. 1998). Polyadenylation of mRNA precursors is carried out in the cell nucleus by a complex set of factors (for review, see Colgan and Manley 1997; Minvielle-Sebastia and Keller 1999; Zhao et al. 1999). These include cleavage polyadenylation-specific complex (CPSF), cleavage stimulatory factor complex (CstF), cleavage factors I and II (CF1, CFII), symplekin, and poly(A) polymerase (PAP). PAP is a single-subunit, template-independent polymerase belonging to the large superfamily of nucleotidyl transferases (Raabe et al. 1991; Martin and Keller 1996). All eukaryotes were initially thought to possess a single PAP-encoding gene. While it is now known that most eukaryotes contain additional genes encoding PAP-related enzymes (Kashiwabara et al. 2000; Topalian et al. 2001; Kwak et al. 2004), the initially characterized PAP appears to play the predominant role in polyadenylation of mRNA precursors.
The functional domains of both mammalian and yeast PAP have been studied extensively (Raabe et al. 1994; Zhelkovsky et al. 1995; Martin and Keller 1996). The N-terminal domain, highly conserved in mammals and yeast, contains the three aspartate residues essential for catalytic activity. An RNA-binding domain overlaps one of two bipartite nuclear localization signals (NLSs). The C-terminal domain, while dispensable for activity in vitro, constitutes a regulatory domain in vertebrate PAP that is absent in yeast PAP. In this domain, the two NLSs surround a serine/threonine-rich region that contains multiple cyclin-dependent kinase (cdk) phosphorylation sites. The extreme C-terminal 20 amino acids are involved in interactions with several proteins, including U1A, U2AF65, a 14–3–3 protein, and the 25-kDa subunit of CFI (Vagner et al. 2000; Ko and Gunderson 2002; Kim and Lee 2001; Kim et al. 2003). Along with other polyadenylation factors, PAP is localized almost exclusively to the nucleus (Raabe et al. 1994; Schul et al. 1998), although minimal cytoplasmic localization has been reported (Thuresson et al. 1994; Kyriakopoulou et al. 2001). Both bipartite NLSs are required for efficient nuclear localization.
PAP regulation occurs at both post-transcriptional as well as post-translational levels. At the post-transcriptional level, PAP is subject to a complex pattern of alternative splicing. Although not well understood, this is thought to provide both a form of autoregulation and a mechanism to modulate the properties of the C-terminal domain (Zhao and Manley 1996). At the post-translational level, regulation by phosphorylation has been the subject of extensive study. Phosphorylated forms of PAP are evident throughout the cell cycle; however, during mitosis, PAP is hyperphosphorylated by cdc2–cyclinB, resulting in repression of PAP activity (Colgan et al. 1996, 1998). This regulation is necessary for proper cell growth, as DT40 cells harboring a cdk site mutant PAP shows impaired growth features (Zhao and Manley 1998). The HIV-1 Vpr accessory protein inactivates the cdc2/cyclin B complex and leads to hypophosphorylation of PAP, which in turn leads to PAP hyperactivity and contributes to HIV pathogenesis (Mouland et al. 2002). The 14–3–3ε protein interacts with the C-terminal region of PAP in a phosphorylation-dependent manner, inhibiting PAP activity and modulating subcellular localization (Kim et al. 2003). PAP is also subject to acetylation in its C-terminal region. Acetylation was shown to inhibit nuclear localization as well as interaction with the CFIm 25-kDa subunit, which is also subject to acetylation (Shimazu et al. 2007). Yeast PAP is also phosphorylated, but unlike mammalian PAP, this occurs preferentially during S phase. A ubiquitinated form has also been detected, although the significance of both modifications is not known (Mizrahi and Moore 2000).
Modification of proteins by the small ubiquitin-like modifier SUMO affects numerous cellular pathways. Sumoylation occurs in a manner mechanistically similar to ubiquitination, using a heterodimeric E1 activating enzyme (SAE1/2 in mammals or uba2/aos1 in yeast), a single E2 (ubc9), and several E3 ligases. SUMO proteases function to process SUMO into mature forms and to remove SUMO from conjugates (Johnson 2004; Hay 2005). Of the three characterized SUMO isoforms, SUMO-1 shares 45% similarity with SUMO-2 and SUMO-3, which are 96% similar and thought to be functionally equivalent. An important difference between SUMO-1 and SUMO-2/3 is the ability of SUMO-2/3, but not SUMO-1, to form polymeric chains (Saitoh and Hinchey 2000; Tatham et al. 2001).
SUMO can have varied effects on the function of the protein it modifies. For example, sumoylation appears to increase the stability of many substrates. In some cases, this occurs by competing with and blocking ubiquitination at the same lysine residue (Desterro et al. 1998; Lin et al. 2003). Sumoylation can also alter subcellular localization of target proteins (for review, see Pichler and Melchior 2002; Gill 2004). For example, SP100, Daxx, and thymine DNA glycosylase are targeted to PML bodies by virtue of sumoylation (Seeler and Dejean 2001). Sumoylation is also associated with the nucleocytoplasmic transport and/or nuclear accumulation of several proteins (Endter et al. 2001; Huang et al. 2003; Chen et al. 2006). SUMO has been shown to act both by enhancing nuclear import and by promoting nuclear retention through inhibiting nuclear export. SUMO can also localize proteins to the cytoplasm. For example, the cytoplasmic localization of RanGAP1 is controlled by SUMO modification, as SUMO-modified RanGAP1 interacts with the nucleoporin RanBP2 at the cytoplasmic fibrils of the NPC (Matunis et al. 1998).
SUMO has been implicated in several cellular processes, most prominent of which are transcription, DNA replication and repair, sister chromatid cohesion, and signal transduction. This list is growing with the discovery of novel targets through proteomic analysis of sumoylated proteins in both yeast and mammalian cells. Some of these studies have identified sumoylation targets as possibly involved in RNA processing (Panse et al. 2004; Wohlschlegel et al. 2004), although this has not been established. Other studies have reported direct sumoylation of two RNA-binding proteins, hnRNPs C and M, the RNA-editing enzyme ADAR1, and the polyadenylation factors symplekin and CPSF-73 (Li et al. 2004; Vassileva and Matunis 2004; Desterro et al. 2005; Vethantham et al. 2007). An interaction between yeast PAP and uba2, the SUMO E1, has also been reported (del Olmo et al. 1997). Depletion of uba2 caused an increase in polyadenylation activity, although it is unknown if PAP was in fact sumoylated.
In this study, we analyzed sumoylation of mammalian PAP. Unexpectedly, sumoylated PAP was found to be extremely abundant in certain mouse tissues and cell lines. SUMO modification of PAP was confirmed by in vitro assays and cotransfection experiments, and the sites of modification were mapped principally to the C-terminal regulatory domain. Sumoylation was found to have multiple effects on PAP function. Most strikingly, nuclear localization of PAP was found to be dependent on sumoylation. Interference with PAP sumoylation also resulted in protein destabilization, suggesting that SUMO can affect PAP stability. Finally, in vitro sumoylation was found to inhibit the activity of purified PAP. SUMO is thus a novel post-translational regulator capable of affecting multiple aspects of PAP function.
Results
Higher-molecular-weight (HMW) forms of PAP accumulate in various tissues and cell lines
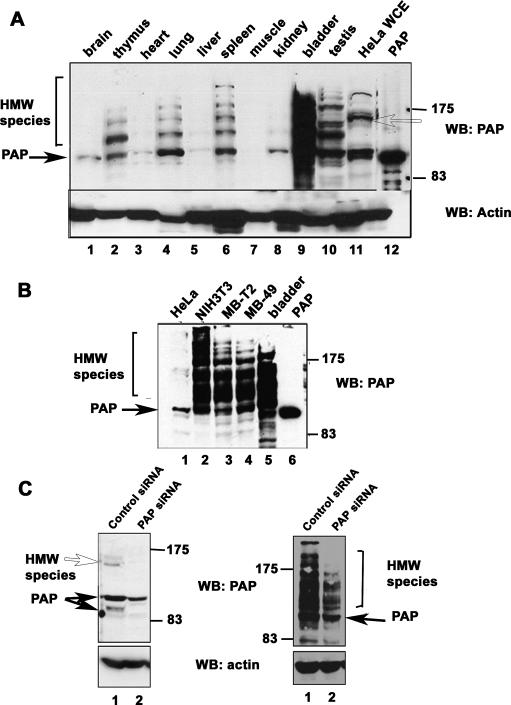
We and others described previously a second PAP gene, which encodes a PAP called Neo-PAP or PAP-γ that is extremely similar to the well-studied PAP described above (Kyriakopoulou et al. 2001; Topalian et al. 2001). In order to begin to study the physiological roles of these two PAPs, we produced anti-peptide antibodies that differentiate between them. As a first experiment, we used the antibodies to analyze different mouse tissues by Western blotting. Neo-PAP was expressed in a limited number of tissues and, consistent with previous results (Topalian et al. 2001), there was no evidence of post-translational modification (results not shown). However, the pattern observed with PAP itself was strikingly different. PAP typically migrates between 90 and 105 kDa on SDS gels, reflecting the presence of phosphorylated forms (Ballantyne et al. 1995; Colgan et al. 1996). However, an unexpected pattern of HMW species in addition to those of the expected size was detected in several mouse tissues. These species were detected at high levels in a striking ladder-like pattern in samples from tissues including spleen, lung, and, most strikingly, bladder (Fig. 1A). As many as four HMW species were observed extending to an apparent molecular size of ∼200 kDa. In tissues such as heart and kidney, where the levels of 100-kDa PAP were low, the HMW species were also proportionately decreased, providing evidence that these species were PAP related.
Figure 1.
Abundant HMW species of PAP are detected in tissues and cell lines. (A) Anti-PAP immunoblotting of mouse tissue lysates. Equivalent amounts of total protein from various mouse tissue extracts, HeLa whole-cell lysate, and 50 ng of purified recombinant PAP protein were analyzed by Western blotting with anti-PAP and anti-actin antibodies. (B) Anti-PAP immunoblotting of cell lines. Whole-cell lysates prepared from the indicated cell lines, recombinant PAP, and bladder tissue extracts were subject to Western blotting as above. (C) Whole-cell lysates were prepared from HeLa or NIH 3T3 cells transfected with PAP-specific siRNAs (lanes 2,4, respectively) or control siRNAs (lanes 1,3, respectively) and subject to Western blotting with anti-PAP and anti-actin antibodies. The positions of unmodified PAP and the HMW species are indicated by closed arrows and open arrows or brackets, respectively. The positions of molecular size markers are indicated on the right.
We next analyzed lysates prepared from several cell lines (Fig. 1B). In keeping with previous studies (Thuresson et al. 1994; Zhao and Manley 1996), HeLa cells showed predominantly 100-kDa PAP although HMW species of less intensity at 150 kDa were also observed (Fig. 1A [lane 11], B [lane 1]). A similar pattern was observed with 293 cells (data not shown). Strikingly, NIH 3T3 cells (Fig. 1B, lane 2) displayed abundant HMW forms very similar to those observed in bladder tissues. Given the abundance of HMW forms in bladder, we analyzed two mouse bladder carcinoma lines, MBT-2 and MB-49 (Fig. 1B, lanes 3,4). Both of these cell lines displayed patterns very similar to bladder (Fig. 1B, lane 5).
We next wished to confirm that the HMW species were PAP related and not cross-reacting proteins. To this end, siRNAs directed against PAP were transfected into both HeLa and NIH 3T3 cells. As expected, not only did the 100- to 106-kDa bands decrease in intensity in the knockdown cells as compared with control siRNA-treated cells (Fig. 1C), but accumulation of the single 150-kDa species in HeLa cells (Fig. 1C, lanes 1,2) and the HMW species in NIH 3T3 (Fig. 1C, lanes 3,4) diminished with transfection of PAP siRNA. We therefore conclude that these proteins are indeed PAP related.
As mentioned above, PAP exists in various phosphorylated states in the cell (Ballantyne et al. 1995; Colgan et al. 1996, 1998). Although the very low mobility of the modified forms does not suggest phosphorylation, we nonetheless tested to determine whether the HMW PAP isoforms we observed were hyperphosphorylated by treating cell extracts with phosphatase. The HMW forms were not affected by phosphatase treatment, although the 100-kDa band decreased in intensity (data not shown). Therefore, the HMW isoforms likely result from a covalent modification other than phosphorylation.
The HMW forms are SUMO-2/3-modified forms of PAP
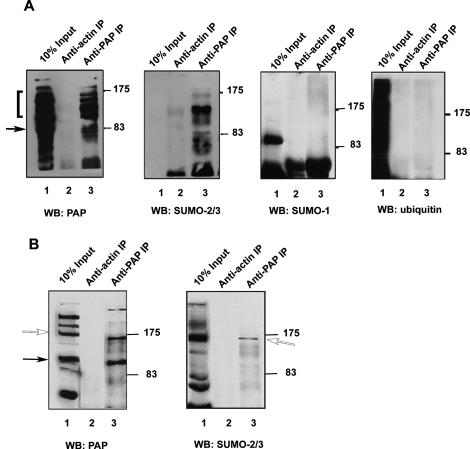
We next wished to determine the nature of the PAP modifications. We observed that the bands in the ladder shown in Figure 1A increased in steps corresponding to a size of ∼20 kDa. Significantly, modification by SUMO also increases the apparent molecular weight of substrates by 20 kDa (Johnson 2004). To determine whether the HMW PAP isoforms were indeed SUMO modified, we first prepared extracts from mouse bladder and immunoprecipitated them with anti-PAP antibodies (Fig. 2A). Along with the 100-kDa PAP, all the HMW forms were immunoprecipitated as shown by immunoblotting with the anti-PAP antibody (Fig. 2A, left-most panel). We then analyzed the immunoprecipitations (IPs) by Western blotting with anti-ubiquitin (Fig. 2A, right-most panel), anti-SUMO-1 (Fig. 2A, second panel from the right), and anti-SUMO-2/3 (Fig. 2A, second panel from the left) antibodies. Strikingly, the anti-SUMO-2/3 antibodies, but none of the others, recognized the HMW PAP isoforms. In nuclear extract prepared from HeLa cells, in which the HMW isoform was more abundant than in the whole-cell lysate analyzed above, the PAP antibody immunoprecipitated the 150-kDa species in addition to lower-molecular-weight forms of PAP (Fig. 2B, left panel, lane 3). As with the bladder extracts, the SUMO-2/3 antibody specifically recognized the HMW isoform (Fig. 2B, right panel, lane 3). These results were confirmed with a second anti-SUMO-2/3 antibody (Supplemental Fig. 1). Taken together, these results indicate that PAP can be extensively modified by SUMO-2/3 in a manner that varies in a tissue- and cell-type-dependent manner.
Figure 2.
The HMW forms of PAP are modified by SUMO-2/3. (A) Proteins were precipitated from bladder extracts with antibodies directed against PAP or actin as described in Materials and Methods and analyzed on SDS-PAGE. Lanes 1, 2, and 3 in all panels are the bladder lysate input (10%), anti-actin IP, and anti-PAP IP, respectively. Positions of protein size standards are marked on the right. Immunoblotting was carried out with anti-PAP (left panel), anti-SUMO2/3 (left middle panel), anti-SUMO-1 (right middle panel), or anti-ubiquitin antibodies (right panel). Closed arrow and bracket indicate unmodified and HMW PAP forms, respectively. (B) Antibodies directed against PAP or nonspecific IgG were used to immunoprecipitate proteins from HeLa NE prepared in the presence of NEM as described in Materials and Methods. The immunoprecipitated proteins (lanes 2,3) along with 10% of the input (lanes 1) were analyzed by Western blotting with anti-PAP (left panel) or anti-SUMO-2/3 (right panel) antibodies. Closed and open arrows indicate unmodified and modified PAP, respectively.
PAP interacts directly with ubc9 and is a substrate for in vitro modification by SUMO.
The above data provide strong evidence that PAP is sumoylated. However, two observations suggest that PAP may be an unusual SUMO substrate. First, in some tissues and cell types, a large fraction of total PAP was detected in sumoylated forms. More typically, only small proportions of substrate proteins appear to be sumoylated (see Discussion). Second, SUMO is frequently conjugated to lysines present in a consensus motif, ΨKXE (Melchior 2000; Yeh et al. 2000). However, neither mouse nor human PAP contains a match to this consensus. We therefore next wished to determine whether PAP is sumoylated in vitro by the characterized sumoylation pathway.
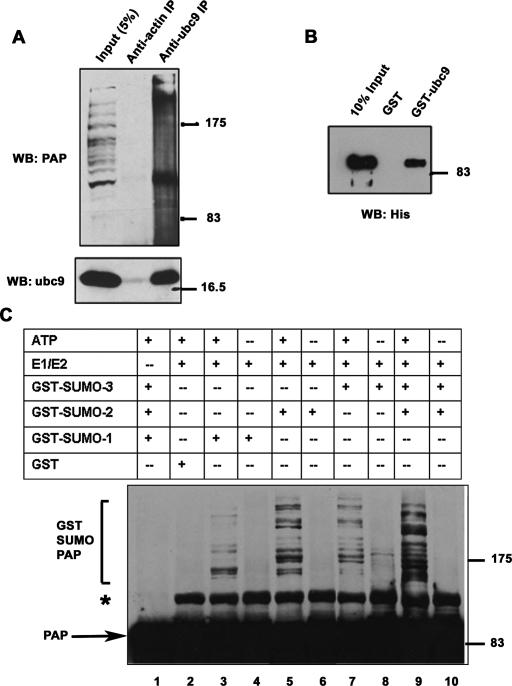
Most SUMO substrates interact directly with the E2 enzyme ubc9 and can be sumoylated using in vitro assays containing recombinant E1, E2, and SUMO (e.g., Sampson et al. 2001). In many cases, the interaction of a protein with ubc9 is itself a strong indication that it is a substrate for SUMO modification (Melchior 2000). To determine whether PAP interacts with ubc9, we first carried out IPs with anti-ubc9 antibodies using NIH 3T3 extracts. The ubc9 antibody selectively immunoprecipitated the unmodified PAP isoform from these extracts (Fig. 3A, top panel). The reverse IP confirmed these results; PAP antibodies immunoprecipitated ubc9 from NIH 3T3 extracts (data not shown). To determine whether the interaction between PAP and ubc9 was direct, we used an in vitro binding assay with purified his-tagged PAP and GST-ubc9. The results (Fig. 3B) indeed revealed a direct interaction between the two proteins.
Figure 3.
PAP can be sumoylated in vitro and interacts directly with ubc9. (A) Proteins from NIH 3T3 cell extracts were precipitated with anti-ubc9 or anti-actin IgG and analyzed by Western blotting with anti-ubc9 (bottom panel) or anti-PAP (top panel) antibodies as indicated. (Lane 1) Ten percent of the extracts used in the binding experiments were loaded as input. (B) His-tagged PAP was incubated with equal amounts of GST or GST-Ubc9 bound to glutathione Sepharose beads. Bound proteins (lanes 2,3) were eluted by boiling in SDS sample buffer and resolved on SDS-PAGE along with 10% of the input (lane 1) and analyzed by Western blotting with anti-His antibodies. (C) Purified His-tagged PAP was incubated for 2 h at 37°C with GST-SAE2/1 (E1) GST-hUBC9 (E2), in the presence of GST-SUMO-1, GST-SUMO-2, or GST-SUMO-3 as indicated, and in the presence (+) or absence (−) of ATP (shown in lanes 3–10). Lane 1 is a control reaction carried out in the absence of E1 and E2, and lane 2 is a control reaction containing GST instead of GST-SUMO-1, GST-SUMO-2, or GST-SUMO-3. Reactions were terminated by adding SDS sample buffer and were analyzed by Western blotting with anti-His antibodies. Unmodified PAP and GST SUMO-PAP are indicated. An ATP-independent species that appears in the presence of E1 is marked with an asterisk.
We next wished to determine whether PAP can be sumoylated in vitro with purified components. To this end, we used an in vitro assay with recombinant His-tagged PAP, GST-E1, GST-E2, and GST-SUMO-1, GST-SUMO-2, and GST-SUMO-3 in the presence of an ATP regenerating system (Tatham et al. 2001). HMW forms of PAP were detected with an anti-his antibody in reactions containing E1/E2 and GST-SUMO-1, GST-SUMO-2, or GST-SUMO-3 in the presence of ATP (Fig. 3C, lanes 3,5,7). No HMW bands were detected in the absence of ATP (Fig. 3C, lanes 4,6,8,10), in control reactions with GST instead of GST-SUMO-1, GST-SUMO-2, or GST-SUMO-3 (Fig. 3C, lane 1) or in reactions lacking E1/E2 (Fig. 3C, lane 2), indicating that the bands observed reflect SUMO-modified forms of PAP. PAP can thus be modified by all three SUMO isoforms in vitro. Significantly, though, we observed that addition of SUMO-2 and SUMO-3 together resulted in more efficient formation of HMW species, resembling more closely the in vivo pattern (Fig. 3C, lane 9).
PAP sumoylation sites are localized in its C-terminal regulatory domain
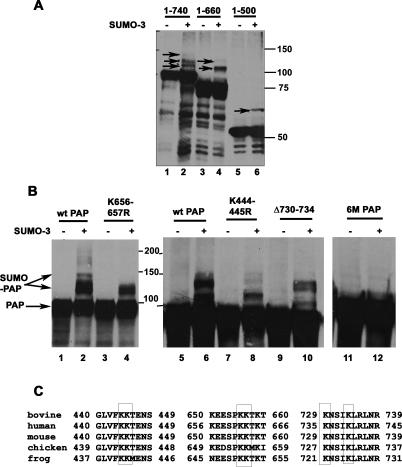
We next wished to identify the lysines in PAP that are modified by SUMO. For this, we developed a cotransfection assay in which HA-tagged mouse PAP is coexpressed with His-tagged SUMO-3 in human 293 cells. However, in initial experiments, we were unable to detect modified forms of HA-PAP in Western blots of whole-cell lysates, indicating that the exogenous PAP was poorly modified. In order to enrich the lysates for SUMO-modified forms of PAP, we carried out the cotransfection as above and then partially purified SUMO conjugates under denaturing conditions using Ni-NTA resin (e.g., Salinas et al. 2004). Western blotting of these lysates with anti-HA antibodies revealed three distinct modified PAP forms (Fig. 4A, lane 2; Fig. 4B, lanes 2,6). Occasionally, more extensively modified forms were also apparent (Fig. 4B, lane 2).
Figure 4.
The sumoylation sites in PAP are localized in the C-terminal regulatory region. (A) Plasmids expressing HA-tagged full-length PAP (lanes 1,2), PAP 1–660 amino acids (lanes 3,4), and PAP 1–500 amino acids (lanes 5,6) were transfected into 293 cells along with empty vector (−) or SUMO-3 expression plasmid (+). Cell extracts were prepared under denaturing conditions and partially purified by binding to Ni-NTA beads as described in Materials and Methods. The bound fractions were analyzed by Western blotting with anti-HA antibodies. (B) HA-PAP expression constructs were transfected into 293 cells in the presence of empty vector (−), or SUMO-3-expressing (+) plasmid and processed as above. Bound proteins from cells transfected with plasmids expressing wild-type PAP (lanes 1,2,5,6), K656–657R PAP (lanes 3,4), K444–445R PAP (lanes 7,8), Δ730–734 PAP (lanes 9,10), and 6M-PAP (lanes 11,12) were analyzed by Western blotting with anti-HA antibodies. Sumoylated PAP species are indicated by closed arrows and positions of molecular weight markers are indicated on the right. (C) A ClustalW alignment of bovine, human, chicken, mouse, and Xenopus PAP is shown. The SUMO acceptor lysines are boxed.
We next used this assay to map the sites of sumoylation in PAP. First, regions in PAP containing the potential lysines were mapped by analyzing truncated versions of PAP containing specific functional domains (e.g., Martin et al. 2000). The N-terminal domain containing residues 1–350 was not modified detectably in the cotransfection assay (data not shown). A single modified species was detected with PAP 1–500 (Fig. 4A, lane 6), and two modified forms were detected with PAP 1–660 (Fig. 4A, lane 4). Based on this, we next systematically mutated all of the 31 lysines between residues 350 and 730 to arginine. In most of the cases, individual lysines were mutated; however, closely spaced Lys444–445, Lys506–507, and Lys656–657 were mutated together, and Lys730–734 were deleted. PAP containing the mutations K444–445R, K656–657R, or lacking Lys730 and Lys734 (Fig. 4B, lanes 4,8,10) showed differences in sumoylation from the wild-type protein (Fig. 4B, lanes 2,6). A PAP construct containing all these mutations (mutant 6M) was completely deficient in sumoylation (Fig. 4B, lane 12). The 6M mutant also could not be sumoylated in vitro, indicating that the same lysines are targeted for sumoylation in vitro and in vivo (Supplemental Fig. 4). These six lysines are conserved across several vertebrate species (Fig. 4C), although they are not conserved in yeast PAP. Interestingly, they are also not conserved in Neo-PAP, which appears not to be sumoylated (results not shown). It is also noteworthy that four of the six lysines are localized in the C-terminal regulatory region, with Lys656–657 overlapping the second PAP NLS (NLS2).
Nuclear localization of PAP is SUMO-dependent
We next wished to investigate the functional consequences of PAP sumoylation. Sumoylation is known to influence the nucleocytoplasmic transport of several substrates (e.g., Rangasamy et al. 2000; Lin et al. 2003). Especially since PAP is sumoylated at lysines overlapping NLS2, we set out to test whether sumoylation affects PAP nuclear localization. We first wished to confirm that sumoylated PAP is indeed nuclear. We compared the intensities of the HMW species of PAP in HeLa whole-cell and nuclear extracts by Western blot analysis. The intensity of the HMW species was considerably higher in nuclear compared with whole-cell extracts (Supplemental Fig. 2A). In addition, immunofluoresence assays of PAP in NIH 3T3 cells, where PAP HMW species are abundant (see Fig. 1), revealed essentially exclusive nuclear staining (Supplemental Fig. 3). These results indicate that sumoylated PAP is indeed enriched in the nucleus.
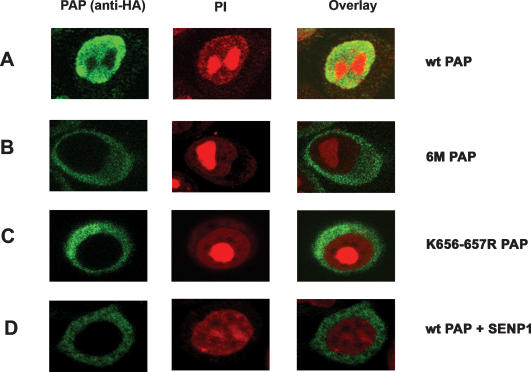
To determine if sumoylation plays a direct role in nuclear localization, we first expressed HA-tagged wild-type and 6M mutant PAPs in HeLa cells. Cells were fixed, stained for PAP using HA antibodies, and visualized by confocal microscopy. Nuclei were visualized by propidium iodide staining. Wild-type PAP localized to the nucleoplasm as expected (Fig. 5A; Raabe et al. 1994). Remarkably, however, the 6M mutant was localized in the cytoplasm in all transfected cells examined (Fig. 5B). We next wished to determine whether loss of sumoylation at Lys656–657 was sufficient to relocalize PAP into the cytoplasm. Therefore, we examined localization of the K656–657R mutant. Strikingly, this mutant behaved like the 6M mutant and was localized largely if not exclusively to the cytoplasm (Fig. 5C). The K444–445R and K730–734R mutant PAPs were localized to the nucleus, indistinguishably from wild-type protein (data not shown). Similar results were obtained with transfected NIH 3T3 cells (data not shown).
Figure 5.
Nuclear localization of PAP is SUMO-dependent. HeLa cells were transfected with HA-tagged wild-type PAP expression vector (A), 6M PAP expression vector (B), K656–657R PAP expression vector (C), and wild-type PAP and Flag-SENP1 expression vectors (D). Forty-eight hours after transfections, cells were fixed and subjected to immunofluorescence with anti-HA antibodies and AlexaFluor 488-conjugated anti-mouse secondary antibody as described in Materials and Methods. Nuclei were counterstained with propidium iodide. Anti-HA staining (green) is shown in the left panels, middle panels show propidium iodide staining, and right panels show a merged image of propidium iodide and antibody staining.
We next wished to confirm that the effect of the lysine-to-arginine mutations was in fact due to loss of sumoylation. To this end, localization of wild-type PAP was examined in HeLa cells coexpressing SENP1 SUMO protease. The subcellular localization of various SUMO proteases often determines their substrate specificity. SENP1 is localized to the nucleoplasm, like PAP (Gong et al. 2000; Bailey and O’Hare 2002). Following transfection and analysis as above, we observed that wild-type PAP was relocalized to the cytoplasm from the nucleus in cells that coexpressed SENP1 (Fig. 5D). Coexpression of a catalytically inactive mutant of SENP1 with PAP did not alter the localization pattern, confirming that the desumoylation activity of SENP1 was required to relocalize PAP (results not shown). These results indicate, firstly, that SENP1 targets PAP for desumoylation in vivo, and secondly, that loss of sumoylation is indeed responsible for the change in PAP localization.
Sumoylation enhances PAP stability
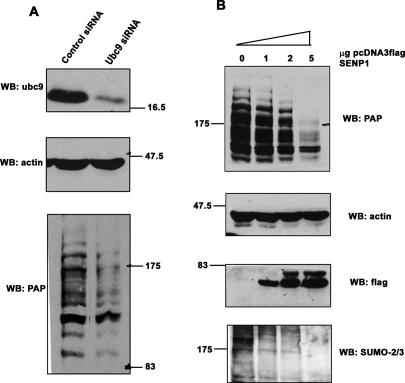
Sumoylation is also known to affect protein stability. For example, SUMO modification of the NFkB repressor IκΒα renders it resistant to degradation, and sumoylation of the tumor suppressor smad4 enhances the stability (Desterro et al. 1998; Lin et al. 2003). Therefore, we next sought to investigate whether SUMO modification affects PAP protein levels by examining the effect of sumoylation on the stability of endogenous PAP. In these experiments we down-regulated the SUMO pathway using two approaches. In the first approach, NIH 3T3 cells were transfected with siRNAs targeting ubc9 or with control siRNAs. After 72–96 h, cells were harvested and lysates analyzed by Western blotting with anti-ubc9, anti-PAP, and anti-actin antibodies. The blot confirmed efficient ubc9 knockdown (Fig. 6A, top panel). Depletion of ubc9 resulted in loss of the HMW PAP forms. However, there was no accumulation of unmodified PAP with the disappearance of modified forms; instead, an overall reduction in total PAP levels was observed (Fig. 6B, bottom panel). Anti-actin blotting confirmed equal loading of the lanes (Fig. 6A, middle panel). We also confirmed by RT–PCR that PAP mRNA levels were unaffected by ubc9 siRNA transfections (data not shown); thus, reduction in PAP occurs at the protein level.
Figure 6.
Sumoylation enhances PAP stability. (A) NIH 3T3 cells were transfected with ubc9 siRNA or control siRNA, and whole-cell lysates were analyzed by Western blotting with anti-ubc9, anti-PAP, or anti-actin antibodies. Positions of protein size standards are indicated. (B) NIH 3T3 cells were transfected with 0, 1, 2, or 5 μg of Flag-SENP1 expression vector using AMAXA nucleofector II. Twenty-four hours after transfection, lysates from equivalent number of cells were analyzed by Western blotting with anti-PAP, anti-actin, anti-Flag, and anti-SUMO2/3 antibodies.
We next used a different approach to reduce overall levels of sumoylation, which was to overexpress SENP1. NIH 3T3 cells transfected with increasing amounts of a plasmid expressing Flag-tagged SENP1 were analyzed for levels of endogenous PAP by Western blotting with anti-Flag, anti-SUMO-2/3, and anti-PAP antibodies. Increasing amounts of SENP1 expression (Fig. 6B, third panel from top) resulted in loss of total SUMO-2/3-conjugated proteins in the transfected cells (Fig. 6B, bottom panel) and, more significantly, of the HMW forms of PAP (Fig. 6B, top panel, lanes 2–4). As was observed in the ubc9 siRNA-treated cells, there was no accumulation of unmodified PAP; instead, the levels of PAP were significantly reduced. Together, these results suggest that disruption of the SUMO pathway in NIH 3T3 cells compromises PAP.
Sumoylation of PAP attenuates polyadenylation activity
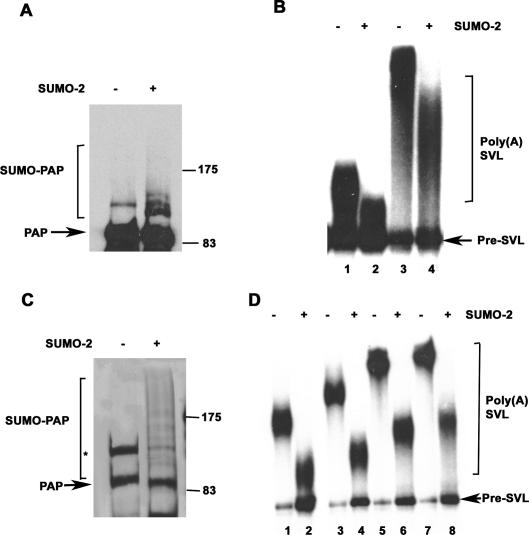
Hyperphosphorylation of the PAP regulatory domain is known to repress PAP activity (Colgan et al. 1996, 1998). We therefore sought to determine whether sumoylation can also affect the enzymatic activity of PAP. Toward this end, we examined the effect of sumoylating purified His-PAP in vitro as described above, except that we replaced GST-tagged forms of SUMO with untagged SUMO to avoid possible artifacts due to the GST tag. His-tagged PAP was added to reactions containing recombinant E1, E2, and an ATP regenerating system, in the presence or absence of SUMO-2. Western blot analysis revealed the appearance of SUMO-2-modified PAP (Fig. 7A, left panel). We then examined activity in nonspecific poly(A) synthesis assays (e.g., Ryner et al. 1989), in which PAP adds adenosine residues in an AAUAAA-independent manner to a 32P-labeled substrate RNA in the presence of Mn2+ ions. Increasing amounts of sumoylated and mock-treated PAP were added to polyadenylation assays, and RNA was purified and analyzed by denaturing PAGE. The results (Fig. 7B) show that both the amount of substrate polyadenylated as well as the poly(A) tail length were lower in reactions containing sumoylated PAP versus reactions containing unmodified PAP, indicating that in vitro sumoylation inhibits polyadenylation activity.
Figure 7.
Sumoylation attenuates enzymatic activity of PAP. (A) In vitro sumoylation reactions were carried out with his-tagged PAP, in the absence (−) or presence (+) of untagged SUMO-2, and analyzed by Western blotting with anti-PAP antibodies. The positions of protein size standards are indicated. (B) Nonspecific polyadenylation assays. Assays containing 5 ng (lanes 1,2) or 10 ng (lanes 3,4) of PAP from A were carried out as described in Materials and Methods. Following incubation, RNAs were isolated, resolved by denaturing PAGE, and subjected to autoradiography. (C) In vitro sumoylation reactions were performed as in A with reaction mixtures scaled up 10-fold, and His-PAP was purified by binding to Ni-NTA beads. A silver-stained SDS gel of the purified proteins is shown. Unmodified PAP, SUMO-PAP, and positions of protein size standards are indicated. The nonspecific band described in Figure 3 is marked with an asterisk. (D) Nonspecific polyadenylation assays with purified in vitro sumoylated PAP. Reactions were carried out with 2 ng (lanes 1,2), 4 ng (lanes 3,4), 10 ng (lanes 5,6), and 20 ng (lanes 7,8) of PAP isolated from reactions containing (lanes 2,4,6,8) or lacking (lanes 1,3,5,7) SUMO-2.
To confirm these results, we repeated the above assay but first repurified the his-tagged PAP from sumoylation reaction mixtures with nickel agarose beads. A silver-stained SDS gel (Fig. 7C) revealed several HMW forms of PAP isolated from the SUMO-2-containing (Fig. 7C, lane 2) but not the control (Fig. 7C, lane 1) reaction lacking SUMO-2. Addition of increasing amounts of repurified control or sumoylated PAP to poly(A) synthesis assays revealed that sumoylated PAP was less active than the control at all concentrations tested (Fig. 7D). These results demonstrate that sumoylation inhibits PAP enzymatic activity.
Discussion
In this study, we identify sumoylation as a novel regulator of PAP function. The presence of very high levels of SUMO-2/3-modified PAP in specific tissues and cell lines was unanticipated and has not been observed previously with other SUMO substrates. This, in addition to the absence of consensus sites and the strong interaction with ubc9, suggests that PAP is an unusual SUMO substrate. SUMO was found to influence PAP function at multiple levels. Most strikingly, we discovered that nuclear localization of PAP is SUMO-dependent. SUMO was also found to enhance PAP stability, particularly in cell lines with high endogenous levels of modified PAP. Finally, we observed that SUMO has an inhibitory effect on PAP enzymatic activity. Below we discuss the underlying mechanisms and physiological relevance of this regulation.
Many SUMO targets can be sumoylated in vitro in the absence of E3 ligases. In vivo, however, E3 ligases play important roles in regulating sumoylation (Johnson 2004). In the case of PAP, the direct interaction with ubc9 does not preclude roles for (an) E3 ligase(s) in vivo. There are several indications to suggest this is the case. For one, unlike the striking endogenous ladder-like pattern of modification that we observed in several mouse tissues, coexpression of exogenous PAP with SUMO yielded mainly three distinct modified species. In addition, the level of exogenous PAP modification upon coexpression with SUMO was much lower than that of the endogenous protein, suggestive of a limiting factor in the transfection experiments, perhaps the putative E3 ligase. E3 ligases not only increase sumoylation of substrates, but have also been known to catalyze polysumoylation (e.g., Rosas-Acosta et al. 2005). Secondly, while all three SUMO paralogs could be conjugated to PAP in vitro, we only detected SUMO-2/3-modified PAP in vivo. E3 ligases have also been shown to confer SUMO paralog specificity to several substrates such as RanGAP1 and SP100 (Reverter and Lima 2005; Tatham et al. 2005), which like PAP can be equally conjugated to all three isoforms in vitro (Reverter and Lima 2004). Finally, E3 ligases have been shown to play a particularly important role in SUMO modification of substrates that lack consensus modification sites. For example, CtBP2, which lacks consensus sites, is only sumoylated when the E3 ligase Pc2 is included in cotransfection experiments (Kagey et al. 2003). The putative PAP E3 ligase may play an important role in regulation of PAP sumoylation, perhaps contributing to the variations in sumoylation levels that we observed in different tissues.
Our finding that SUMO is capable of influencing not one but several aspects of PAP function was unexpected. Perhaps the most profound effect was on PAP localization. Sumoylation has been found to enhance nuclear accumulation of several substrates, although in no case as directly as we showed for PAP. For example, sumoylation-deficient mutants of the transcription factor CtBP1 and the bovine papillomavirus E1 protein are mislocalized at least partially to the cytoplasm, but the sumoylation sites are not located near an NLS (Rangasamy et al. 2000; Lin et al. 2003). In the case of the transcription factor Daxx, sumoylation sites are located adjacent to an NLS, but coexpression of SUMO with a ΔNLS mutant of Daxx still relocalized the protein to the nucleus, indicating that noncovalent interactions of SUMO may be involved (Chen et al. 2006). The effect of sumoylation on PAP localization is particularly striking not only because sumoylation at NLS2 is necessary for correct localization, but also because abolishing sumoylation generally, by overexpression of SENP1, also causes mislocalization.
The mechanism by which PAP sumoylation influences localization is unclear. PAP does not contain recognizable nuclear export signals, and experiments with leptomycin ruled out nuclear export mechanisms influencing PAP localization (V. Vethantham, N. Rao, and J.L. Manley, unpubl.). A role for SUMO in import therefore seems more likely. A recent study showed that PAP acetylation at NLS2 (Lys644) inhibits interactions with the importin α/β complex, thereby inhibiting nuclear localization (Shimazu et al. 2007). Sumoylation could compete with and prevent acetylation at NLS2, thus promoting import. However, we cannot rule out other explanations such as SUMO inducing conformational changes that expose the NLS and/or promoting interactions with the nuclear import machinery.
Despite the fact that only a small fraction of exogenous PAP was detectably modified, loss of sumoylation had large effects on localization. This behavior is not unusual among SUMO substrates. For example, the strong repressive effects of sumoylation on transcription factors such as p300 and Elk-1 are almost entirely lost when sumoylation sites are mutated, although only very small proportions of these proteins are detected in sumoylated forms (Girdwood et al. 2003; Salinas et al. 2004; for review, see Hay 2005). It has been postulated that once the transcription factor has entered the repressive environment for which sumoylation is required, the modification may no longer be necessary to maintain those interactions. Thus a protein with a past history of modification behaves differently from a protein that has never been modified. Similarly, sumoylation of PAP may be required to initiate interactions that facilitate correct nuclear localization, such as incorporation into import complexes (see Melchior et al. 2003), but once this is achieved, sumoylation is no longer required.
Sumoylation, unlike ubiquitination, is often associated with increased stability of its targets. The effect of SUMO in promoting stability can occur by blocking ubiquitination at the same lysine (Desterro et al. 1998; Steffan et al. 2004). However, there is currently no evidence that metazoan PAP is ubiquitinated, nor is it currently known whether instability is a direct consequence of loss of PAP sumoylation.
Our studies revealed an inhibitory effect of SUMO on the activity of purified PAP. In this respect, the effect of sumoylation resembles PAP phosphorylation (Colgan et al. 1996, 1998). The absence of any other factors in our in vitro assay suggests that a conformational change induced by SUMO is responsible for the inhibition. Sumoylation has been known to reduce the RNA-binding activity of hnRNPs C and M (Vassileva and Matunis 2004) and to reduce the RNA-editing activity of ADAR1 (Desterro et al. 2005). The inhibitory effect of sumoylation on PAP activity is thus in keeping with these previous findings. However, it contrasts with our recent observations that SUMO plays a positive role in enhancing 3′ processing complex assembly and activity. We showed that CPSF-73 and symplekin are sumoylation targets and that desumoylation of HeLa nuclear extracts inhibited 3′ processing by preventing formation of polyadenylation-specific complexes (Vethantham et al. 2007). These contradictory findings can be reconciled by considering the fact that PAP enzymatic activity was measured in the absence of other factors in the complex, under conditions in which the enzyme behaves in a distributive and not a processive manner (Ryner et al. 1989; Martin et al. 2004). Additionally, the levels of sumoylated PAP are low in HeLa nuclear extracts, and the inhibitory effect of desumoylation on complex assembly may thus overshadow the small increase in PAP activity that would be predicted to arise from its desumoylation. It is likely that in a cellular context, where polyadenylation is carried out by a multisubunit complex, several factors come into play. These include the levels of sumoylated PAP in the nucleus, how PAP sumoylation modulates its interactions with other factors, and the combined effects of sumoylation of other factors in the 3′ processing complex.
The multiple effects of SUMO on PAP function indicate that SUMO regulates PAP in a complex manner. Future studies will be instrumental in determining how these different effects can be integrated into a single model to better comprehend the impact of SUMO on PAP function. For example, is the localization of PAP related to its susceptibility to degradation pathways? Does PAP undergo cycles of sumoylation and desumoylation within the nucleus with respect to its incorporation into the 3′ processing machinery? The presence of high levels of PAP in certain cell types and tissues is intriguing, and we speculate that in these tissues, SUMO may play additional roles in regulating PAP function.
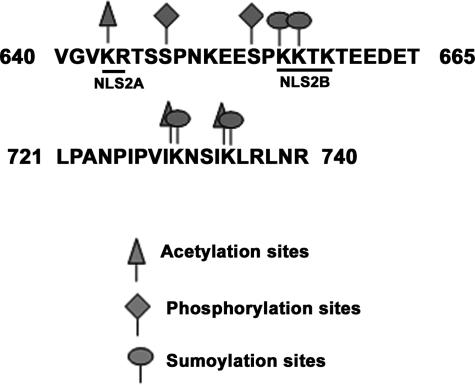
Regulation of PAP by post-translational modifications is emerging as a complex picture. PAP is now known to be subject to phosphorylation, acetylation, and sumoylation. This is consistent with the need to regulate PAP activity tightly (Zhao and Manley 1998). It is conceivable that the different modifications interact to control PAP under various conditions, since sites of phosphorylation, acetylation, and sumoylation are situated very close together or even overlap (Fig. 8). The consensus cdk sites of phosphorylation at Ser654 and Ser648 are close to sumoylation sites. Mutation of both of these serines to alanines did not affect sumoylation (V. Vethantham, N. Rao, and J.L. Manley, unpubl.), indicating that sumoylation is not dependent on phosphorylation. In addition, while phosphorylation of PAP is regulated in a cell cycle-dependent manner (Ballantyne et al. 1995; Colgan et al. 1996), we did not detect changes in PAP sumoylation levels during the cell cycle (V. Vethantham, N. Rao, and J.L. Manley, unpubl.). While phosphorylation of several proteins has been shown to positively influence sumoylation (Hietakangas et al. 2003; Hayakawa and Privalsky 2004), this appears not to be the case for PAP. It is possible that under certain physiological conditions, given the close proximity of sites, the two modifications may interact. Sumoylation sites at 656–657 and acetylation sites at 644 both overlap the bipartite NLS (Fig. 8). Lys730–734 are both sumoylated as well as acetylated, making it likely that acetylation may interact with sumoylation to regulate PAP function.
Figure 8.
PAP modifications cluster in the C-terminal domain. Amino acid sequence of two regions in the C-terminal domain showing NLS2 (underlined as NLS2A and NLS2B) and modification sites. Sumoylation sites are marked with an open circle, acetylation sites are denoted by open triangles, and phosphorylation sites are indicated by rectangles.
Our discovery of PAP sumoylation has revealed an important aspect of gene regulation at the post-transcriptional level. SUMO has a substantial impact on PAP function, and our data indicate that alterations in PAP sumoylation have the potential to profoundly affect gene expression. For example, misregulation of PAP activity has been associated with disease (for review, see Scorilas 2002). PAP activity is known to reflect the proliferative capacity of cells, as several neoplastic and leukaemic cells show increased PAP activity with or without increased PAP expression (Papamichail et al. 1983). Such increases in PAP activity have been correlated with malignancy in breast cancer and leukemia (Scorilas et al. 1998). Conversely, reduced PAP activity is correlated with apoptosis (Atabasides et al. 1998). Sumoylation-mediated regulation of PAP thus may have important consequences not only in the normal cellular physiological context, but also in malignancy and disease.
Materials and methods
Plasmids, siRNAs, and site-directed mutagenesis
pcDNA3-mousePAP and pcDNA3-FlagSENP1 were generated from RT–PCR products amplified from total RNA of mouse or human, respectively. Expression vectors were generated from SAE2/1, SUMO-2, and SUMO-3 constructs obtained from Dr. R.T. Hay and SUMO-1 and Ubc9 constructs from Dr. D. Wotton. PAP mutations were generated by PCR using appropriate mutagenic primers, and mutations were verified by sequencing. siRNA against mouse ubc9 was targeted to the sequence CAAT GAACCTGATGAACTG. siRNAs against mouse PAP and human PAP were targeted to the sequences GACATGTTGATC GAAGT and AAAATCCCGACAAGGAAGAAT, respectively. All siRNAs were purchased from Dharmacon.
Antibodies
To make anti-PAP antibodies, the N-terminal peptide of PAPII conjugated to KLH was used for immunization of rabbit. Purified antibodies were obtained by affinity purification of the crude serum against purified recombinant bovine PAPII protein as described earlier (Zhao and Manley 1996). Anti-ubc9, anti-His, anti-ubiquitin (Santa Cruz Biotechnology), anti-actin, anti-Flag (Sigma), anti-SUMO-1, anti-SUMO-2/3 (Invitrogen), and anti-HA (Covance) antibodies were purchased from the indicated companies.
Cell culture and transfections
HeLa and 293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), and NIH 3T3 cells maintained in DMEM supplemented with 10% bovine calf serum. Plasmid and siRNA transfections in HeLa and 293 cells were carried out with Lipofectamine 2000. Transfections of SENP1 expression plasmids in NIH 3T3 cells were carried out with the Amaxa nucleofector II kit. siRNA transfections in NIH 3T3 cells were carried out in six-well plates using RNAifect reagent (Qiagen). Transfection protocols followed the reagent manufacturer’s instructions.
IPs and Western blotting
Bladder extracts were prepared by homogenizing bladder tissues in Buffer A (20 mM Tris at pH 7.5, 150 mM NaCl, 2 mM EDTA, 1% NP-40, 20 mM N-ethylmaleimide [NEM] with protease inhibitor cocktail purchased from Sigma). The homogenate was centrifuged at 20,000g for 20 min to remove cell debris, and the supernatant was diluted four times in binding buffer B (phosphate-buffered saline [PBS] containing 0.05% BSA and 0.1% NP40). NIH 3T3 whole-cell extracts were prepared by lysing cells in buffer A, and cell debris was removed by centrifugation. HeLa nuclear extracts were prepared as described earlier (Kleiman and Manley 2001), but with 20 mM NEM included in all buffers. Standard IP protocols were as follows: Extracts were precleared with protein A Sepharose and incubated with protein A Sepharose prebound to 1 μg of antibodies for 3 h at 4°C. Washes were carried out in buffer B + 500 mm NaCl for analyzing endogenous sumoylation. For PAP-ubc9 coimmunoprecipitation experiments, washes were carried out in buffer B. The bound proteins were eluted by boiling in SDS sample buffer, separated by SDS-PAGE, transferred onto nitrocellulose membrane, and immunoblotted with the indicated antibodies.
In vitro sumoylation assays
Sumoylation reactions (20 μL) were assembled on ice in 50 mM Tris (pH 7.5); 5 mM MgCl2; with 1 μg of GST-SAE2/SAE1; 2 μg of GST-Ubc9; 2 μg of GST-SUMO-1, GST-SUMO-2, and GST-SUMO-3; and 1 μg of recombinant His-PAP. Reactions were initiated by the addition of an ATP regeneration system (2 mM ATP, 10 mM creatine phosphate, 3.5 U/mL creatine kinase, and 0.6 U/mL inorganic pyrophosphatase), and incubated for 2.0 h at 37°C. Reactions were terminated by addition of SDS sample buffer and were subjected to SDS-PAGE and Western blotting with anti-PAP or anti-His antibodies.
Polyadenylation assays with in vitro sumoylated PAP
Sumoylation reactions for poly(A) synthesis assays were assembled as described above, with untagged SUMO-2 protein (+) or with buffer (−). 32P-labeled SVL precleaved substrate (pre-SVL) was prepared as described (Ryner et al. 1989). A nonspecific polyadenylation assay reaction mixture consisted of 2.5% PVA, 1 mM MnCl2, 100–125 ng of BSA, 1 mM ATP, 0.5 U of RNasin, 10 mM HEPES (pH 7.9), 25 mM NH4(SO4)2, 0.2 mM PMSF, 0.2 mM DTT, and the indicated amounts of PAP from both reaction mixtures. The nonspecific assays were incubated for 30 min at 30°C, and RNAs were resolved by denaturing PAGE and subjected to autoradiography. Large-scale in vitro sumoylation reactions were carried out by scaling up the above reactions to 200 μL. For repurification of His-PAP, reactions after incubation were diluted four times in buffer A (20 mM Tris at pH 7.5, 150 mM NaCl, 0.05% NP40, 5 mM imidazole), bound to Ni-NTA beads and agitated for 3 h. Washes were carried out in buffer B (buffer A + 10 mM imidazole + 1 M NaCl). Bound proteins were eluted in buffer C (buffer B containing 100 mM imidazole) and quantified by Bradfords assay. The indicated amounts of proteins were used in nonspecific polyadenylation assays.
In vivo sumoylation and denaturing purification
293 cells were transfected with 6 μg of mouse-PAP expression vector and 6 μg of empty vector or His-SUMO-3 expression vector using the calcium phosphate method. Forty-eight hours after transfection, cells were lysed in 6 M guanidine-HCl, 50 mM NaH2PO4 (pH 8.0), 10 mM Tris-HCl (pH 8.0), and 100 mM NaCl. His-tagged proteins were bound to Ni-NTA beads (Qiagen) by rocking for 3 h. Beads were washed several times sequentially in 8 M urea, 50 mM NaH2PO4, 100 mM NaCl (pH 8.0) (buffer A), buffer A with 10 mM imidazole, and buffer A with pH adjusted to 6.3. Bound proteins were eluted by boiling in SDS sample buffer (1% SDS, 100 mM DTT, 50 mM Tris at pH 6.8, 10% glycerol, 100 mM imidazole).
Immunofluorescence
HeLa cells were grown to 90% confluence in six-well plates and transfected with wild-type PAP and various mutants using Lipofectamine 2000. Twenty-four hours after transfection, the cells were trypsinized and split into two six-well plates containing coverslips. Twenty-four hours later, the cells were fixed for 15 min at room temperature with PBS containing 4% paraformaldehyde. Subsequently, the cells were permeabilized in PBS containing 0.2% Triton X-100 and 1% FBS for 10 min on ice. The coverslips were incubated with anti-HA monoclonal antibody (1:500 dilution) and AlexaFluor 488 anti-mouse IgG (1:500 dilution) diluted in PBS containing 1% FBS. All washes were carried out in the antibody dilution buffer except for the final four washes, which were carried out in PBS. Prior to mounting, cell nuclei were counterstained with propidium iodide for 20 min at 37°C. The cells were visualized with the Olympus FluoView FV500 confocal laser scanning microscope.
Acknowledgments
We thank Drs. R.T. Hay and D. Wotton for generous gifts of plasmids, D. di Giammartino for technical assistance, Y. Xu for help with the microscopy, and Y. Shi for helpful discussions. The work was supported by the National Institutes of Health grant R01 GM28983.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1628208
References
- Atabasides H., Tsiapalis C.M., Havredaki M. Dephosphorylation, proteolysis, and reduced activity of poly(A) polymerase associated with U937 cell apoptosis. Exp. Cell Res. 1998;244:433–440. doi: 10.1006/excr.1998.4231. [DOI] [PubMed] [Google Scholar]
- Bailey D., O’Hare P. Herpes simplex virus 1 ICP0 co-localizes with a SUMO-specific protease. J. Gen. Virol. 2002;83:2951–2964. doi: 10.1099/0022-1317-83-12-2951. [DOI] [PubMed] [Google Scholar]
- Ballantyne S., Bilger A., Astrom J., Virtanen A., Wickens M. Poly (A) polymerases in the nucleus and cytoplasm of frog oocytes: Dynamic changes during oocyte maturation and early development. RNA. 1995;1:64–78. [PMC free article] [PubMed] [Google Scholar]
- Chen A., Wang P.Y., Yang Y.C., Huang Y.H., Yeh J.J., Chou Y.H., Cheng J.T., Hong Y.R., Li S.S. SUMO regulates the cytoplasmonuclear transport of its target protein Daxx. J. Cell. Biochem. 2006;98:895–911. doi: 10.1002/jcb.20703. [DOI] [PubMed] [Google Scholar]
- Colgan D.F., Manley J.L. Mechanism and regulation of mRNA polyadenylation. Genes & Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- Colgan D.F., Murthy K.G., Prives C., Manley J.L. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature. 1996;384:282–285. doi: 10.1038/384282a0. [DOI] [PubMed] [Google Scholar]
- Colgan D.F., Murthy K.G., Zhao W., Prives C., Manley J.L. Inhibition of poly(A) polymerase requires p34cdc2/cyclin B phosphorylation of multiple consensus and non-consensus sites. EMBO J. 1998;17:1053–1062. doi: 10.1093/emboj/17.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Olmo M., Mizrahi N., Gross S., Moore C.L. The Uba2 and Ufd1 proteins of Saccharomyces cerevisiae interact with poly(A) polymerase and affect the polyadenylation activity of cell extracts. Mol. Gen. Genet. 1997;255:209–218. doi: 10.1007/s004380050491. [DOI] [PubMed] [Google Scholar]
- Desterro J.M., Rodriguez M.S., Hay R.T. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- Desterro J.M., Keegan L.P., Jaffray E., Hay R.T., O’Connell M.A., Carmo-Fonseca M.2005. SUMO-1 modification alters ADAR1 editing activity. Mol. Biol. Cell 165115–5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endter C., Kzhyshkowska J., Stauber R., Dobner T. SUMO-1 modification required for transformation by adenovirus type 5 early region 1B 55-kDa oncoprotein. Proc. Natl. Acad. Sci. 2001;98:11312–11317. doi: 10.1073/pnas.191361798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G. SUMO and ubiquitin in the nucleus: Different functions, similar mechanisms? Genes & Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- Girdwood D., Bumpass D., Vaughan O.A., Thain A., Anderson L.A., Snowden A.W., Garcia-Wilson E., Perkins N.D., Hay R.T.2003. P300 transcriptional repression is mediated by SUMO modification. Mol. Cell 111043–1054. [DOI] [PubMed] [Google Scholar]
- Gong L., Millas S., Maul G.G., Yeh E.T. Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J. Biol. Chem. 2000;275:3355–3359. doi: 10.1074/jbc.275.5.3355. [DOI] [PubMed] [Google Scholar]
- Hay R.T.2005. SUMO: A history of modification. Mol. Cell 181–12. [DOI] [PubMed] [Google Scholar]
- Hayakawa F., Privalsky M.L. Phosphorylation of PML by mitogen-activated protein kinases plays a key role in arsenic trioxide-mediated apoptosis. Cancer Cell. 2004;5:389–401. doi: 10.1016/s1535-6108(04)00082-0. [DOI] [PubMed] [Google Scholar]
- Hietakangas V., Ahlskog J.K., Jakobsson A.M., Hellesuo M., Sahlberg N.M., Holmberg C.I., Mikhailov A., Palvimo J.J., Pirkkala L., Sistonen L. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol. Cell. Biol. 2003;23:2953–2968. doi: 10.1128/MCB.23.8.2953-2968.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Carmichael G.G. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T.T., Wuerzberger-Davis S.M., Wu Z.H., Miyamoto S. Sequential modification of NEMO/IKKγ by SUMO-1 and ubiquitin mediates NF-κB activation by genotoxic stress. Cell. 2003;115:565–576. doi: 10.1016/s0092-8674(03)00895-x. [DOI] [PubMed] [Google Scholar]
- Johnson E.S.2004. Protein modification by SUMO. Annu. Rev. Biochem. 73355–382. [DOI] [PubMed] [Google Scholar]
- Kagey M.H., Melhuish T.A., Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;113:127–137. doi: 10.1016/s0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- Kashiwabara S., Zhuang T., Yamagata K., Noguchi J., Fukamizu A., Baba T. Identification of a novel isoform of poly(A) polymerase, TPAP, specifically present in the cytoplasm of spermatogenic cells. Dev. Biol. 2000;228:106–115. doi: 10.1006/dbio.2000.9894. [DOI] [PubMed] [Google Scholar]
- Kim H., Lee Y. Interaction of poly(A) polymerase with the 25-kDa subunit of cleavage factor I. Biochem. Biophys. Res. Commun. 2001;289:513–518. doi: 10.1006/bbrc.2001.5992. [DOI] [PubMed] [Google Scholar]
- Kim H., Lee J.H., Lee Y. Regulation of poly(A) polymerase by 14–3–3ε. EMBO J. 2003;22:5208–5219. doi: 10.1093/emboj/cdg486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman F.E., Manley J.L.2001. The BARD1–CstF-50 interaction links mRNA 3′ end formation to DNA damage and tumor suppression. Cell 104743–753. [DOI] [PubMed] [Google Scholar]
- Ko B., Gunderson S.I. Identification of new poly(A) polymerase-inhibitory proteins capable of regulating pre-mRNA polyadenylation. J. Mol. Biol. 2002;318:1189–1206. doi: 10.1016/s0022-2836(02)00240-1. [DOI] [PubMed] [Google Scholar]
- Kwak J.E., Wang L., Ballantyne S., Kimble J., Wickens M.2004. Mammalian GLD-2 homologs are poly(A) polymerases. Proc. Natl. Acad. Sci. 1014407–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakopoulou C.B., Nordvarg H., Virtanen A. A novel nuclear human poly(A) polymerase (PAP), PAPγ. J. Biol. Chem. 2001;276:33504–33511. doi: 10.1074/jbc.M104599200. [DOI] [PubMed] [Google Scholar]
- Li T., Evdokimov E., Shen R.F., Chao C.C., Tekle E., Wang T., Stadtman E.R., Yang D.C., Chock P.B.2004. Sumoylation of heterogeneous nuclear ribonucleoproteins, zinc finger proteins, and nuclear pore complex proteins: A proteomic analysis. Proc. Natl. Acad. Sci. 1018551–8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Liang M., Liang Y.Y., Brunicardi F.C., Feng X.H. SUMO-1/Ubc9 promotes nuclear accumulation and metabolic stability of tumor suppressor Smad4. J. Biol. Chem. 2003;278:31043–31048. doi: 10.1074/jbc.C300112200. [DOI] [PubMed] [Google Scholar]
- Martin G., Keller W. Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. EMBO J. 1996;15:2593–2603. [PMC free article] [PubMed] [Google Scholar]
- Martin G., Keller W., Doublie S. Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP. EMBO J. 2000;19:4193–4203. doi: 10.1093/emboj/19.16.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G., Moglich A., Keller W., Doublie S. Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) polymerase. J. Mol. Biol. 2004;341:911–925. doi: 10.1016/j.jmb.2004.06.047. [DOI] [PubMed] [Google Scholar]
- Matunis M.J., Wu J., Blobel G. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol. 1998;140:499–509. doi: 10.1083/jcb.140.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F. SUMO—Nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 2000;16:591–626. doi: 10.1146/annurev.cellbio.16.1.591. [DOI] [PubMed] [Google Scholar]
- Melchior F., Schergaut M., Pichler A. SUMO: Ligases, isopeptidases and nuclear pores. Trends Biochem. Sci. 2003;28:612–618. doi: 10.1016/j.tibs.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Keller W. mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr. Opin. Cell Biol. 1999;11:352–357. doi: 10.1016/S0955-0674(99)80049-0. [DOI] [PubMed] [Google Scholar]
- Mizrahi N., Moore C. Posttranslational phosphorylation and ubiquitination of the Saccharomyces cerevisiae Poly(A) polymerase at the S/G(2) stage of the cell cycle. Mol. Cell. Biol. 2000;20:2794–2802. doi: 10.1128/mcb.20.8.2794-2802.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouland A.J., Coady M., Yao X.J., Cohen E.A. Hypophosphorylation of poly(A) polymerase and increased polyadenylation activity are associated with human immunodeficiency virus type 1 Vpr expression. Virology. 2002;292:321–330. doi: 10.1006/viro.2001.1261. [DOI] [PubMed] [Google Scholar]
- Panse V.G., Hardeland U., Werner T., Kuster B., Hurt E. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J. Biol. Chem. 2004;279:41346–41351. doi: 10.1074/jbc.M407950200. [DOI] [PubMed] [Google Scholar]
- Papamichail M., Trangas T., Courtis N., Ioannides C., Cosmidou H., Pangalis G.A., Tsiapalis C.M. Poly(A)-polymerase levels in leukemia. Haematol. Blood Transfus. 1983;28:28–29. doi: 10.1007/978-3-642-68761-7_5. [DOI] [PubMed] [Google Scholar]
- Pichler A., Melchior F. Ubiquitin-related modifier SUMO1 and nucleocytoplasmic transport. Traffic. 2002;3:381–387. doi: 10.1034/j.1600-0854.2002.30601.x. [DOI] [PubMed] [Google Scholar]
- Preiss T., Muckenthaler M., Hentze M.W. Poly(A)-tail-promoted translation in yeast: Implications for translational control. RNA. 1998;4:1321–1331. doi: 10.1017/s1355838298980669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raabe T., Bollum F.J., Manley J.L.1991. Primary structure and expression of bovine poly(A) polymerase. Nature 353229–234. [DOI] [PubMed] [Google Scholar]
- Raabe T., Murthy K.G., Manley J.L. Poly(A) polymerase contains multiple functional domains. Mol. Cell. Biol. 1994;14:2946–2957. doi: 10.1128/mcb.14.5.2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangasamy D., Woytek K., Khan S.A., Wilson V.G. SUMO-1 modification of bovine papillomavirus E1 protein is required for intranuclear accumulation. J. Biol. Chem. 2000;275:37999–38004. doi: 10.1074/jbc.M007777200. [DOI] [PubMed] [Google Scholar]
- Reverter D., Lima C.D. A basis for SUMO protease specificity provided by analysis of human Senp2 and a Senp2–SUMO complex. Structure. 2004;12:1519–1531. doi: 10.1016/j.str.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Reverter D., Lima C.D. Insights into E3 ligase activity revealed by a SUMO–RanGAP1–Ubc9–Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Acosta G., Langereis M.A., Deyrieux A., Wilson V.G. Proteins of the PIAS family enhance the sumoylation of the papillomavirus E1 protein. Virology. 2005;331:190–203. doi: 10.1016/j.virol.2004.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryner L.C., Takagaki Y., Manley J.L. Multiple forms of poly(A) polymerases purified from HeLa cells function in specific mRNA 3′-end formation. Mol. Cell. Biol. 1989;9:4229–4238. doi: 10.1128/mcb.9.10.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H., Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- Salinas S., Briancon-Marjollet A., Bossis G., Lopez M.A., Piechaczyk M., Jariel-Encontre I., Debant A., Hipskind R.A. SUMOylation regulates nucleo-cytoplasmic shuttling of Elk-1. J. Cell Biol. 2004;165:767–773. doi: 10.1083/jcb.200310136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson D.A., Wang M., Matunis M.J. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 2001;276:21664–21669. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- Schul W., van Driel R., de Jong L. A subset of poly(A) polymerase is concentrated at sites of RNA synthesis and is associated with domains enriched in splicing factors and poly(A) RNA. Exp. Cell Res. 1998;238:1–12. doi: 10.1006/excr.1997.3808. [DOI] [PubMed] [Google Scholar]
- Scorilas A. Polyadenylate polymerase (PAP) and 3′ end pre-mRNA processing: Function, assays, and association with disease. Crit. Rev. Clin. Lab. Sci. 2002;39:193–224. doi: 10.1080/10408360290795510. [DOI] [PubMed] [Google Scholar]
- Scorilas A., Courtis N., Yotis J., Talieri M., Michailakis M., Trangas T.1998. Poly(A)polymerase activity levels in breast tumour cytosols. J. Exp. Clin. Cancer Res. 17511–518. [PubMed] [Google Scholar]
- Seeler J.S., Dejean A. SUMO: Of branched proteins and nuclear bodies. Oncogene. 2001;20:7243–7249. doi: 10.1038/sj.onc.1204758. [DOI] [PubMed] [Google Scholar]
- Shimazu T., Horinouchi S., Yoshida M. Multiple histone deacetylases and the CREB-binding protein regulate pre-mRNA 3′-end processing. J. Biol. Chem. 2007;282:4470–4478. doi: 10.1074/jbc.M609745200. [DOI] [PubMed] [Google Scholar]
- Steffan J.S., Agrawal N., Pallos J., Rockabrand E., Trotman L.C., Slepko N., Illes K., Lukacsovich T., Zhu Y.Z., Cattaneo E., et al. SUMO modification of Huntingtin and Huntington’s disease pathology. Science. 2004;304:100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- Tatham M.H., Jaffray E., Vaughan O.A., Desterro J.M., Botting C.H., Naismith J.H., Hay R.T.2001. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 27635368–35374. [DOI] [PubMed] [Google Scholar]
- Tatham M.H., Kim S., Jaffray E., Song J., Chen Y., Hay R.T. Unique binding interactions among Ubc9, SUMO and RanBP2 reveal a mechanism for SUMO paralog selection. Nat. Struct. Mol. Biol. 2005;12:67–74. doi: 10.1038/nsmb878. [DOI] [PubMed] [Google Scholar]
- Thuresson A.C., Astrom J., Astrom A., Gronvik K.O., Virtanen A. Multiple forms of poly(A) polymerases in human cells. Proc. Natl. Acad. Sci. 1994;91:979–983. doi: 10.1073/pnas.91.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian S.L., Kaneko S., Gonzales M.I., Bond G.L., Ward Y., Manley J.L. Identification and functional characterization of neo-poly(A) polymerase, an RNA processing enzyme overexpressed in human tumors. Mol. Cell. Biol. 2001;21:5614–5623. doi: 10.1128/MCB.21.16.5614-5623.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner S., Vagner C., Mattaj I.W. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes & Dev. 2000;14:403–413. [PMC free article] [PubMed] [Google Scholar]
- Vassileva M.T., Matunis M.J. SUMO modification of heterogeneous nuclear ribonucleoproteins. Mol. Cell. Biol. 2004;24:3623–3632. doi: 10.1128/MCB.24.9.3623-3632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vethantham V., Rao N., Manley J.L. Sumoylation modulates the assembly and activity of the pre-mRNA 3′ processing complex. Mol. Cell. Biol. 2007;27:8848–8858. doi: 10.1128/MCB.01186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel J.A., Johnson E.S., Reed S.I., Yates J.R. Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:45662–45668. doi: 10.1074/jbc.M409203200. [DOI] [PubMed] [Google Scholar]
- Yeh E.T., Gong L., Kamitani T. Ubiquitin-like proteins: New wines in new bottles. Gene. 2000;248:1–14. doi: 10.1016/s0378-1119(00)00139-6. [DOI] [PubMed] [Google Scholar]
- Zhao W., Manley J.L. Complex alternative RNA processing generates an unexpected diversity of poly(A) polymerase isoforms. Mol. Cell. Biol. 1996;16:2378–2386. doi: 10.1128/mcb.16.5.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Manley J.L. Deregulation of poly(A) polymerase interferes with cell growth. Mol. Cell. Biol. 1998;18:5010–5020. doi: 10.1128/mcb.18.9.5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Hyman L., Moore C. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelkovsky A.M., Kessler M.M., Moore C.L. Structure–function relationships in the Saccharomyces cerevisiae poly(A) polymerase. Identification of a novel RNA binding site and a domain that interacts with specificity factor(s) J. Biol. Chem. 1995;270:26715–26720. doi: 10.1074/jbc.270.44.26715. [DOI] [PubMed] [Google Scholar]