Abstract
Mycolic acids are a major constituent of the mycobacterial cell wall, and they form an effective permeability barrier to protect mycobacteria from antimicrobial agents. Although the chemical structures of mycolic acids are well established, little is known on their biosynthesis. We have isolated a mycolate-deficient mutant strain of Mycobacterium smegmatis mc2-155 by chemical mutagenesis followed by screening for increased sensitivity to novobiocin. This mutant also was hypersensitive to other hydrophobic compounds such as crystal violet, rifampicin, and erythromycin. Entry of hydrophobic probes into mutant cells occurred much more rapidly than that into the wild-type cells. HPLC and TLC analysis of fatty acid composition after saponification showed that the mutant failed to synthesize full-length mycolic acids. Instead, it accumulated a series of long-chain fatty acids, which were not detected in the wild-type strain. Analysis by 1H NMR, electrospray and electron impact mass spectroscopy, and permanganate cleavage of double bonds showed that these compounds corresponded to the incomplete meromycolate chain of mycolic acids, except for the presence of a β-hydroxyl group. This direct identification of meromycolates as precursors of mycolic acids provides a strong support for the previously proposed pathway for mycolic acid biosynthesis involving the separate synthesis of meromycolate chain and the α-branch of mycolic acids, followed by the joining of these two branches.
Mycobacteria include important human pathogens. Tuberculosis, caused by Mycobacterium tuberculosis, is the most deadly infectious disease caused by bacteria. Between 2 and 3 million people die from this disease every year, and the total number of infected individuals is estimated to be 1.7 billion around the world (1). In industrialized countries, a serious problem has been the outbreak of multidrug-resistant tuberculosis (2). M. leprae causes leprosy. In addition, “atypical mycobacteria,” which include the M. avium-intracellulare complex, have become important causative agents of opportunistic infections among AIDS patients (3).
Mycolic acids are large, α-alkyl β-hydroxy fatty acids with the general structure of R1-CH(OH)-CH(R2)-COOH, where R1 is a meromycolate chain containing typically 50–56 carbons and R2 is a shorter α-branch containing 22–26 carbons. They constitute the inner leaflet of the lipid bilayer of the mycobacterial cell wall and form an effective barrier to the penetration of antibiotics and chemotherapeutic agents (4–6).
Understanding the biosynthesis of mycolic acids is important because these lipids are the target for an important chemotherapeutic agent, isoniazid (7), and may offer various attractive targets for the development of novel agents. It appears that the final step of mycolic acid synthesis involves the Claisen-type condensation between the meromycolate chain and the α-alkyl chain, a reaction that was demonstrated in corynebacteria, which synthesize a smaller version of mycolate, corynemycolates (8). We know, however, little about the biosynthesis of meromycolates. Past approaches included the identification of putative meromycolate precursors in mycobacterial cultures (9, 10) and the development of cell-free systems for the biosynthesis of longer fatty acids (11–13). These studies have produced results that are only suggestive. Thus, long-chain fatty acids, including saturated C30-to-C56 fatty acids and mono- and diunsaturated C22-to-C47 acids, were isolated from whole cells of M. tuberculosis H37Rv (9), but there is no evidence that they serve as precursors of mycolate synthesis. One cell-free system resulted in the synthesis of mycolic acids, but the results did not shed light on the details of the pathway (12). In another cell-free system, various long-chain fatty acids were synthesized, but the products were not characterized in detail (11).
M. smegmatis synthesizes α-mycolates as the major mycolate species (14). The α1 and α2 series are full-length mycolic acids most frequently containing 78 and 79 carbons, respectively. The α1-mycolate contains two cis double bonds in the meromycolate chain, whereas α2 contains a cis double bond and a trans double bond with an adjacent methyl branch. M. smegmatis also produces shorter α′-mycolic acids, most often 64 carbons in length. We report here the isolation of a mycolate-deficient mutant of M. smegmatis that accumulates, as precursors of mycolate synthesis, novel long-chain fatty acids that correspond in structure to the incomplete meromycolate chains of mycolic acids.
MATERIALS AND METHODS
Mutagenesis and Screening of Mutants.
A 5-ml culture of M. smegmatis mc2-155 was grown at 37°C in Middlebrook 7H9 broth (Difco) supplemented with 10% Middlebrook OADC enrichment (Difco), 0.2% glycerol, and 0.05% Tween-80 to midexponential phase. The cells were washed and resuspended in 0.1 M sodium-citrate buffer (pH 5.5). 1-Methyl-3-nitro-1-nitrosoguanidine was added to the culture at a final concentration of 0.1 mg/ml. The sample was incubated for 2 hr at 37°C. The cells were collected by centrifugation, washed twice with 0.1 M sodium-phosphate buffer (pH 7.0), and plated onto 7H11 (Difco) plates. The resulting colonies were transferred onto 7H11 master plates and also were replica-plated onto 7H11 plates containing 40 μg/ml of novobiocin. The plates were incubated for 3 days at 30°C. Colonies that did not grow on the novobiocin-containing plates were identified, and the corresponding colony from the master plate was collected for further analysis. Unless indicated otherwise, all subsequent experiments were done with cells grown at 30°C.
Drug Susceptibility Test.
Minimal inhibitory concentrations were determined by 2-fold serial broth dilution. The inoculum was 5 × 104 cells per tube, and the results were read after a 48-hr incubation at 30°C.
Saponification and Derivatization of Fatty Acids.
The cells (10 mg dry weight) first were lyophilized and then were treated for 1 hr at 110°C in 2 ml of saponification mixture (5% KOH in 88% methylcellosolve) (15). The mixture was acidified by adding 1.5 ml of 6 N HCl and was extracted twice with CHCl3. The dried lipids were dissolved in 0.5 ml of CHCl3 containing 2 mg of K2CO3, and 25 μl each of p-bromophenacyl bromide and of dicyclohexyl 18-crown-6-ether catalyst (fatty acid derivatization kit, Alltech Associates) was added. The vials were heated for 45 min at 85°C with constant stirring.
HPLC and TLC Analysis of Fatty Acids.
The fatty acid derivatives were separated by a reverse-phase HPLC. A Beckman C18 Ultrasphere (5-μm spherical) column was used. A solvent gradient of CH2Cl2 in methanol, consisting of four linear segments (0–13 min, 0–10%; 13–14 min, 10–25%; 14–34 min, 25–70%; and 34–49 min, constantly at 70%) was run, with a flow rate of 1 ml/min. The detection was by absorption at 260 nm.
Fatty acid methyl esters were separated by TLC on Whatman silica gel 60 plates with hexane/ethyl acetate (9:1, vol/vol) as solvent and were visualized by charring.
NMR and MS Analysis of Lipids.
Proton NMR was performed on a Bruker AM500 NMR spectrometer by Numega Resonance Labs (San Diego). Electrospray–MS (ES-MS) and electron impact–MS were performed by Mass Consortium (San Diego) and by M-Scan (West Chester, PA), respectively. In the latter procedure, the source temperature was 200°C and the spectra were acquired with electrons accelerated at 70 eV.
Permeability Assay.
Entry of hydrophobic agents into mycobacterial cells was measured as described previously (16). Two hydrophobic probes, [14C]chenodeoxycholate (specific activity, 50 mCi/mmol) and [14C]erythromycin (specific activity, 40 mCi/mol), were tested.
RESULTS
Isolation of Hypersusceptible Mutants.
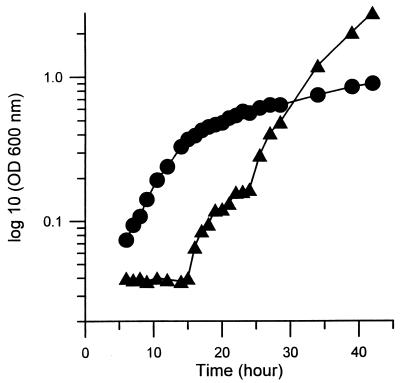
Approximately 2,000 mutagenized colonies of M. smegmatis mc2-155 were screened for increased sensitivity to novobiocin. Because some of these mutations may occur at gene loci that are essential for the viability of the cells, screening of mutants was performed at a lower growth temperature, 30°C, to allow isolation of conditional lethal mutants. Eleven novobiocin-hypersensitive mutants were isolated. One mutant, named as 155NS1, was chosen for further studies. The growth rate of the 155NS1 mutant was not reduced in comparison with the parent in the supplemented 7H9 broth (Materials and Methods) at 30°C, and duration of the initial lag phase was shorter than in the wild-type strain (Fig. 1). It failed to show visible growth, however, at 37°C (not shown), and therefore is a temperature-sensitive mutant. Although there was a slow increase in optical density after 40 hr, this appeared to be due to the growth of revertants, as judged by the properties of the cells that were recovered after several days of growth.
Figure 1.
Growth of the parent mc2-155 (▴) and mutant 155NS1 (●) in 7H9 broth medium at 30°C. Cultures were diluted 40-fold with fresh 7H9 broth supplemented with 10% Difco OADC enrichment. At times indicated, small portions of the cultures were removed and the cell density was determined by OD600.
Drug susceptibility was determined for this mutant and the parent strain (see Materials and Methods). The mutant also showed increased sensitivity to several other hydrophobic compounds, including rifampicin, erythromycin, chloramphenicol, and crystal violet (Table 1); for example, the rifampicin minimal inhibitory concentration of the mutant was 128-fold lower than that of the wild-type strain. This mutant was also extremely sensitive to many β-lactams (data not shown).
Table 1.
Minimal inhibitory concentrations (μg/ml) of some lipophilic inhibitors
| Strain | Novobiocin | Rifampicin | Erythromycin | Chloramphenicol | Crystal violet |
|---|---|---|---|---|---|
| mc2-155 | 128 | 64 | 16 | 16 | 8 |
| 155NS1 | 32 | 0.5 | 4 | 8 | 0.5 |
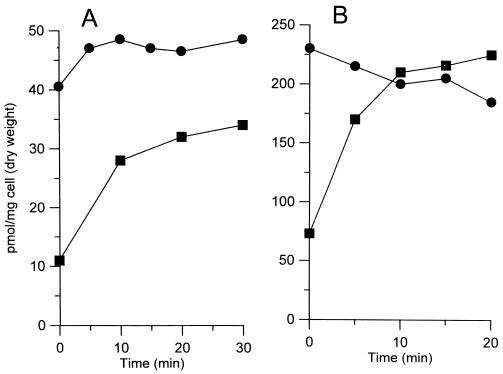
One possible cause for the increased sensitivity of the 155NS1 mutant to various hydrophobic drugs is the increased cell wall permeability. To test this hypothesis, we determined the accumulation kinetics of two hydrophobic agents, [14C]chenodeoxycholate and [14C]erythromycin, by whole cells. Indeed, both probes entered much more rapidly into the mutant cells than into the cells of the wild-type parent strain (Fig. 2). It is difficult to correlate quantitatively the increased uptake of antibiotics with the decrease of minimal inhibitory concentrations, because the contribution of other factors, such as target affinity and efflux pumps, is unknown. Nevertheless, these results strongly suggested that the increased cell wall permeability was responsible, at least partially, for the enhanced sensitivity of the 155NS1 mutant to hydrophobic drugs. The extreme sensitivity of this mutant to many β-lactams apparently was caused by a different mechanism, which is not the subject of this paper.
Figure 2.
Entry of erythromycin (A) or chenodeoxycholate (B) into whole cells of mc2-155 (■) or 155NS1 (●). To 1-ml cell suspensions in 0.1 M K-phosphate buffer (pH 7.0) preincubated at 37°C, [14C]erythromycin or [14C]chenodeoxycholate was added at time zero to a final concentration of 5 μM. At various time points, 50-μl portions of the suspension were removed, filtered, and washed. The radioactivity retained on the filter was determined by scintillation counting.
HPLC and TLC Analysis of Fatty Acids.
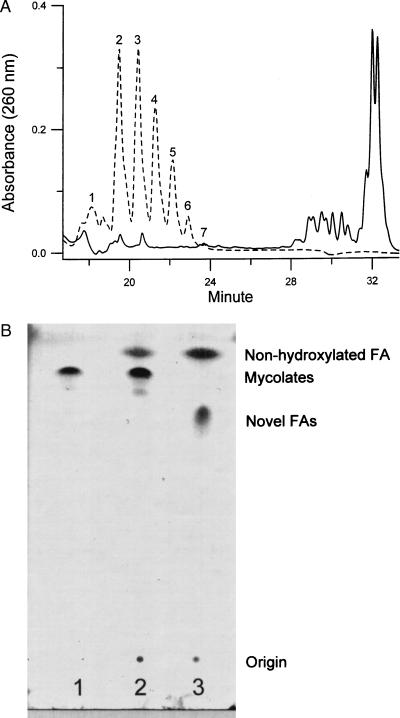
To identify the functions affected by the mutation, we analyzed the fatty acid compositions of the mutant by HPLC. We followed a procedure that was reported to saponify all fatty acid esters in the envelope (15). They then were derivatized with p-bromophenacyl bromide and analyzed in HPLC by using a gradient system. Addition of fatty acid derivatives as internal standards established that the derivatives eluted during the first 13 min contained short-chain fatty acids (C14–C20) (data not shown). Mycolic acids were eluted between 28 and 33 min (Fig. 3A, solid line), as confirmed by 1H-NMR and ES-MS (data not shown). Comparing the fatty acid profiles of the mutant with that of the parent strain, we noted that the mutant was devoid of mycolic acids (Fig. 3A, broken line). At the same time, a cluster of novel fatty acids appeared at significant levels in the mutant, whereas such fatty acids were not detected in the parent strain (Fig. 3A). These novel fatty acids were eluted between 18 and 24 min and apparently were shorter than mycolic acids but significantly longer than the C14–C20 fatty acids. This result was highly reproducible, and the relative intensity of individual peaks was always the same.
Figure 3.
HPLC and TLC analysis of fatty acids. (A) Fractionation of the p-bromophenacyl esters of fatty acids from the mc2-155 (solid line) or the 155NS1 mutant (dashed line) by HPLC. Cells (10 mg dry weight) were saponified and derivatized with p-bromophenacyl bromide as described in Materials and Methods. The fatty acid esters were dissolved in 100 μl of CH2Cl2, and 10 μl of this solution was applied to the HPLC column. Only fractions eluted between 18 and 34 min are shown. (B) TLC analysis of fatty acid methyl esters. Lanes: 1, methyl esters of mycolates purified from mc2-155; 2, esters of fatty acids from mc2-155; 3, esters of fatty acids from 155NS1. TLC was developed with hexane/ethyl acetate (9:1) as described in Materials and Methods. The weak spot underneath the mycolate ester spot in lane 2 contains mostly epoxy-mycolate, based on two-dimensional TLC analysis (not shown).
TLC of fatty acid methyl esters also showed that with the sample from 155NS1 the spot corresponding to mycolate esters has disappeared completely and was replaced with a new spot migrating more slowly (Fig. 3B). Thus, 155NS1 appears to be a mycolate-deficient mutant and the observed novel fatty acids likely represent precursors of mycolic acids.
NMR and MS Analysis of the Novel Fatty Acids.
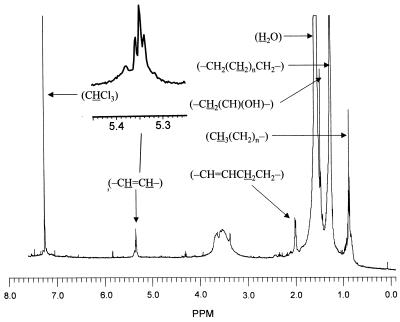
To identify these novel lipids, three fractions from HPLC, fractions A (peak 1), B (peak 2), and C (peaks 3–6) (Fig. 3A, broken line), were collected and subjected to analysis. 1H NMR spectra were determined both before and after the saponification of the samples. There was little difference between the spectra of the three fractions. We show in Fig. 4, as an example, the spectrum of the fraction B that was saponified to remove the p-bromophenacyl group. Besides the contaminating water signal around 1.55 ppm and the CHCl3 signal at 7.23 ppm used as the internal standard, several resonances were observed and were quantified by using the phenyl ring protons of the p-bromophenacyl group at 7.65 and 7.78 ppm in the spectra of unsaponified esters (not shown). The assignment of resonances was assisted greatly by the reported chemical shift values for various protons in mycolic acids (17). Besides the large number of protons belonging to (-CH2(CH2)nCH2-) group at 1.25 ppm, between 3 and 4 protons of (-CH⩵CH-) group were seen at 5.35 ppm, and about 8 protons belonging to (-CH⩵CHCH2CH2-) were seen at 2.00 ppm. These signals thus show the presence of two double bonds, which appear to be cis because the resonance at 5.35 ppm appears as a broad triplet (enlarged in Fig. 4) that is most easily interpreted as a triplet of doublets with only small J-splitting values (less than 5 Hz). Further, if these were trans double bonds, α-methyl branches that always accompany trans double bonds in mycolic acids should have been present, but there was no indication of a doublet at 0.95 ppm. As shown below, MS analysis suggested the presence of a hydroxyl group in these fatty acids, and the NMR data were consistent with this idea, showing a small signal at 3.38 ppm possibly corresponding to the methine proton in (-CH2(CH)(OH)-), as well as a sharp signal at 1.47 ppm presumably corresponding to the methylene protons (-CH2(CH)(OH)-) in this structure.
Figure 4.
Proton NMR spectrum of fatty acids purified from mutant 155NS1. p-Bromophenacyl esters of fatty acids were collected from the HPLC fractions and were saponified again to remove the p-bromophenacyl group. Free fatty acids were extracted into CDCl3 under acidic conditions and were analyzed with a 500-MHz spectrometer. We show here the spectrum of fatty acids derived from fraction B from HPLC (peak 2 of Fig. 3A) as an example. Assignment of several resonances is shown.
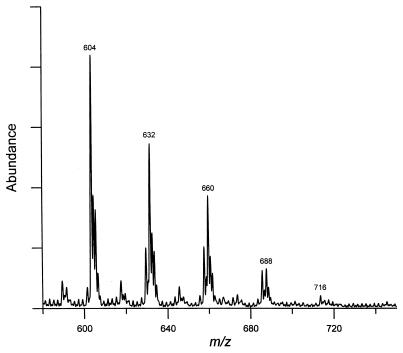
For ES-MS analysis, all three fractions collected from HPLC were resaponified to remove the p-bromophenacyl groups. ES-MS was performed with a negative ionization mode. The most abundant ions in the mass spectra of fractions A and B were at m/z 548 and 576, respectively (data not shown). Fraction C, containing 5 peaks from HPLC, showed major ions at m/z 604, 632, 660, 688, and 716 (Fig. 5). Thus, the peaks eluted later in the HPLC separation produced major ions with higher m/z values. (A control experiment with α-mycolic acids from the parent strain produced a major peak at m/z 1122, as expected.) We conclude that the novel fatty acids observed in the 155NS1 mutant are composed of seven major species with molecular weights of 549, 577, 605, 633, 661, 689, and 717.
Figure 5.
ES-MS of fatty acids from the mutant 155NS1. Fatty acid p-bromophenacyl esters from HPLC were collected and resaponified to remove the p-bromophenacyl group. The resultant free fatty acids were subjected to EM-MS analysis. We show here the spectrum of fatty acids derived from fraction C of HPLC (peaks 3–6 of Fig. 3A).
The simplest interpretation, based on the NMR and ES-MS data, is that these fatty acids belong to a homologous series differing only in the chain length, because the molecular weight difference between two adjacent species was always 28, the size of -(CH2)2-. Assuming that these compounds are straight chain fatty acids containing two double bonds, the observed molecular weights were consistent only with those fatty acids containing one hydroxyl group.
To determine the location of the double bonds, fraction A was oxidized by permanganate/periodate (18) and, after acidification, the fragments were extracted by t-butanol and analyzed by ES-MS. Two new peaks at m/z 241 and 285 were obtained (data not shown), representing the mono- and dicarboxylic fragments CH3(CH2)13COOH and HOOC(CH2)14COOH, respectively. Identical results were obtained with fraction C. These results also indicated that the 31-carbon segment containing the methyl terminus of the molecule did not contain the presumed single hydroxyl group. Based on these results, as well as the electron impact–MS data described below, we propose the structures of the novel fatty acids in the 155NS1 mutant as CH3(CH2)13CH⩵CH(CH2)14CH⩵CH(CH2)2n+1CH(OH)CH2COOH, where n is from 0 to 6. Their methyl esters showed lower Rf values on TLC in comparison with mycolate methyl esters (Fig. 3B); this is as expected because esters of hydroxy fatty acids showed lower Rf values than those of nonhydroxylated fatty acids, and, among the former, the esters of shorter acids showed slower migration than mycolic acid esters (19).
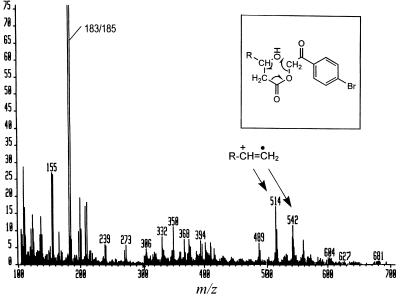
To confirm the presence of the hydroxyl group and its location, p-bromophenacyl esters (combined peaks 2 and 3 of Fig. 3A) were analyzed by electron impact–MS. As shown in Fig. 6, the major fragmentation products at m/z 514 and 542, presumably corresponding to [CH3(CH2)13CH⩵CH(CH2)14CH⩵CH(CH2)3 or 5CH⩵CH2]+⋅, were those generated by the specific fragmentation process caused by the presence of a 3-OH group as was shown earlier for p-bromophenacyl esters of α-mycolates (20), which also contain 3-OH groups (Fig. 6 Inset).
Figure 6.
Electron impact–MS of fatty acids accumulated in 155NS1. A mixture of peaks 2 and 3 from Fig. 3 was analyzed. Fragmentation shown in the Inset (from ref. 19) generates the ions with m/z of 514 and 542 (see text), CO2, and p-bromophenacylalcohol, the last compound fragmented further to produce the most prominent doublet at m/z 183/185, as also was seen in ref. 19.
DISCUSSION
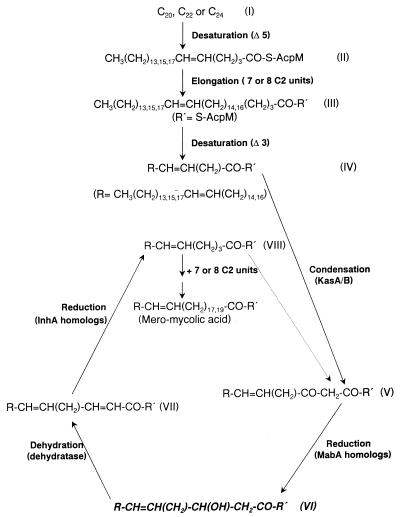
As described in the Introduction, many of the steps in the biosynthesis of mycolate are still obscure. In this study, we used a novel approach, the isolation of mycolic acid-deficient mutant strains. Our previous studies showed that mycolic acids play a critical role in establishing the low fluidity and the low permeability of the mycobacterial cell wall and that they contribute to the intrinsic resistance of mycobacteria to hydrophobic drugs (5, 16, 21). We reasoned that mutants that are hypersensitive to various hydrophobic drugs are likely to contain defects in their cell wall components, especially mycolic acids. Indeed, a novobiocin-hypersensitive mutant of M. smegmatis, 155NS1, also was shown to be hypersusceptible to several other hydrophobic agents and to take up hydrophobic probes much more rapidly than did the wild-type parent. It lacked mycolic acids nearly completely, and this supports the hypothesis that mycolates are responsible for producing the low-fluidity permeation barrier in the cell wall (4–6). The mutant also accumulated significant amounts of putative precursors, i.e., long-chain fatty acids that were not detected in the wild-type strain. Detailed chemical characterizations of these novel fatty acids by NMR, ES-MS, and electron impact–MS showed that they contained 36–48 carbon atoms and had the same basic structure as the meromycolate chain of mycolic acids, although they were shorter than the 52 carbon meromycolate needed for the synthesis of the major C78 α-mycolate species and contained a β-hydroxyl group. These results thus indicate that meromycolic acids are the precursors of mycolic acids. Although tracer amounts of long-chain fatty acids were synthesized successfully earlier in a cell-free system (11), this study represents the isolation and characterization of meromycolates as precursors of mycolic acids. We note that a mycolate-deficient mutant of M. smegmatis has been reported earlier (22). Although the mutant was hypersusceptible to penicillin, and TLC confirmed the absence of the mycolate spot, the HPLC separation of p-bromophenacyl esters of fatty acids showed a poorly resolved peak with the retention time identical to that of mycolate esters, and the nature of these fatty acids remained unclear (22). In contrast, the structural elucidation of the accumulated fatty acids in this study allows us to discuss a likely scheme for the biosynthesis of the meromycolate chain (Fig. 7).
Figure 7.
Presumed pathway for the biosynthesis of the meromycolate chain of mycolic acids in M. smegmatis. The β-hydroxy fatty acids accumulated in the 155NS1 mutant are highlighted (structure VI). Although the second desaturation step here is depicted to occur at the 3 position, there currently is no strong evidence for this proposal. R′ indicates acyl carrier protein, probably -S-AcpM.
There is evidence that the point of the divergence of mycolic acid biosynthesis from that of conventional fatty acids is at the C22–C26 level. A series of Δ5-unsaturated C22, C24, and C26 fatty acids were detected at low concentrations in lipid extracts of mycobacteria, and these acids are structurally similar to the methyl terminus of the meromycolate chain (10). This led to the proposal that a Δ5-desaturase operates on the saturated C24 acid to yield C24:1, followed by its elongation. This pathway was supported by additional experimental data (23, 24), although direct incorporation of these proposed precursors into mycolic acids has not yet been demonstrated and the accumulation of a hexacosanoyl-ACP in isoniazid-treated M. tuberculosis (25) suggests that the desaturation may occur later in the pathway.
The steps leading from the 24:1 cis Δ5 fatty acids to the complete meromycolate chain, the C50–C56 fatty acids, remain unknown. However, it generally is assumed that two similar reaction cycles, each of which is composed of desaturation followed by rounds of elongation steps, are involved (see ref. 6 and Fig. 7). The first rounds of elongation steps would take place with Δ5-unsaturated C20–C24 fatty acids as the initial substrates (I→II), resulting in the addition of seven or eight C2 units (II→III). At this point, the second desaturation catalyzed presumably by a Δ3-desaturase could occur (III→IV), which is followed by additional rounds of elongation. The details of the presumed elongation steps in the second round are shown in Fig. 6. They involve a condensation reaction (IV→V) catalyzed by a β-ketoacyl-ACP synthase, reduction by a 3-oxoacyl-ACP reductase (V→VI), dehydration by a β-hydroxyacyl-ACP dehydratase (VI→VII), and another reduction step by an enoyl-ACP reductase (VII→VIII). Each cycle will add two-carbon units, and this cycle is repeated until the meromycolate is synthesized. A β-hydroxy fatty acid is generated in each cycle, and we assume that this corresponds to the novel long-chain fatty acids identified in this study (highlighted in Fig. 7). Thus, the mutation in 155NS1 presumably is at the β-hydroxyacyl-ACP dehydratase gene. Likely, the activity of this enzyme is not totally absent, and its low activity produces progressively lower levels of the elongated products, as seen in Fig. 3A. In fact, the putative C48 β-hydroxymeromycolate, with the formula weight of 717, was only barely detectable. Because the production of the most abundant species of mycolate in M. smegmatis, the C78 species (14), requires the condensation between the C26 fatty acid (containing the C24 α-chain) and C52 meromycolate, this progressive diminution of the larger meromycolate species clearly makes the condensation difficult, causing the virtual absence of α-mycolic acids. It is less clear why the content of α′-mycolate, most commonly containing 64 carbons (14), also was diminished. However, because α′-mycolates are only minor species even in the parent strain (ref. 14; also see Fig. 3A), the decreased supply of meromycolates could have a more pronounced effect on the synthesis of α′-mycolate.
Pathogenic mycobacteria, such as M. tuberculosis and the M. avium-intracellulare complex, produce slightly different α-mycolates. Instead of the double bonds, they contain two cyclopropane groups (26). It is noteworthy that the two major long-chain fatty acids detected from M. tuberculosis, a cis, cis-3,4-, and 15,16-dimethylenetetratriacontanoic acid (C36) and a C37 acid containing two cyclopropane rings and an α,β unsaturation (10), are analogs of the structures IV and VII in Fig. 7, respectively. These pathogenic species also synthesize oxygenated mycolic acids, such as ketomycolates and methoxymycolates. The biosynthesis of these functional groups, including cyclopropane and methoxy, has been elucidated recently (17, 27–29).
Fatty acid synthesis in mycobacteria is probably similar, in terms of its mechanistic principles, to that in other bacteria. However, there are often multiple proteins that carry out the same reaction, and, because of differences in substrate specificity, each may play a unique role in the physiological regulation of the spectrum of products. At least two types of enzyme systems, fatty acid synthase (FAS) I and FAS II, are present in mycobacteria (6). Short-chain fatty acids, e.g., C16, are synthesized by FAS I, which is a single polypeptide with multiple catalytic activities. Gene fas, coding for FAS I, was identified in the M. tuberculosis genome (29). In contrast, FAS II appears to consist of dissociable enzyme components. The M. tuberculosis genome contains an operon with four genes presumably encoding components of the FAS II, fabD-acpM-kasA-kasB (30). AcpM is an acyl carrier protein that may be involved in the synthesis of longer-chain fatty acids, including meromycolates (25). In response to isoniazid treatment, M. tuberculosis accumulated a protein species that consists of a covalent complex of isoniazid, AcpM, and KasA (25). Mutations in kasA were found in isoniazid-resistant clinical isolates (25). These results suggested that KasA, a homolog of known condensing enzymes, is a part of FAS II and is involved in the chain elongation in meromycolate synthesis. KasA and KasB are highly related (66% identity) but their elongation capacities might be different. The identities of the proteins catalyzing the other steps of the elongation cycle currently are unknown. Two other genes, mabA-inhA, which are cotranscribed, are present in M. tuberculosis (17). Although InhA was shown to have enoyl-ACP reductase activity (31), its activity is limited to relatively short-chain acids and it is unlikely to be responsible for the synthesis of longer-chain meromycolates. Again, multiple homologs of mabA (fabG1), a putative β-ketoacyl-ACP reductase gene, were found in the genome of M. tuberculosis, fabG2 through fab5 (30), and it is unclear which of these genes are involved in meromycolate synthesis.
Finally, it was surprising that a mutant nearly totally deficient in mycolic acid synthesis could grow at apparently normal rates at least at 30°C, in view of the important role mycolates are supposed to play in the architecture of the cell wall (16, 20). Possibly the meromycolates can substitute for mycolates in the cell wall to a limited extent. The morphology and physiology of the mutant will be studied in detail, but it is interesting that M. avium recently was shown to be able to survive even when the mycolic acid synthesis was inhibited completely by isoniazid (32).
Acknowledgments
This work was supported by a University of California AIDS Fellowship (F96-B-021), Grant MT-15107 from the Medical Research Council of Canada (to J.L.), and Grants AI-09644 and AI-33702 from the National Institute of Allergy and Infectious Diseases (to H.N.). We thank Dr. C. E. Barry III for advice on analytical methods.
ABBREVIATION
- ES
electrospray
References
- 1.Bloom B R, Murray C J L. Science. 1992;257:1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 2.Frieden T R, Sterling T, Pablos-Mendez A, Kilburn J O, Cauthen G M, Dooley S W. New Engl J Med. 1993;328:521–526. doi: 10.1056/NEJM199302253280801. [DOI] [PubMed] [Google Scholar]
- 3.Chan I S F, Neaton J D, Saravolatz L D, Crane L R, Osterberger J. AIDS. 1995;9:1145–1151. doi: 10.1097/00002030-199510000-00005. [DOI] [PubMed] [Google Scholar]
- 4.Jarlier V, Nikaido H. FEMS Microbiol Lett. 1994;123:1–8. doi: 10.1111/j.1574-6968.1994.tb07194.x. [DOI] [PubMed] [Google Scholar]
- 5.Liu, J., Barry, C. E., III & Nikaido, H. in Mycobacteria: Molecular Biology and Virulence, eds. Ratledge, C. & Dale, J. W. (Blackwell Scientific, Oxford), in press.
- 6.Brennan P J, Nikaido H. Annu Rev Biochem. 1995;64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- 7.Barry C E, III, Slayden R A, Mdluli K. Drug Resist Updates. 1998;1:128–134. doi: 10.1016/s1368-7646(98)80028-9. [DOI] [PubMed] [Google Scholar]
- 8.Walker R W, Prome J-C, Lacave C S. Biochim Biophys Acta. 1973;326:52–62. doi: 10.1016/0005-2760(73)90027-1. [DOI] [PubMed] [Google Scholar]
- 9.Qureshi N, Schnoes H K, Takayama K. J Biol Chem. 1980;255:182–189. [PubMed] [Google Scholar]
- 10.Asselineau C, Lacave C S, Montrozier H L, Promé J C. Eur J Biochem. 1970;14:406–410. doi: 10.1111/j.1432-1033.1970.tb00304.x. [DOI] [PubMed] [Google Scholar]
- 11.Qureshi N, Sathyamoorthy N, Takayama K. J Bacteriol. 1984;157:46–52. doi: 10.1128/jb.157.1.46-52.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lacave C, Lanéele M A, Lanéele G. Biochim Biophys Acta. 1990;1042:315–323. doi: 10.1016/0005-2760(90)90159-u. [DOI] [PubMed] [Google Scholar]
- 13.Lopez-Marin L M, Quémard A, Lanéelle D, Lacave C. Biochim Biophys Acta. 1991;1086:22–28. doi: 10.1016/0005-2760(91)90150-g. [DOI] [PubMed] [Google Scholar]
- 14.Kaneda K, Imaizumi S, Mizuno S, Baba T, Tsukamura M, Yano I. J Gen Microbiol. 1988;134:2213–2229. doi: 10.1099/00221287-134-8-2213. [DOI] [PubMed] [Google Scholar]
- 15.Daffé M, Lanéelle M A, Asselineau C, Lévy-Frébault V, David H. Ann Microbiol (Paris) 1983;134 B:241–256. [PubMed] [Google Scholar]
- 16.Liu J, Barry C E, III, Besra G S, Nikaido H. J Biol Chem. 1996;271:29545–29551. doi: 10.1074/jbc.271.47.29545. [DOI] [PubMed] [Google Scholar]
- 17.Yuan Y, Barry C E., III Proc Natl Acad Sci USA. 1996;93:12828–12833. doi: 10.1073/pnas.93.23.12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Downing D T, Greene R S. Lipids. 1967;3:96–100. doi: 10.1007/BF02530977. [DOI] [PubMed] [Google Scholar]
- 19.Hamid M E, Minnikin D, Goodfellow M. J Gen Microbiol. 1993;139:2203–2213. doi: 10.1099/00221287-139-9-2203. [DOI] [PubMed] [Google Scholar]
- 20.Qureshi N, Takayama K, Jordi H C, Schnoes H K. J Biol Chem. 1978;253:5411–5417. [PubMed] [Google Scholar]
- 21.Liu J, Rosenberg E Y, Nikaido H. Proc Natl Acad Sci USA. 1995;92:11254–11258. doi: 10.1073/pnas.92.24.11254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kundu M, Basu J, Chakrabarti P. J Gen Microbiol. 1991;137:2197–2200. doi: 10.1099/00221287-137-9-2197. [DOI] [PubMed] [Google Scholar]
- 23.Lacave C, Quémard A, Lanéelle G. Biochim Biophys Acta. 1990;1045:58–68. doi: 10.1016/0005-2760(90)90203-a. [DOI] [PubMed] [Google Scholar]
- 24.Wheeler P R, Besra G S, Minnikin D E, Ratledge C. Biochim Biophys Acta. 1993;1167:182–188. doi: 10.1016/0005-2760(93)90160-b. [DOI] [PubMed] [Google Scholar]
- 25.Mdluli K, Slayden R A, Zhu Y Q, Ramaswamy S, Pan X, Mead D, Crane D D, Musser J M, Barry C E., III Science. 1998;280:1607–1610. doi: 10.1126/science.280.5369.1607. [DOI] [PubMed] [Google Scholar]
- 26.Minnikin D E. In: The Biology of the Mycobacteria. Ratledge C, Stanford J, editors. San Diego: Academic; 1982. pp. 95–184. [Google Scholar]
- 27.Yuan Y, Lee R E, Besra G S, Belisle J T, Barry C E., III Proc Natl Acad Sci USA. 1995;92:6630–6634. doi: 10.1073/pnas.92.14.6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.George K M, Yuan Y, Sherman D R, Barry C E., III J Biol Chem. 1995;270:27292–27298. doi: 10.1074/jbc.270.45.27292. [DOI] [PubMed] [Google Scholar]
- 29.Yuan Y, Crane D C, Musser J M, Sreevatsan S, Barry C E., III J Biol Chem. 1997;272:10041–10049. doi: 10.1074/jbc.272.15.10041. [DOI] [PubMed] [Google Scholar]
- 30.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, III, et al. Nature (London) 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 31.Quemard A, Sacchettini K C, Dessen A, Vilcheze C, Bittman R, Jacobs W R, Jr, Blanchard J S. Biochemistry. 1995;34:8235–8241. doi: 10.1021/bi00026a004. [DOI] [PubMed] [Google Scholar]
- 32.Mdluli K, Swanson J, Fischer E, Lee R E, Barry C E., III Mol Microbiol. 1998;27:1223–1233. doi: 10.1046/j.1365-2958.1998.00774.x. [DOI] [PubMed] [Google Scholar]