Abstract
The BioMagResBank (BMRB: www.bmrb.wisc.edu) is a repository for experimental and derived data gathered from nuclear magnetic resonance (NMR) spectroscopic studies of biological molecules. BMRB is a partner in the Worldwide Protein Data Bank (wwPDB). The BMRB archive consists of four main data depositories: (i) quantitative NMR spectral parameters for proteins, peptides, nucleic acids, carbohydrates and ligands or cofactors (assigned chemical shifts, coupling constants and peak lists) and derived data (relaxation parameters, residual dipolar couplings, hydrogen exchange rates, pKa values, etc.), (ii) databases for NMR restraints processed from original author depositions available from the Protein Data Bank, (iii) time-domain (raw) spectral data from NMR experiments used to assign spectral resonances and determine the structures of biological macromolecules and (iv) a database of one- and two-dimensional 1H and 13C one- and two-dimensional NMR spectra for over 250 metabolites. The BMRB website provides free access to all of these data. BMRB has tools for querying the archive and retrieving information and an ftp site (ftp.bmrb.wisc.edu) where data in the archive can be downloaded in bulk. Two BMRB mirror sites exist: one at the PDBj, Protein Research Institute, Osaka University, Osaka, Japan (bmrb.protein.osaka-u.ac.jp) and the other at CERM, University of Florence, Florence, Italy (bmrb.postgenomicnmr.net/). The site at Osaka also accepts and processes data depositions.
INTRODUCTION
Nuclear magnetic resonance (NMR) spectroscopy is unique among biophysical approaches in its ability to provide a broad range of atomic-level information relevant to the structural, dynamic and chemical properties of biological macromolecules. Newly developed NMR techniques (1–3) are enabling the collection of NMR data from increasingly larger systems, including membrane bound biopolymers. BioMagResBank (4,5) (BMRB, www.bmrb.wisc.edu) is designed to empower structural biologists, macromolecular chemists and other scientists who use these data to address questions concerning a variety of macromolecular phenomena, including macromolecular recognition, enzyme reaction mechanisms, macromolecular ligand interactions, protein conformational stability, protein folding pathways and nucleic acid conformations. The database consists of four large archives: (i) over 4500 entries containing NMR spectral values and derived information (assigned chemical shifts, coupling constants, relaxation parameters, residual dipolar couplings, etc.); these are cross-referenced to 3D structures in the PDB as available. (ii) The NMR Restraints Grid, a relational database for NMR restraints sets that is continually updated as new entries are released at the PDB (6,7). (iii) An archive of multi-dimensional, time-domain NMR data associated with structural studies of proteins and nucleic acids. (iv) A database containing time-domain NMR spectral data, spectral images, spectral peak lists and assigned chemical shifts for more than 250 metabolites.
The goals in several laboratories around the world are to streamline procedures for structure determination from NMR data and to develop ways to improve the quality of structures. Promising approaches include the automation of NMR peak assignments (8–10) and computerized sorting of ambiguous structural restraints (11–13). Dipolar couplings induced by field-dependent magnetic ordering without (14,15) and with (16) the addition of agents that promote molecular ordering are providing a new source of structural information (in addition to NOEs and J couplings)—and new parameters to be included in BMRB entries.
The BMRB website has been expanded recently to provide easy access to the available information and to incorporate additional reference and educational information for the NMR community and those outside the community interested in using NMR spectroscopy in their research. A new ADIT-NMR deposition system has been developed in collaboration with the RCSB-PDB that provides NMR spectroscopists a one-stop site for submitting atomic coordinates, restraints and experimental data to both the RCSB-PDB and BMRB. This deposition system is accessible from the BMRB (http://deposit.bmrb.wisc.edu/bmrb-adit/), PDBj-BMRB (http://nmradit.protein.osaka-u.ac.jp/bmrb-adit/) and RCSB-PDB (http://deposit.rcsb.org/) websites. The BMRB has worked with the NMR community to develop a data model for NMR experimental results called NMR-STAR. NMR-STAR v3 is used to drive the new ADIT-NMR data deposition system and data validation systems used at BMRB. A draft version of the data model has been released on the BMRB and RCSB-PDB websites and a final version will be published in the near future.
BMRB is a member of the Research Collaboratory for Structural Bioinformatics (RCSB) (6) and the wwPDB (7). BMRB mirror sites exist at Osaka University, Japan, and at CERM in Florence, Italy, with the Osaka facility also being a data deposition and processing site. In addition to the Osaka and Florence collaborations, BMRB collaborates closely with the RCSB-PDB, the EBI, the CCPN group at the University of Cambridge, the NMR protein structural genomics groups especially the NESG, NMR metabolomic/metabonomic groups and the National Magnetic Resonance Facility at Madison (NMRFAM) and with many other groups in the NMR community.
DATA ACQUISITION AND METHODS
ADIT-NMR data deposition system
Authors submit data to BMRB primarily through the ADIT-NMR data deposition system. The system was developed using the ADIT software platform provided by the RCSB-PDB as the starting point. Extensive modifications were made to the underlying code to create a one-stop deposition that allows NMR spectroscopists to submit in one session all data required for both a wwPDB atomic coordinate deposition and a BMRB NMR experimental data entry. While the overall look and feel of the ADIT interface was maintained, a number of features were added, including default values for many fields, drop-down enumerated lists for easy entry selection and linked fields to facilitate data entry and decrease the time required to complete a deposition. Users can deposit information interchangeably into the PDB or BMRB sides of the system; information common to both sides is transferred automatically to the other side. Our statistics show that many users have required 2 h or less to fill out an original deposition to either the BMRB or PDB side of the system, and then required only an additional 15 min to complete the deposition to the other side. In addition to the standard information (molecular system studied, sample and sample condition descriptions, NMR experiments carried out, atomic coordinates and restraints files, assigned chemical shifts) more than 15 other kinds of NMR data can be deposited, including derived relaxation parameters, thermodynamic and kinetic data. Uploaded files containing atomic coordinates are pre-checked automatically for format issues and, if the depositor wishes, can be validated by ProCheck prior to actual deposition. This choice generates reports that the depositor can use to evaluate the quality of the structure; at this point, the depositor can choose to continue with the deposition or to refine the structure further.
Upon receipt of a completed deposition, the depositor is provided immediately with a PDB or BMRB ID. A finished PDB entry along with the atomic coordinate and restraints data are processed and annotated at either the RCSB-PDB or PDBj. Depositions containing NMR experimental data are processed at either BMRB or PDBj-BMRB. The BMRB attempts to adhere to the recommendations of the IUPAC-IUBMB-IUPAB Inter-Union Task Group published in 1998 (17) in setting standards for the deposition of NMR spectroscopic data and the use of protein and nucleic acid residue and atom nomenclature. BMRB annotators process entries using software developed at BMRB and tools donated by other groups including the AVS system created by Moseley et al. (18).
Tools are available from the BMRB website for generating template NMR-STAR files that can be populated with assigned chemical shifts, coupling constants, hydrogen exchange data and a variety of relaxation data. A number of third-party software applications read and write NMR-STAR compliant files. Data in this format can be easily processed by BMRB annotators to generate finished BMRB entries. However, NMR researchers make use of a variety of software packages that output data in other formats. The STARch (STAR conversion handler) tool has been developed at BMRB for converting files generated by a variety of software tools into the NMR-STAR format (http://www.bmrb.wisc.edu/tools/). Depositors are encouraged to use this or other similar tools to convert their data into the NMR-STAR format before submission to BMRB. One advantage of having assigned chemical shift data in the NMR-STAR format is that they can be visualized and checked (for example by LACS (19) for consistent chemical shift referencing) through uploading to the BMRB website at (http://www.bmrb.wisc.edu/vis_serv/).
NMR-STAR data model
The ADIT-NMR deposition system, as well as many other BMRB tools, is driven by the NMR-STAR data dictionary. The dictionary has evolved over time with the current version 3.1 published at (http://www.bmrb.wisc.edu/formats.html). The NMR-STAR dictionary and the resulting NMR-STAR files exported by BMRB conform to the STAR (Self-defining Text Archival and Retrieval) specifications defined by Hall and coworkers (20–22). The PDB exchange dictionary used by the RCSB-PDB to drive the ADIT deposition system and to construct mmCIF files (23,24) is another application of the STAR specification. The NMR-STAR data model contains over 4600 defined data tags organized into more than 300 categories and 80 category groups. Data defined by category groups represent real objects such as molecular assemblies, molecular entities (polymers and non-polymers), samples and sample conditions, NMR experiments, chemical shift referencing and many kinds of NMR spectral data and derived data (assigned chemical shifts, coupling constants, relaxation parameters, etc.). NMR-STAR and the derived NMRIF (NMR information file) are compatible with the PDB exchange dictionary and extend the data model to include information relevant to biological NMR spectroscopy. NMR-STAR is extensible and can accommodate the addition of new NMR parameters of interest to the biomolecular NMR community (recent additions have included solid-state NMR and information from NMR experiments on metabolomic standard compounds). The tag-value nature of the NMR-STAR format and tabular organization of the NMR-STAR data model make it easy to create a corresponding relational data model or XML format.
CONTENT AND ACCESS
Assigned chemical shifts
Assigned NMR chemical shifts constitute the bulk of the data submitted by depositors directly to BMRB. The first BMRB entries (∼1200) were constructed by annotators from data extracted from journal publications. Nearly all BMRB entries released since 1996 are the result of author depositions. The archive now contains over 4400 entries representing more than 2.5 million chemical shifts for individual proteins, peptides, DNA and RNA molecules, and a variety of macromolecular complexes.
BMRB provides statistical tables containing the mean, minimum, maximum and standard deviation for the reported chemical shifts for each 1H, 13C, 15N and 31P atom found in the common amino and nucleic acids. These tables provide results both from the full BMRB archive and also from the subset of the archive that is believed to represent diamagnetic molecular systems filtered to remove suspected erroneous chemical shift values. Additional tables provide lists of chemical shifts that fall outside 3 SD from the mean for each atom in the 20 common amino acids. These shifts are often of great interest and are linked to the source entry to allow users to quickly determine the type of molecular system involved. For those interested in obtaining lists of chemical shifts for a particular atom from any of the common biopolymer residues (protein, DNA or RNA), a query system is provided that will generate either an HTML page or a tab-delimited file that can be downloaded for further analysis.
The BMRB Query Grid Interface (http://www.bmrb.wisc.edu/search/query_grid/initial_grid.html) can be used to perform a number of common queries. Examples are: (i) select entries with molecular systems containing proteins, DNA or RNA; (ii) select protein only entries; (iii) select entries with matched PDB entries; (iv) select protein entries that reported the use of IUPAC recommended chemical shift referencing (17) and (v) select protein entries containing at least one reported ligand (metal ion and/or organic compound). The Query Grid Tool also can be used to locate BMRB entries that contain other kinds of NMR spectral or derived data including, coupling constants, T1, T2, heteronuclear NOE and order parameters, residual dipolar couplings and hydrogen exchange rates.
From the ‘Search Archive’ page (http://www.bmrb.wisc.edu/search/), individual entries can be retrieved by BMRB accession number, BMRB deposition code or a related PDB code. Other query tools located on the page allow users to search the archive for entries by particular author, biopolymer name, range of pH or temperature and BMRB or PDB accession code (http://www.bmrb.wisc.edu/cgi-bin/nmrbrowse.cgi). A FASTA search engine allows entries to be retrieved on the basis of sequence relatedness to a query sequence.
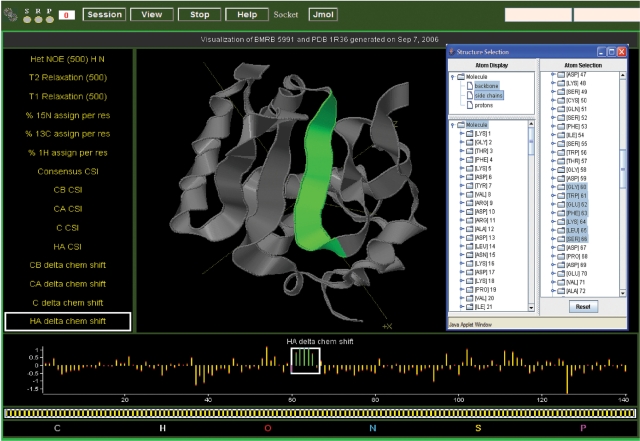
The DEVise data visualization tool (25,26) has been integrated into the BMRB website as both a query tool and a presentation tool for data in or derived from individual entries. The system provides web-based interactive plots for a wide variety of data either as individual plots or as multiple plots linked to a 3D structure (data derived from PDB). DEVise histogram visualizations of chemical shift data for selected amino acid atoms can be used to locate outlier chemical shifts; a shift-click on a bar in the histogram retrieves a list of the entries containing the chemical shift values that contribute to the bar. Hyperlinks allow direct access to the entries. The user can select whether the histogram visualizations utilize statistics from the full database or from the ‘diamagnetic’, filtered database. For individual entries depending on data availability, plots can be chosen that display the chemical shift index (CSI) (27,28), the experimental chemical shifts versus CSI delta, coupling constants, heteronuclear NOEs, T1 values, T2 values or order parameters. In cases where a BMRB entry can be matched to a PDB entry, these plots are combined with an interactive 3D Jmol (http://www.jmol.org/) model of the structure. The data plots and 3D image are linked, making it possible to easily visualize the residues in the structure that have the data characteristics selected from the plots (Figure 1). For entries that contain 13C chemical shifts for amino acid Hα, Cα and Cβ atoms, LACS (19) visualizations are available that provide a statistical identification of outlier chemical shift values and an indication of the correctness of the chemical shift referencing used in the entry.
Figure 1.
DEVise interactive data visualization for data from BMRB entry 5991 [(39); BMRB accession code 5991] with a model from PDB entry 1R36 [(39); PDB accession code 1R36]. In the figure, the kinds of data that can be plotted are listed on the left, with the ‘HA delta chem shift’ selected, and the plot shown at the bottom of the visualization. A stretch of strong positive delta chemical shift values (indicative of a β-strand) is selected by placing the box around the values in the bar graph plot. These values are highlighted in green, and the corresponding residues are colored in green in the ribbon diagram of the PDB model (rendered using Jmol) present at the center of the visualization. The selected residues are highlighted in the window that displays the protein sequence.
Restraints
The NMR derived restraints used to calculate 3D models of biological macromolecules are the key to understanding the quality of the reported structures. In the past, the restraints files available from the PDB depositions were not annotated with respect to the deposited coordinates and the restraint data were present in a variety of formats that were difficult to access. BMRB, through close collaboration with members of the CCPN group, the EBI-MSD team and the Universities of Utrecht and Nijmegen, recently developed tools (i) to parse the vast majority of the restraints data from the PDB into the NMR-STAR format, (ii) to make the atom nomenclature in the parsed files consistent with the corresponding atomic coordinates and (iii) to filter the restraints to remove redundant, duplicate, inconsistent and other values that do not contribute to the structure calculation (29,30). The derived data, currently representing over 3600 PDB entries, are available at BMRB in the ‘parsed’ database, Database of Converted Restraints (DOCR) and Filtered Restraints Database (FRED), collectively called the ‘NMR Restraints Grid.’ Table 1 contains statistics for the PDB entries used to construct the databases. The NMR Restraints Grid provides an interface for locating entries with specific kinds of restraints (distance, torsion angle, residual dipolar coupling, etc.) and links to the PDB entry that is the source of the data. Data from previous implementations of the FRED database have been used by several groups to recalculate 3D structures (31–34). In each case, the resulting structures were compared with the originals to assess current NMR structure calculation protocols.
Table 1.
NMR Restraints Grid statisticsa
| PDB entry sets | Entry counts |
|---|---|
| Total PDB entries | 42 073 |
| Total PDB NMR entries | 6587 |
| Total PDB NMR entries in DOCR and FRED | 3620 |
| NMR entries with parsable restraintsb | 3233 |
| NMR entries with parsable distance restraintsb | 3202 |
| Total NMR entries with restraints parsing and matching issuesc | 419 |
| <80% restraints linked to coordinates (DOCR) | 184 |
| <33% restraints remaining after filtering (FRED) | 100 |
| Maximum distance violations >2 Å | 209 |
| RMS distance violations >0.25 Å | 151 |
aThe reported statistics were collected on 13 September 2007.
bRestraints files in the formats used by the software packages X-PLOR, CYANA, Discover, Amber and EMBOSS can be parsed.
cEntries with parsing and matching issues are the result of limitations of the software used to carry out these functions and not indicative of the quality of the data in the files. The total number of these entries is smaller than the sum of the entries for the subheadings as many entries have more than one type of issue.
Metabolomics standard compounds
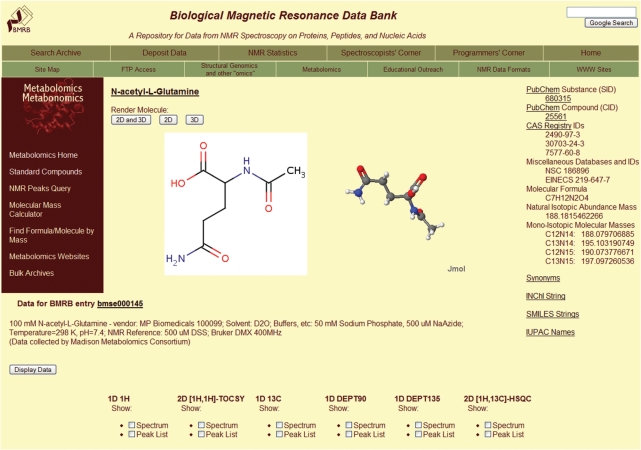
BMRB maintains an archive of NMR spectral data for metabolomic compounds. The current standard compound data have been provided by the Madison Metabolomic Consortium (MMC). Annotated data for over 270 compounds are available with 1H 1D, 13C 1D, 13C 90° DEPT. 13C 135° DEPT, 1H-1H TOCSY and 1H-13C HSQC natural abundance spectra present for most compounds (Figure 2). Peak lists, spectral transition lists, images of the spectra and assigned peak lists are being added to the archive. BMRB will accept NMR data for metabolites from any group interested in making a deposition.
Figure 2.
Summary page for the metabolite N-acetyl-l-glutamine from the BMRB metabolomics website.
The archive can be queried for data on specific compounds by name, KEGG ID, PubChem compound or substance ID and PDB ID. 1H, 13C or paired 1H-13C spectral peak lists taken from spectral of biological samples can be used to query for compounds with matching properties. An ordered list of matching compounds is returned prioritized with the best match at the top of the list and linked to a visual presentation of the chemical shift alignments for the subject data and the compounds returned by the query. A list of mass values also can be used to query the archive and retrieve compounds that have mass values within defined limits.
Time-domain data
For the past few years, BMRB has been archiving and making available sets of raw spectral time-domain data collected from the NMR experiments carried out to derive assigned chemical shifts provided in BMRB entries and the restraints used to calculate atomic structures deposited at the wwPDB. Data sets of this kind, which now number more than 65 entries, are welcome from any research group. Most of the original entries were contributed by two groups funded by the NIH Protein Structure Initiative: the Northeast Structural Genomics Consortium (NESG) and the Center for Eukaryotic Structural Genomics (CESG). Recently, the first set of time-domain data for NOESY spectra used in the determination of an RNA structure were deposited by Butcher and coworkers [(35); BMRB accession code 15417]. All of these datasets are listed on the BMRB website (http://www.bmrb.wisc.edu/data_library/timedomain/), and links can be found on the summary page for those entries that have associated time-domain data. Data for over 500 NMR experiments are included, representing more than 100 unique NMR experiments. Often the pulse sequence used and the acquisition parameter files are available for the experiments. In some cases, processing parameters and peak lists have been provided along with the experimental parameters. Three of the entries, those corresponding to references [(36–38); BMRB accession codes 6128, 6176 and 6318] contain the full input and output data sets for the software used to derive the peaklists, automated peak assignments, NOE peak lists and restraints used to calculate the 3D structures of the proteins studied.
Educational resources
BMRB contains educational resources for NMR spectroscopists and software programmers interested in probing the archive. A special section of the BMRB website highlights biomolecules that have been studied extensively by NMR spectroscopy. A brief description of each molecule is given, along with its historical significance and links to associated BMRB and PDB entries and literature references.
ACKNOWLEDGEMENTS
We gratefully acknowledge our many fruitful interactions with members of the scientific community and, in particular, Wim Vranken in developing the NMR Restraints Grid, the PDB, the Protein Data Bank Japan (PDBj), the National Magnetic Resonance Facility at Madison (NMRFAM), the Collaborative Computational Program on NMR (CCPN), the European Bioinformatics Institute (EBI) and the Condor team. This work was funded by National Library of Medicine, National Institutes of Health (LM05799). Funding to pay the Open Access publication charges for this article was provided by the National Library of Medicine, National Institutes of Health (LM05799).
Conflict of interest statement. None declared.
REFERENCES
- 1.Pervushin KV, Riek R, Wider G, Wüthrich K. Transverse relaxation-optimized spectroscopy (TROSY) for NMR studies of aromatic spin systems in 13C-labeled proteins. J. Am. Chem. Soc. 1998;120:6394–6400. [Google Scholar]
- 2.Pervushin KV, Wider G, Wüthrich K. Single transition-to-single transition polarization transfer (ST2-PT) in [15N,1H]-TROSY. J. Biomol. NMR. 1998;12:345–348. doi: 10.1023/A:1008268930690. [DOI] [PubMed] [Google Scholar]
- 3.Pervushin KV, Wider G, Riek R, Wüthrich K. The 3D NOESY-[H-1,N-15,H-1]-ZQ-TROSY NMR experiment with diagonal peak suppression. Proc. Natl Acad. Sci. USA. 1999;96:9607–9612. doi: 10.1073/pnas.96.17.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ulrich EL, Markley JL, Kyogoku Y. Creation of a nuclear magnetic resonance data repository and literature database. Prot. Seq. Data Anal. 1989;2:23–37. [PubMed] [Google Scholar]
- 5.Seavey BR, Farr EA, Westler WM, Markley JL. A relational database for sequence-specific protein NMR data. J. Biomol. NMR. 1991;1:217–236. doi: 10.1007/BF01875516. [DOI] [PubMed] [Google Scholar]
- 6.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berman HM, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 8.Eghbalnia HR, Bahrami A, Wang L, Assadi A, Markley JL. Probabilistic identification of spin systems and their assignments including coil-helix inference as output (PISTACHIO) J. Biomol. NMR. 2005;32:219–233. doi: 10.1007/s10858-005-7944-6. [DOI] [PubMed] [Google Scholar]
- 9.Moseley HN, Monleon D, Montelione GT. Automatic determination of protein backbone resonance assignments from triple resonance nuclear magnetic resonance data. Meth. Enzymol. 2001;339:91–108. doi: 10.1016/s0076-6879(01)39311-4. [DOI] [PubMed] [Google Scholar]
- 10.Bartels C, Billeter M, Güntert P, Wüthrich K. Automated sequence-specific NMR assignment of homologous proteins using the program GARANT. J. Biomol. NMR. 1996;7:207–213. doi: 10.1007/BF00202037. [DOI] [PubMed] [Google Scholar]
- 11.Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 12.Lopez-Mendez B, Güntert P. Automated protein structure determination from NMR spectra. J. Am. Chem. Soc. 2006;128:13112–13122. doi: 10.1021/ja061136l. [DOI] [PubMed] [Google Scholar]
- 13.Rieping W, Habeck M, Bardiaux B, Bernard A, Malliavin TE, Nilges M. ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007;23:381–382. doi: 10.1093/bioinformatics/btl589. [DOI] [PubMed] [Google Scholar]
- 14.Prestegard JH. New techniques in structural NMR – anisotropic interactions. Nat. Struct. Biol. 1998;5:517–522. doi: 10.1038/756. [DOI] [PubMed] [Google Scholar]
- 15.Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. NMR evidence for slow collective motions in cyanometmyoglobin. Nat. Struct. Biol. 1997;4:292–297. doi: 10.1038/nsb0497-292. [DOI] [PubMed] [Google Scholar]
- 16.Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 17.Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wüthrich K. Recommendations for the presentation of NMR structures of proteins and nucleic acids. Pure Appl. Chem. 1998;70:117–142. doi: 10.1006/jmbi.1998.1852. [DOI] [PubMed] [Google Scholar]
- 18.Moseley HN, Sahota G, Montelione GT. Assignment validation software suite for the evaluation and presentation of protein resonance assignment data. J. Biomol. NMR. 2004;28:341–355. doi: 10.1023/B:JNMR.0000015420.44364.06. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Eghbalnia HR, Bahrami A, Markley JL. Linear analysis of carbon-13 chemical shift differences and its application to the detection and correction of errors in referencing and spin system identifications. J. Biomol. NMR. 2005;32:13–22. doi: 10.1007/s10858-005-1717-0. [DOI] [PubMed] [Google Scholar]
- 20.Hall SR. The STAR File: a new format for electronic data transfer and archiving. J. Chem. Inf. Comput. Sci. 1991;31:326–333. [Google Scholar]
- 21.Hall SR, Spadaccini N. The STAR File: detailed specifications. J. Chem. Inf. Comput. Sci. 1994;34:505–508. [Google Scholar]
- 22.Hall SR, Cook A.PF. STAR dictionary definition language: initial specification. J. Chem. Inf. Comput. Sci. 1995;35:819–825. [Google Scholar]
- 23.Bourne PE, Berman HM, McMahon B, Watenpaugh KD, Westbrook J, Fitzgerald P.MD. The macromolecular crystallographic information file (mmCIF) Meth. Enzymol. 1997;277:571–590. doi: 10.1016/s0076-6879(97)77032-0. [DOI] [PubMed] [Google Scholar]
- 24.Westbrook JD, Bourne PE. STAR/mmCIF: an extensive ontology for macromolecular structure and beyond. Bioinformatics. 2000;16:159–168. doi: 10.1093/bioinformatics/16.2.159. [DOI] [PubMed] [Google Scholar]
- 25.Yao H, Wenger K, Livny M. DEVise and the JavaScreen: visualization on the Web'; San Jose, California. 2000. Proceedings of the SPIE Conference on Visual Data Exploration and Analysis. [Google Scholar]
- 26.Livny M, Ramakrishnan R, Beyer K, Chen G, Donjerkovic D, Lawande S, Myllymaki J, Wenger K. Proceedings of ACM SIGMOD. Tucson, Arizona: 1997. DEVise: integrated querying and visual exploration of large datasets. [Google Scholar]
- 27.Wishart DS, Sykes BD, Richards FM. Relationship between Nuclear Magnetic Resonance chemical shift and protein secondary structure. J. Mol. Biol. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- 28.Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical shift data. J. Biomol. NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 29.Doreleijers JF, Nederveen A, Vranken W, Lin J, Bonvin A.M.JJ, Kaptein R, Markley JL, Ulrich EL. BioMagResBank databases DOCR and FRED containing converted and filtered sets of experimental NMR restraints and coordinates from over 500 protein PDB structures. J. Biomol. NMR. 2005;32:1–12. doi: 10.1007/s10858-005-2195-0. [DOI] [PubMed] [Google Scholar]
- 30.Doreleijers JF, Mading S, Maziuk D, Sojourner K, Yin L, Zhu J, Markley JL, Ulrich EL. BioMagResBank database with sets of experimental NMR constraints corresponding to the structures of over 1400 biomolecules deposited in the Protein Data Bank. J. Biomol. NMR. 2003;26:139–146. doi: 10.1023/a:1023514106644. [DOI] [PubMed] [Google Scholar]
- 31.Nederveen A, Doreleijers JF, Vranken W, Miller Z, Spronk C.A.EM, Nabuurs SB, Guntert P, Livny M, Markley JL, et al. RECOORD: a recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins. 2005;59:662–672. doi: 10.1002/prot.20408. [DOI] [PubMed] [Google Scholar]
- 32.Bardiaux B, Malliavin TE, Nilges M, Mazur AK. Comparison of different torsion angle approaches for NMR structure determination. J. Biomol. NMR. 2006;34:153–166. doi: 10.1007/s10858-006-6889-8. [DOI] [PubMed] [Google Scholar]
- 33.Nabuurs SB, Nederveen AJ, Vranken W, Doreleijers JF, Bonvin AM, Vuister GW, Vriend G, Spronk CA. DRESS: a database of REfined solution NMR structures. Proteins. 2004;55:483–486. doi: 10.1002/prot.20118. [DOI] [PubMed] [Google Scholar]
- 34.Lee SY, Zhang Y, Skolnick J. TASSER-based refinement of NMR structures. Proteins. 2006;63:451–456. doi: 10.1002/prot.20902. [DOI] [PubMed] [Google Scholar]
- 35.Marcheschi RJ, Staple DW, Butcher SE. Programmed ribosomal frameshifting in SIV is induced by a highly structured RNA stem-loop. J. Mol. Biol. 2007;26:652–663. doi: 10.1016/j.jmb.2007.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vinarov DA, Lytle BL, Peterson FC, Tyler E, Volkman BF, Markley JL. Cell-free protein production and labeling protocol for NMR-based structural proteomics. Nat. Methods. 2004;1:149–153. doi: 10.1038/nmeth716. [DOI] [PubMed] [Google Scholar]
- 37.Lytle BL, Peterson FC, Qui SH, Luo M, Zhao Q, Markley JL, Volkman BF. Solution structure of a ubiquitin-like domain of tubulin-folding cofactor B. J. Biol. Chem. 2004;279:46787–46793. doi: 10.1074/jbc.M409422200. [DOI] [PubMed] [Google Scholar]
- 38.Peterson FC, Lytle BL, Sampath S, Vinarov DA, Tyler EM, Shahan M, Markley JL, Volkman BF. Solution structure of thioredoxin h1 from Arabidopsis thaliana. Protein Sci. 2004;14:2195–2200. doi: 10.1110/ps.051477905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee GM, Donaldson LW, Pufall MA, Kang H-S, Pot I, Graves BJ, McIntosh LP. The structural and dynamic basis of Ets-1 DNA binding autoinhibition. J. Biol. Chem. 2005;280:7088–7099. doi: 10.1074/jbc.M410722200. [DOI] [PubMed] [Google Scholar]