Abstract
Telomerase is a ribonucleoprotein enzyme that extends DNA at the chromosome ends in most eukaryotes. Since 1985, telomerase has been studied intensively and components of the telomerase complex have been identified from over 160 eukaryotic species. In the last two decades, there has been a growing interest in studying telomerase owing to its vital role in chromosome stability and cellular immortality. To keep up with the remarkable explosion of knowledge about telomerase, we compiled information related to telomerase in an exhaustive database called the Telomerase Database (http://telomerase.asu.edu/). The Telomerase Database provides comprehensive information about (i) sequences of the RNA and protein subunits of telomerase, (ii) sequence alignments based on the phylogenetic relationship and structure, (iii) secondary structures of the RNA component and tertiary structures of various subunits of telomerase, (iv) mutations of telomerase components found in human patients and (v) active researchers who contributed to the wealth of current knowledge on telomerase. The information is hierarchically organized by the components, i.e. the telomerase reverse transcriptase (TERT), telomerase RNA (TR) and other telomerase-associated proteins. The Telomerase Database is a useful resource especially for researchers who are interested in investigating the structure, function, evolution and medical relevance of the telomerase enzyme.
INTRODUCTION
Eukaryotic chromosomes are linear DNA molecules that contain special structures, called telomeres, which cap the ends of chromosomes to protect against end-to-end fusion or apoptosis (1,2). With a few exceptions from some insects, the telomeric DNA sequences are usually simple tandem repeats, e.g. (TTAGGG)n as in humans. During DNA replication, telomeric DNA shortens progressively mainly due to the end-replication problem (3,4). To overcome this problem, eukaryotic cells have evolved with a specialized reverse transcriptase (RT) enzyme, called telomerase, which functions to counteract this continuous degradation of telomeres by adding telomeric DNA repeats to the 3′-ends of chromosomes (5).
Telomerase is a ribonucleoprotein complex composed of two essential core components, the telomerase reverse transcriptase (TERT) protein and the telomerase RNA (TR), as well as several telomerase-associated accessorial proteins. The catalytic TERT protein synthesizes telomeric DNA repeats using a short sequence in the TR component as a template (6,7). The TERT gene was first identified in 1997 from the ciliate, Euplotes aediculatus and the yeast Saccharomyces cerevisiae (8). Since then, homologs of TERT have been identified in 105 species. TR sequences have currently been identified in 28 ciliates (9–11), 14 yeasts (12,13) and 43 vertebrates (14). The size of TR varies dramatically from ∼150 nt in ciliates, to 312–556 nt in vertebrates, and to over 1500 nt in yeasts. Remarkably, there is no similarity in TR sequence between these three groups of species. As a result, secondary structure models of TR have been independently established for each group of species using phylogenetic comparative analysis. The structures of TR from ciliates, yeasts and vertebrates share a similar pseudoknot structure near the template region, but vary substantially due to additional species-specific structural elements. For example, in addition to the universal pseudoknot domain, vertebrate TR contains the CR4–CR5 domain which is essential for enzymatic activity and the sno/scaRNA domain which is critical for TR biogenesis.
Due to its unusual evolutionary divergence, the telomerase holoenzyme varies significantly in its composition among different groups of species. A large number of putative telomerase-associated proteins have been identified in different groups of species through biochemical analyses such as UV cross-linking or immunoprecipitation. Interestingly, most of these proteins interact with telomerase in a species-specific manner. For example, the dyskerin protein complex appears to be associated with vertebrate telomerase (15), but not with yeast or ciliate telomerases. Since these associated proteins are not necessary to reconstitute telomerase activity in vitro, they likely function in the regulation and/or biogenesis of telomerase in vivo. However, the actual roles of many of these proteins remain unknown.
Structural studies of telomerase using NMR and crystallography have been limited to small structural domains or elements of the core components, TERT and TR. For the TERT protein, only the N-terminal TEN domain of Tetrahymena TERT has been successfully crystallized and the structure determined (16). However, the central portion of TERT protein contains seven motifs called RT motifs (1, 2, A, B, C, D, E) that are highly conserved among all reverse transcriptases. The RT domain of TERT likely folds into a structure similar to the available crystal structures of HIV and MMLV RTs (17,18). For this reason, the structure of HIV RT domain has often been used as a structural model for the TERT RT domain. For the TR component, while the secondary structure is known, the tertiary structure of the full-length RNA has not been determined. Nevertheless, NMR solution structures are available for helix II and stem-loop IV of Tetrahymena TR (19–21), as well as the pseudoknot (P2b/P3), P6 and P6.1 helices of human TR (22–24). While structural studies of small RNA and protein fragments have provided useful information, the structure of a catalytically active TR–protein complex will ultimately help reveal the mechanism of how this unique DNA polymerase functions.
Telomerase plays a vital role in the cellular immortality of stem cells. Mutations in telomerase genes affect the proliferation capacity of stem cells and have been linked to three human diseases, dyskeratosis congenita (DKC), aplastic anemia (AA) and idiopathic pulmonary fibrosis (IPF) (25–27). The reduction of telomerase activity correlates to telomere shortening and reduced proliferative capacity in cells from patients. The three types of DKC: (i) the X-linked-recessive, (ii) autosomal dominant and (iii) autosomal recessive, have been linked to mutations in the dyskerin gene (DKC1), TERT and TR genes and Nop10 gene, respectively (28–31). AA and IPF have been linked to mutations in both TERT and TR genes (26,27,32,33). It is puzzling that mutations in the TERT or TR gene at different sites can cause an array of clinical presentations associated with DKC, AA and/or IPF. The molecular mechanism explaining the unusual phenomenon of which mutations in the same gene cause these diseases remains to be elucidated.
The unusual evolutionary divergence of telomerase and its important role in cancer, aging and human diseases have attracted a great number of researchers who are devoted to telomerase research. This database aims to facilitate and expedite the research on telomerase biology by providing a comprehensive collection of information as a useful and accessible resource.
CONTENT OF THE DATABASE
The purpose of the Telomerase Database is to compile and organize known information about the telomerase ribonucleoprotein. Currently, components of the telomerase complex have been identified from over 160 different eukaryotic species. Structural information has also been available for several domains of the RNA and several protein subunits. Furthermore, multiple human diseases such as DKC, AA and IPF have been linked to a large number of mutations found in telomerase component genes. In the database, information is organized into six major pages: (i) Home/Overview, (ii) Sequences, (iii) Alignments, (iv) Structures, (v) Diseases and (vi) Researchers. The detailed features of these six main pages and other sub-level pages are described individually below.
Overview
The overview page within the main ‘Home’ page gives a broad and historical introduction to telomerase research for the general public and research scientists who are interested in telomerase. It also describes the organization of the database content, nomenclature of each telomerase component and other general information.
Sequences
The sequence page includes gene and protein sequences of telomerase components from over 160 eukaryotic species. Most sequences were obtained from GenBank (http://www.ncbi.nlm.nih.gov/) and have GenBank accession numbers assigned, while some sequences have not been deposited and were derived from original publications. The GenBank records and literature references for each sequence are provided through hyperlinks to NCBI. Partial or hypothetical sequences derived from genome sequencing or EST projects are indicated accordingly.
The main ‘Sequences’ page contains a table summarizing all the sequence data collected in the database. Sequence data were grouped in taxonomical order into eight individual tables: vertebrates, invertebrates, fungi, plants, algae, ciliates, other protists and viruses. Each table contains sequence data for TERT, TR, other telomerase-associated components and telomere repeats, presented as four individual columns. For the TERT and TR sequences, GenBank accession numbers are shown and linked directly to the original GenBank records. While the TERT genes have been identified in 105 species spanning across almost every major group of species, the TR gene has only been identified in 85 species from three groups of species, 28 ciliates, 14 yeasts and 43 vertebrates. To date, TR sequences have not been identified in invertebrates, plants, algae and non-ciliate protists. Interestingly, only 26 among these 164 species have had both their TERT and TR components identified. Other telomerase-associated protein components from human, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Tetrahymena thermophila and Euplotes aediculatus are listed. Known telomere repeat sequences from 89 species are shown.
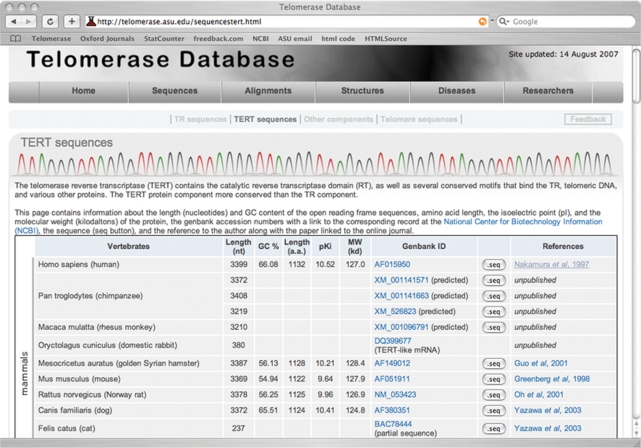
Four sub-level pages, designated as the ‘TR sequences’, ‘TERT sequences’, ‘Other components’ and ‘Telomere sequences’, provide more detailed information derived from the primary sequences. For example, in addition to the GenBank accession numbers, the sub-level TERT sequences page includes information such as the length, GC content, sequence in text format and literature reference of each sequence (Figure 1).
Figure 1.
A snapshot of the TERT sequences page of the database. The TERT sequence table includes detailed information for each sequence, i.e. nucleotide length, GC content, amino acid length, pKi, molecular weight, GenBank accession numbers, sequences in text format and references.
Alignments
The ‘Alignments’ page provides multiple sequence alignments for the telomerase core components, TERT and TR and telomerase-associated proteins such as dyskerin, Nop10, Gar1, NHP2, Est1 and Est3. To ensure accuracy of these results, partial and putative sequences were excluded from the sequence alignment. Due to the lack of sequence similarity between groups of species, TR sequences from vertebrates, Saccharomyces, Kluyveromyces, Tetrahymena, Paramecium and Euplotes were aligned independently. The alignments of TR sequences were performed manually based on structural information and sequence conservation. Compared to TR, the amino acid sequences of TERT are relatively conserved across all eukaryotes, especially in the central RT domain. Nonetheless, the N-terminal and C-terminal domains of TERT from several species, such as nematodes, Plasmodium and sea squirts, are variable containing truncations, insertions and sequence variations. To achieve a more accurate and meaningful alignment, full-length TERT sequences from different groups, i.e. vertebrates, invertebrates, fungi, plants/algae, ciliates and other protists, were aligned independently. The sequences of the conserved RT domain from all known TERT proteins were aligned. The aligned sequences can either be viewed online in a separate window or be downloaded in the rich text or plain text file format. For each sequence alignment, a phylogenetic tree was constructed using either the MEGA 3.1 program or the DNAML program from the Phylip package v.3.66 to reveal the evolutionary relationship between sequences. The phylogenetic trees can either be viewed online in a separate window or downloaded in the JPG or PDF file format.
Structures
The ‘Structures’ page includes secondary structures for the TR and tertiary structures of the TR, TERT, telomerase-associated proteins and telomere-binding proteins. The secondary structure section includes TRs from vertebrates, fungi and ciliates. These secondary structure models were generated based on the sequence alignments as well as published structures. The RNA secondary structures are available for download in both JPG and PDF formats and can also be viewed online.
The tertiary structures of different telomerase components, including TR and TERT fragments, are grouped by component and sorted by species. The PDB files that describe atomic coordinates of the structures are available for download directly from this database or through links to the Protein Data Bank (http://www.rcsb.org/pdb/).
Diseases
The ‘Diseases’ page summarizes disease-related mutations found in genes encoding various telomerase components, TR, TERT, dyskerin encoded by the DKC1 gene and Nop10. The disease background section briefly describes the association of telomerase mutations with the three human diseases, DKC, AA and IPF. DKC has three distinctive forms, X-linked recessive, autosomal dominant and autosomal recessive. To date, 25 mutations in TR, 18 mutations in TERT, 44 mutations in the DKC1 gene and 1 Nop10 mutation have been identified in patients with one or more of the three diseases. The positions of each mutation are indicated on the secondary structure of the TR component, or on the figures that illustrate the organization of protein motifs or the positions of exons and introns of the protein genes. Detailed information such as the nucleotide position, identity or nature of the mutations, the presentation of the disease in patients, and the literature references that describe the mutation are listed in individual tables.
Researchers
The last page is the ‘Researchers’ page listing the names and affiliations of over 150 principle investigators who have and continue to contribute to the wealth of knowledge on the telomerase enzyme. The objective of the list is to facilitate communications and interactions among researchers, which will then promote collaborations, e.g. sharing of reagents, unpublished results or ideas. The large number of researchers in the telomerase field reflects the importance and medical potential of this unique enzyme.
DATABASE ACCESS
The Telomerase Database is accessible via the World Wide Web at http://telomerase.asu.edu. The database was constructed with the intention of cross platform/browser accessibility. The database contents can be easily browsed using Internet Explorer for PC, Safari for Mac or Firefox for PC/Mac and the files can be downloaded in several file formats. To create a more uniform user interface, elements available for download are displayed as hyperlinks for each of the file formats, while buttons generate a separate window for online viewing of sequences, alignments and structures. Literature references are linked to PubMed, where the abstracts can be retrieved immediately. A feedback button has been included in most pages to allow users to actively participate in the improvement of the database.
FUTURE WORK
The sequence pages will be expanded as more TR and TERT sequences are identified. The sequence alignments will continuously be refined as new sequences are added to the database. The collection of RNA secondary structures will be expanded to ultimately include all identified TRs. In the next release, we plan to include more comprehensive information on telomerase-associated proteins and telomere-binding proteins. The database will be updated regularly to include newly published sequences and new mutations identified from patients.
ACKNOWLEDGEMENTS
We thank Tracy Niday for critical reading of the manuscript. Research on telomerase in the authors’ laboratory is supported by National Science Foundation (CAREER Award—MCB0642857 to J.-L.C.). J.D.P. and R.V.O. are supported by National Science Foundation via the Research Experience for Undergraduates (REU) program. Funding to pay the Open Access publication charges for this article was provided by National Science Foundation.
Conflict of interest statement. None declared
REFERENCES
- 1.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 2.de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- 3.Watson JD. Origin of concatameric T7 DNA. Nat. New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 4.Olovnikov AM. A theory of marginotomy. J. Theoret. Biol. 1973;41:181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- 5.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 6.Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- 7.Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 8.Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 9.Romero DP, Blackburn EH. A conserved secondary structure for telomerase RNA. Cell. 1991;67:343–353. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- 10.Lingner J, Hendrick LL, Cech TR. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes Dev. 1994;8:1984–1998. doi: 10.1101/gad.8.16.1984. [DOI] [PubMed] [Google Scholar]
- 11.McCormick-Graham M, Romero DP. Ciliate telomerase RNA structural features. Nucleic Acids Res. 1995;23:1091–1097. doi: 10.1093/nar/23.7.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seto AG, Livengood AJ, Tzfati Y, Blackburn EH, Cech TR. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev. 2002;16:2800–2812. doi: 10.1101/gad.1029302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dandjinou AT, Levesque N, Larose S, Lucier JF, Abou Elela S, Wellinger RJ. A phylogenetically based secondary structure for the yeast telomerase RNA. Curr. Biol. 2004;14:1148–1158. doi: 10.1016/j.cub.2004.05.054. [DOI] [PubMed] [Google Scholar]
- 14.Chen J-L, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 15.Dez C, Henras A, Faucon B, Lafontaine D, Caizergues-Ferrer M, Henry Y. Stable expression in yeast of the mature form of human telomerase RNA depends on its association with the box H/ACA small nucleolar RNP proteins Cbf5p, Nhp2p and Nop10p. Nucleic Acids Res. 2001;29:598–603. doi: 10.1093/nar/29.3.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobs SA, Podell ER, Cech TR. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat. Struct. Mol. Biol. 2006;13:218–225. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- 17.Georgiadis MM, Jessen SM, Ogata CM, Telesnitsky A, Goff SP, Hendrickson WA. Mechanistic implications from the structure of a catalytic fragment of Moloney murine leukemia virus reverse transcriptase. Structure. 1995;3:879–892. doi: 10.1016/S0969-2126(01)00223-4. [DOI] [PubMed] [Google Scholar]
- 18.Sarafianos SG, Das K, Tantillo C, Clark A.D., Jr, Ding J, Whitcomb JM, Boyer PL, Hughes SH, Arnold E. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 2001;20:1449–1461. doi: 10.1093/emboj/20.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Fender J, Legassie JD, Jarstfer MB, Bryan TM, Varani G. Structure of stem-loop IV of Tetrahymena telomerase RNA. EMBO J. 2006;25:3156–3166. doi: 10.1038/sj.emboj.7601195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richards RJ, Theimer CA, Finger LD, Feigon J. Structure of the Tetrahymena thermophila telomerase RNA helix II template boundary element. Nucleic Acids Res. 2006;34:816–825. doi: 10.1093/nar/gkj481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richards RJ, Wu H, Trantirek L, O’Connor CM, Collins K, Feigon J. Structural study of elements of Tetrahymena telomerase RNA stem-loop IV domain important for function. RNA. 2006;12:1475–1485. doi: 10.1261/rna.112306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leeper T, Leulliot N, Varani G. The solution structure of an essential stem-loop of human telomerase RNA. Nucleic Acids Res. 2003;31:2614–2621. doi: 10.1093/nar/gkg351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leeper TC, Varani G. The structure of an enzyme-activating fragment of human telomerase RNA. RNA. 2005;11:394–403. doi: 10.1261/rna.7222505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Theimer CA, Blois CA, Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol. Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 25.Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 26.Fogarty PF, Yamaguchi H, Wiestner A, Baerlocher GM, Sloand E, Zeng WS, Read EJ, Lansdorp PM, Young NS. Late presentation of dyskeratosis congenita as apparently acquired aplastic anaemia due to mutations in telomerase RNA. Lancet. 2003;362:1628–1630. doi: 10.1016/S0140-6736(03)14797-6. [DOI] [PubMed] [Google Scholar]
- 27.Armanios MY, Chen JJ, Cogan JD, Alder JK, Ingersoll RG, Markin C, Lawson WE, Xie M, Vulto I, et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N. Engl. J. Med. 2007;356:1317–1326. doi: 10.1056/NEJMoa066157. [DOI] [PubMed] [Google Scholar]
- 28.Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat. Genet. 1998;19:32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- 29.Vulliamy T, Marrone A, Goldman F, Dearlove A, Bessler M, Mason PJ, Dokal I. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413:432–435. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- 30.Armanios M, Chen JL, Chang YP, Brodsky RA, Hawkins A, Griffin CA, Eshleman JR, Cohen AR, Chakravarti A, et al. Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. Proc. Natl Acad. Sci. USA. 2005;102:15960–15964. doi: 10.1073/pnas.0508124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walne AJ, Vulliamy T, Marrone A, Beswick R, Kirwan M, Masunari Y, Al-Qurashi FH, Aljurf M, Dokal I. Genetic heterogeneity in autosomal recessive dyskeratosis congenita with one subtype due to mutations in the telomerase-associated protein NOP10. Hum. Mol. Genet. 2007;16:1619–1629. doi: 10.1093/hmg/ddm111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamaguchi H, Calado RT, Ly H, Kajigaya S, Baerlocher GM, Chanock SJ, Lansdorp PM, Young NS. Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N. Engl. J. Med. 2005;352:1413–1424. doi: 10.1056/NEJMoa042980. [DOI] [PubMed] [Google Scholar]
- 33.Tsakiri KD, Cronkhite JT, Kuan PJ, Xing C, Raghu G, Weissler JC, Rosenblatt RL, Shay JW, Garcia CK. Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proc. Natl Acad. Sci. USA. 2007;104:7552–7557. doi: 10.1073/pnas.0701009104. [DOI] [PMC free article] [PubMed] [Google Scholar]