Abstract
Eps15 homology (EH) domain-containing proteins play a key regulatory role in intracellular membrane trafficking and cell signalling. EH domains serve as interaction platforms for short peptide motifs comprising the residues NPF within natively unstructured regions of accessory proteins. The EH–NPF interactions described thus far are of very low affinity and specificity. Here, we identify the presynaptic endocytic sorting adaptor stonin2 as a high-affinity ligand for the second EH domain (EH2) of the clathrin accessory protein Eps15. Calorimetric data indicate that both NPF motifs within stonin2 interact with EH2 simultaneously and with sub-micromolar affinity. The solution structure of this complex reveals that the first NPF motif binds to the conserved site on the EH domain, whereas the second motif inserts into a novel hydrophobic pocket. Our data show how combination of two EH-attachment sites provides a means for modulating specificity and allows discrimination from a large pool of potential binding partners containing NPF motifs.
Keywords: endocytosis, Eps15, protein–protein interactions, specificity, stonin2
Introduction
Clathrin-mediated endocytosis (CME) constitutes a major pathway for the internalization of many cell-signalling receptors as well as recycling of presynaptic vesicles in the mammalian brain (Heuser, 1989; Waterman and Yarden, 2001). CME involves a complex dynamic network of endocytic proteins and lipids that are thought to drive the different stages of vesicle formation. Protein–protein interactions play a central role in the assembly of endocytic protein complexes, and are often mediated by small modular domains that recognize short peptide stretches within their binding partners. Many such protein interaction domains, including SH3 and Eps15 homology (EH) domains, have been characterized in molecular detail and were found to bind targets with only moderate (i.e., low micromolar) affinity and specificity (reviewed in Mayer and Eck, 1995; Salcini et al, 1997; Mayer, 2001; Confalonieri and Di Fiore, 2002). Further complexity arises from the existence of highly overlapping binding motifs and an apparent redundancy of the corresponding recognition domains. Therefore, a central challenge is to understand how binding affinity is modulated and specificity is achieved in protein–protein interactions within complex networks involving multiple potential binding partners.
The endocytic EH domain-containing proteins, including Eps15, intersectin (ITSN) and EHD1–4, are of particular interest for studying domain specificity. EH domains are involved in protein-complex assembly during endocytosis or other trafficking processes (Di Fiore et al, 1997; Montesinos et al, 2005; Naslavsky and Caplan, 2005) and have been shown to interact with ligands containing NPF motifs (Salcini et al, 1997; Paoluzi et al, 1998; de Beer et al, 2000; Morgan et al, 2003). Recently, the EH domains of EHD1 and Eps15 were shown to be capable of binding phosphoinositides as well (Naslavsky et al, 2007).
EH domains generally displayed extremely low affinities in binding to short peptides, usually in the high micromolar range, with no or little apparent preference for a certain sequence outside the core NPF motif (de Beer et al, 1998; Paoluzi et al, 1998; Yamabhai et al, 1998; Kim et al, 2001). This apparent lack of specificity is hard to reconcile with the physiological role of EH domains and their ligands in regulating protein assembly in trafficking networks.
As many EH-binding proteins contain multiple NPF motifs, one possible explanation for the low affinity observed with single domains could be that full-length proteins within the cell may interact simultaneously with several EH domains. Moreover, endocytic proteins such as Eps15 and ITSN comprise multiple EH domains, often in combination with coiled-coil regions that could promote oligomerization (Tebar et al, 1997), providing an opportunity for the formation of high-avidity complexes.
Eps15 binds to a number of endocytic proteins containing NPF motifs, such as epsin, synaptojanin-p170 and stonin2 (Haffner et al, 1997; Chen et al, 1998; Martina et al, 2001). Eps15 and stonin2 have both been shown to directly associate with the clathrin adaptor complex AP-2 and to localize to clathrin-coated pits (CCPs) (Benmerah et al, 1995; van Delft et al, 1997; Martina et al, 2001; Walther et al, 2004). In addition, stonin2 was recently identified as a specific sorting adaptor for the synaptic vesicle protein synaptotagmin, and may thus regulate synaptic vesicle recycling (Diril et al, 2006). Here, we unravel a novel mechanism by which high affinity and specificity between EH domains and their ligands can be achieved: the recognition of two NPF motifs by a single EH domain. We show that the second EH domain (EH2) of Eps15 binds stonin2, containing two NPF motifs, with an exceptional affinity, two to three orders of magnitude higher than EH domain–NPF interactions reported previously. Furthermore, using mutational analyses and isothermal titration calorimetry (ITC) we demonstrate that both NPF motifs bind EH2 simultaneously and are both required for a tight interaction. We present the solution structure of a complex between the second EH domain of Eps15 and the NPF region of stonin2, revealing a novel motif recognition mode for EH domains with two distinct binding sites for NPF motifs within one EH domain. The first NPF motif of stonin2 inserts into the conserved binding groove, whereas the second NPF sequence binds into an adjacent, hydrophobic pocket. This new binding site could represent a specificity pocket in EH domains and provide a molecular explanation for the relatively high frequency of multiple NPF motifs in EH-binding partners.
Results
Targeting of Eps15 EH domains in living cells depends on stonin2 NPF motifs
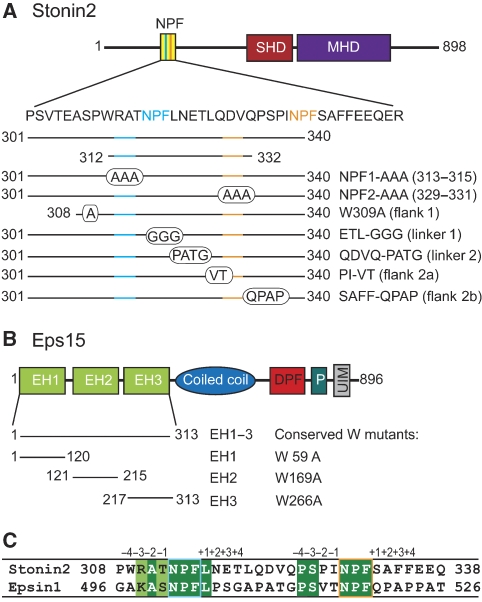
As a model to study the specific interaction between an EH domain and its cognate NPF-motif protein, we analysed the Eps15–stonin2 complex in vitro and in living cells. The domain structure of the two proteins and details of the constructs used are given in Figure 1.
Figure 1.
Domain structure of Eps15, stonin2 and constructs thereof. (A) Domain structure of stonin2 with the NPF region, the stonin2 homology domain (SHD) and μ-homology domain (MHD). The constructs and mutants used for this study are indicated. (B) Domain structure of Eps15 containing three EH domains, a central coiled-coil region, AP-2-interaction motifs (DPF), a proline-rich motif (P) and ubiquitin-interacting motifs (UIM). (C) Sequence alignment of human stonin2 and rat epsin1. The two NPF motifs are indicated by cyan and orange boxes, respectively. Identical and homologous residues are highlighted in green or light green.
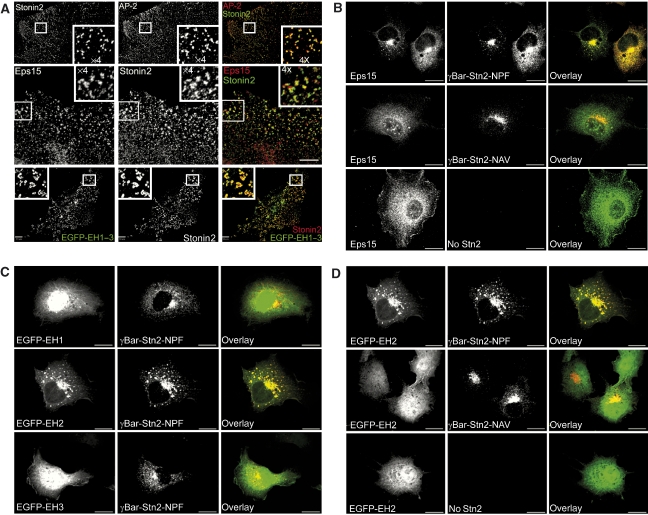
Associations of stonin2 with Eps15 and with the clathrin adaptor complex AP-2 have been observed previously in pull-down and co-immunoprecipitation (IP) assays (Martina et al, 2001; Walther et al, 2004), suggesting that these three proteins can form a ternary complex. In line with these data, endogenous stonin2 colocalized with Eps15 in AP-2-containing CCPs in primary astrocytes (Figure 2A, top and central panels), and all three proteins were present in immunoprecipitates from detergent-extracted rat brain homogenates using antisera against stonin2 (Supplementary Figure 1A). To determine whether the EH domains of Eps15 might contribute to CCP targeting, we constructed a fusion protein comprising EH domains 1–3 and enhanced green fluorescent protein (EGFP-EH1–3). As seen in Figure 2A (bottom panel), EGFP-EH1–3 co-localized with endogenous stonin2 in CCPs in astrocytes, suggesting that interactions between the Eps15–EH domains and NPF motifs within stonin2 might underlie this phenotype. Similar results were obtained in N1E neuroblastoma cells overexpressing the synaptic vesicle membrane protein synaptotagmin (syt1), a bona fide cargo for stonin2-dependent clathrin-mediated endocytosis. In these cells, stonin2 is recruited to the plasma membrane through direct interaction with synaptotagmin (Diril et al, 2006). As expected, EGFP-EH1–3 colocalized partially with wild-type stonin2 and synaptotagmin at the plasmalemma. By contrast, overexpression of a stonin2 NPF mutant did not facilitate plasma membrane targeting of EGFP-EH1–3 (Supplementary Figure 1B). These data imply that membrane-targeted stonin2 might contribute to the recruitment of Eps15 to synaptotagmin-containing membrane sites in situ.
Figure 2.
Eps15 and stonin2 colocalize and interact in situ. (A) Endogenous stonin2 colocalizes with AP-2 and Eps15 in CCPs in primary astrocytes. Primary hippocampal astrocytes (at 12 days in vitro (DIV)) were analysed by indirect deconvolution immunofluorescence microscopy with antibodies against Eps15, stonin2 or AP-2α. Eps15 colocalizes with stonin2 in CCPs immunopositive for the clathrin adaptor AP-2. Insets depict fourfold-magnified views of the boxed area. Scale bars, 10 μm. Bottom panel: The EH domains of Eps15 are sufficient for the recruitment of Eps15 to stonin2-containing CCPs. Primary astrocytes (8 DIV) were transfected with an EGFP-EH1–3 expression plasmid. At 12 DIV, cells were analysed by indirect deconvolution immunofluorescence microscopy with antibodies against stonin2. Inset depicts fourfold-magnified view of the boxed area. Scale bar, 10 μm. (B–D) γBAR–stonin2-mediated recruitment of endogenous Eps15 or fusion proteins of individual Eps15–EH domains (EH1, EH2, EH3) and EGFP to the TGN. Cos7 fibroblasts overexpressing a chimaeric protein comprising the stonin2 NPF region fused to a TGN-localized fragment of γBAR (γBAR-Stn2; internally tagged with a c-myc epitope), were analysed by spinning-disc confocal microscopy, using antibodies against c-myc (red). Endogenous Eps15 and EH2, but not EH1 or EH3, displayed robust and quantitative targeting to the TGN, which was dependent on stonin2 NPF motifs. Scale bar, 10 μm.
To further investigate the potential role of NPF–EH domain interactions in endocytic protein localization, and to delineate the molecular determinants involved, we constructed a chimaeric protein composed of the NPF region of stonin2 (comprising both NPF motifs at positions 313–315 and 329–331) and the N-terminal domain (amino acids 1–100) of γBAR, a recently described peripheral AP-1-binding membrane protein localized to the trans-Golgi network (TGN) (Neubrand et al, 2005). Similar to wild-type γBAR, this chimaeric protein (γBAR–stonin2–NPF) localizes to the TGN in transfected fibroblasts (Figure 2B–D). Strikingly, we observed that a major fraction of endogenous Eps15 was recruited to the TGN area in cells co-expressing γBAR–stonin2 (Figure 2B, top panel). This localization was dependent on the presence of the NPF motifs within the γBAR–stonin2 chimaera, as their mutation to NAV abolished targeting to the TGN (Figure 2B, central panel). Similar results were obtained if the distribution of EGFP-EH1–3 was analysed in fibroblasts co-expressing γBAR–stonin2–NPF (Supplementary Figure 2).
When individual EH domains fused to EGFP were analysed for their interaction with γBAR–stonin2–NPF, only constructs containing EH2 but not EH1 or EH3 were effectively targeted to the TGN (Figure 2C). Colocalization of EH2 and γBAR–stonin2 was disrupted when the NPF motifs were mutated to NAV (Figure 2D). These experiments indicate that the second EH domain of Eps15 is the major determinant for NPF-motif-dependent interactions with stonin2 in living cells.
Characterization of a high-affinity Eps15–stonin2 complex
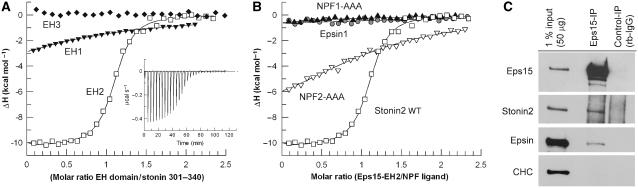
To determine the affinity of the interaction between Eps15–EH domains and the NPF region of stonin2, we first characterized complex formation by ITC (summarized in Table I). Binding of EH1–3 of Eps15 to stonin2 could be analysed using a model for two independent binding events, indicating that at least two EH domains can bind stonin2 (ITC no. 1; Table I). Analysis resulted in a high- and a low-affinity interaction with dissociation constants of 0.16 and 30 μM, respectively. We then measured the affinity between single EH domains of Eps15 and stonin2–NPF, to identify the domains involved in this interaction. EH1 binds with a low affinity of 75 μM (ITC no. 2), whereas complex formation between EH3 and stonin2 was not observed (ITC no. 3; Figure 3A). Consistent with the colocalization experiments, we found that EH2 alone is necessary and sufficient for the high-affinity interaction (Kd of 0.15 μM) (ITC no. 4). This exceptionally high affinity, observed with all constructs containing EH2, exceeds the previously observed affinities for EH domain binding to short peptides by a factor of 100–3000 (de Beer et al, 1998; Paoluzi et al, 1998; Kim et al, 2001). A stoichiometry of one EH domain binding to a single NPF motif has been reported so far (Salcini et al, 1997; Paoluzi et al, 1998; Yamabhai et al, 1998; de Beer et al, 2000; Kim et al, 2001; Confalonieri and Di Fiore, 2002). To determine which of the two NPF motifs might be responsible for binding to EH2, we mutated the individual NPF residues to alanines (Figure 3B; Table I).
Table 1.
Binding affinities of Eps15 EH domains for stonin2 constructs
| ITC number | EH domaina | NPF liganda | Kd (μM)b | ΔHb | NNPF/EHb |
|---|---|---|---|---|---|
| 1c | Eps15 EH1–3 | Stonin2 301–340 | 29.5±8.7 | −4.6±2.7 | 1.2±0.6 |
| 0.16±0.04 | −13.1±0.5 | 1.1±0.1 | |||
| 2 | Eps15 EH1 | Stonin2 301–340 | 75.2±1.6 | −8.3±3.0 | 1.2±0.3 |
| 3 | Eps15 EH3 | Stonin2 301–340 | ND | ND | ND |
| 4 | Eps15 EH2 | Stonin2 301–340 | 0.15±0.02 | −10.9±0.3 | 1.2±0.1 |
| 5 | Eps15 EH2 | Stn301–340 NPF1AAA | ND | ND | ND |
| 6 | Eps15 EH2 | Stn301–340 NPF2AAA | 7.5±1.0 | −7.8±2.2 | 1.0±0.3 |
| 7 | Eps15 EH2 | Stonin2 312–332 | 6.6±0.2 | −7.3±0.01 | 0.65±0.03 |
| 8 | Eps15 EH1–3 | Epsin1 496–575 | 90 | −3.5 | 1 |
| 9 | Eps15 EH2 | Stn301–340 | 0.42±0.03 | −12.8±3.2 | 1.2±0.1 |
| ETL-GGG, Linker1 | |||||
| 10 | Eps15 EH2 | Stn301–340 | 0.30±0.2 | −13.0±4.4 | 1.3±0.5 |
| QDVQ-PATG, Linker2 | |||||
| 11 | Eps15 EH2 | Stn308–340 | 0.9±0.2 | −8.8±3.2 | 1.0±0.4 |
| W309A, Flanking 1 | |||||
| 12 | Eps15 EH2 | Stn301–340 | 1.1±0.1 | −11.7±0.9 | 1.2±0.1 |
| PI327/328VT, Flanking 2a | |||||
| 13 | Eps15 EH2 | Stn301–340 | 2.8±1.3 | −8.4±1.2 | 1.3±0.04 |
| SAFF332-335QPAP, Flanking 2b | |||||
| 14 | Eps15R EH2 | Stonin2 301–340 | 0.9±0.02 | −5.8±0.2 | 1.1±0.2 |
| EH, Eps15 homology; ITC, isothermal titration calorimetry; ND, not detected. | |||||
| aAll titrations were carried out between 2 and 4 times, with the exception of epsin1 binding to EH2, which was measured once due to protein limitations. | |||||
| bThe indicated error represents the standard deviation between measurements. N=stoichiometry (NPF ligand to EH domain). | |||||
| cTitration 1 was analysed with a model for two independent binding sites, yielding a high- and a low-affinity interaction. All other titrations were fitted using a simple one binding event model. | |||||
Figure 3.
High-affinity interaction of Eps15 with stonin2 requires two NPF motifs. (A) Isothermal titrations of Eps15 EH domains with the NPF region of stonin2 (residues 301–340). EH1 (closed triangles) binds stonin2 with low affinity, whereas EH2 (open squares) interacts strongly with the NPF region. Analysis of the titration isotherms resulted in a Kd of 75 μM for EH1 and 0.15 μM for EH2. No binding could be observed for EH3 (closed diamonds). The inset shows raw titration data for binding of EH2 to stonin2 (301–340). (B) Binding of NPF mutants. Titrations of EH2 with stonin2 wild type (open squares) and mutant NPF1-AAA (closed triangles) or NPF2-AAA (open triangles), compared with the epsin1 NPF region (residues 491–526) (grey circles). Least-square fits of the data gave a Kd of 0.15 μM for wild-type stonin2 and 6 μM for stonin2 NPF2-AAA. Binding for the stonin2-mutant NPF1-AAA could not be detected. The enthalpy for epsin1 binding was at the detection limit. (C) Eps15 co-immunoprecipitates with both stonin2 and epsin1. Eps15 antibody or preimmune serum-coupled protein A/G beads were incubated with rat brain extract. Immunoprecipitated proteins were detected by immunoblotting for Eps15, stonin2, epsin1 and clathrin heavy chain (CHC) as control. A total of 1% of the input was loaded for comparison. Note the de-enrichment of epsin1 in Eps15 immunoprecipitates compared with the input.
Surprisingly, both NPF motifs in stonin2 were necessary for a tight interaction. Mutation of the first motif abolished binding completely and exchange of the second NPF motif reduced the affinity about 50-fold (Kd of 8 μM) (ITC nos. 5 and 6). We conclude that the first NPF motif is indispensable for the interaction, whereas the second NPF motif adds to affinity and specificity. We observed no difference in binding between fragments comprising amino acids 204–340 (not shown) and 301–340 (ITC no. 4). However, complex formation was significantly reduced if the stonin2 NPF region was further truncated (Kd of 6.6 μM for residues 312–332) (ITC no. 7).
Epsin1 represents another endocytic accessory protein reported to bind to Eps15 via EH–NPF interactions (Chen et al, 1998). Epsin1 contains three NPF motifs, two of which are separated by exactly the same spacing as found in stonin2 (Figure 1C). Although epsin1 has been identified as a major interaction partner of Eps15, the affinity of Eps15–EH2 to an epsin1 fragment (residues 491–526 equivalent to stonin2 301–340) was at the detection limit of the ITC, particularly due to a low binding enthalpy (Figure 3B). An NMR titration yielded a Kd value of 42±8 μM (data not shown). Extension of the epsin1 construct to comprise all three of epsin's NPF motifs (residues 496–575) (ITC no. 8; Table I) did not result in an increased affinity when titrated with a construct comprising all three EH domains of Eps15. To further analyse complex formation between Eps15 and its NPF partners in vivo, we performed co-IP experiments from detergent-extracted rat brain homogenates. In agreement with the published data, both stonin2 and epsin1 were present in Eps15 immunoprecipitates, albeit with different degrees of relative enrichment (Figure 3C), in line with the affinity measurements described above. Taken together, these data suggest that NPF motifs alone are necessary but at least in some cases might not be sufficient for a tight interaction with EH domains. We conclude that the regions flanking the NPF motifs contribute considerably to complex formation between Eps15 and stonin2, in contrast to previously published EH domain interactions.
Structure determination of the Eps15–stonin2 complex
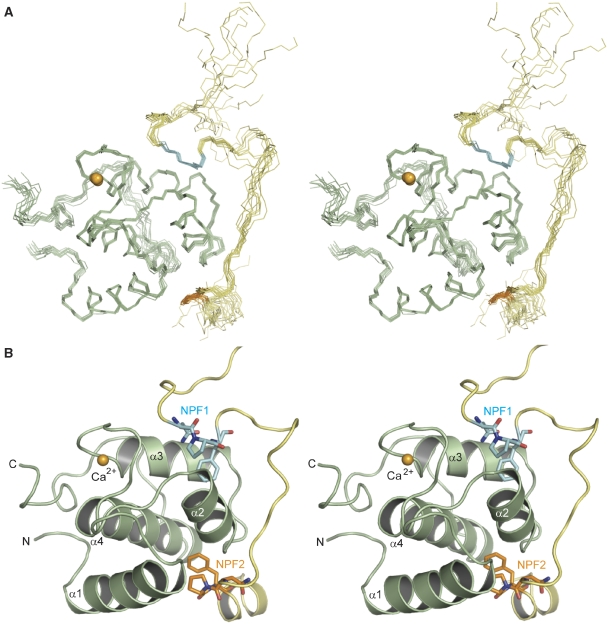
In order to understand the basis for Eps15–stonin2 complex formation at the molecular level, we solved the three-dimensional structure of a complex between the second EH domain of Eps15 and the NPF region of stonin2 (residues 301–340) by heteronuclear multidimensional NMR spectroscopy. The NMR frequencies of free and stonin2-bound states of the EH domain are in slow exchange regime relative to the chemical shift time scale, indicative of a high-affinity interaction. A stereo-image of an ensemble of 10 superimposed NMR structures and a ribbon representation of the lowest energy structure are shown in Figure 4A and B, respectively. In agreement with the published EH domain structures (de Beer et al, 1998, 2000; Koshiba et al, 1999; Enmon et al, 2000; Kim et al, 2001), Eps15–EH2 in our complex structure exhibits the canonical fold of EH domains—two EF hands consisting of the characteristic two helix–loop–helix motifs each. The stonin2 peptide wraps around one side of the EH domain starting from helix α3, around helix α2 and ends between helices α1 and α4. The peptide adopts an extended conformation with the exception of two turns, which serve to position the two NPF motifs within their respective hydrophobic pockets on the EH domain, whereas the linker between NPF1 and NPF2 is flexible. Furthermore, residues Ser332–Glu336 form a short α-helix at the C-terminus of the peptide.
Figure 4.
Solution structure of Eps15–EH2 in complex with the stonin2 NPF region. (A) Stereoview of the Eps15–stonin2 complex structure. Backbone trace of 10 superimposed lowest energy structures (out of 100 calculated structures), with Eps15–EH2 shown in light green and stonin2 depicted in yellow. (B) Cartoon diagram of the complex between Eps15–EH2 (light green) with stonin2 (as yellow sticks) in stereoview. The two NPF regions are highlighted in cyan (NPF1) and orange (NPF2), and the calcium ion is illustrated as bright orange sphere. Both the stonin2 NPF motifs interact with the EH domain: the N-terminal motif (NPF1, cyan) binds to the canonical binding pocket between helices α2 and α3, lined by highly conserved hydrophobic residues, as described in references de Beer et al (1998) and de Beer et al (2000). The second NPF motif (NPF2, orange) inserts into an adjacent hydrophobic groove, flanked by helices α2, α4 and α1 of the EH domain.
In the previously described complex structure of EH2 with a short Hrb1 peptide (11-mer), only four residues of the peptide were involved in complex formation based on the analysis of nuclear overhauser effects (NOEs) (de Beer et al, 2000). By contrast, we observe many more intermolecular contacts (400 versus 64 NOEs) produced by 19 stonin2-peptide residues directly interacting with the EH domain. This explains the much higher affinity of EH2 for stonin2 (0.15 μM) compared with the previously published Hrb1 peptide (600 μM) (de Beer et al, 1998). 15N relaxation data (Supplementary Figure 3) demonstrate that residues within and immediately adjacent to the NPF motifs are more ordered, whereas the linker region between the two motifs is flexible. Exchange of the linker region to epsin1 residues had a negligible effect on binding to EH2 (ITC nos. 9 and 10; Table I), indicating this region does not appreciably contribute to the interaction.
Close-up of the two individual binding sites
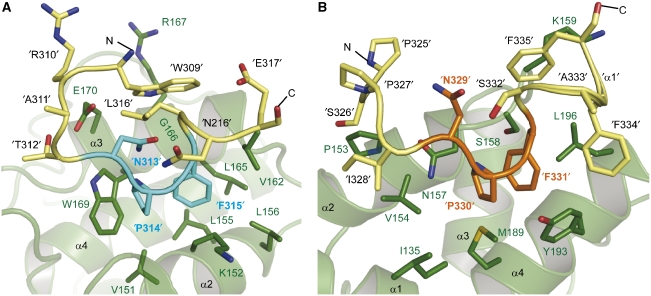
The main residues involved in the recognition of NPF1 are the conserved Trp169, Leu165, Lys152 and Val151 on EH2 (Figure 5A). These form a deep hydrophobic groove in which the NPF1 tripeptide is almost completely buried. The first NPF motif of stonin2 superposes well with the respective residues of the Hrb1 peptide in the Eps15 EH2-complex structure of de Beer et al (2000). Thus, Asn313, Pro314, Phe315 (NPF1) and Leu316 form a β-turn, which is stabilized by hydrogen bonds between the backbone and side-chain oxygen atoms of Asn313 with the amide protons of Leu316 and Phe315, respectively. This is indicated by the short distance between the acyl oxygen of Asn313 and the amide proton of Leu316 (residue +3), and backbone torsion angles of Pro314 and Phe315, consistent with a beta I-type backbone conformation (Suzuki, 1989). Pro314 packs tightly against Trp169, which provides a typical environment for Pro recognition (Zarrinpar et al, 2003). Phe315 is positioned deeply within the hydrophobic groove lined by Lys152, Leu155, Leu156, Val162, Leu165 and Trp169.
Figure 5.
Close-up view of the two EH domain-binding sites containing the NPF motifs of stonin2. (A) The first NPF motif (NPF1) binds into the conserved hydrophobic pocket. The side chains of the EH2 residues that interact with NPF1 (cyan) and adjacent residues (yellow) are depicted as sticks (green). (B) The second NPF motif inserts into a novel site on the EH domain. Residues of NPF2 (orange) and surrounding amino acids (yellow) point into a hydrophobic groove formed by α-helices α2, α4 and α1 (shown as green cartoon). Side chains of residues lining this pocket and interacting with stonin2 are represented as green sticks and labelled in green (EH2), whereas side chains of peptide residues are indicated by apostrophes.
The conformation of the NPF1 motif in the first binding site is partially stabilized by Trp309 in position −4 relative to NPF1. The aromatic ring system is positioned parallel to EH-helix-α3 flanking the first binding pocket, based on NOEs between Trp309 and Glu170, Asp163, Arg167 and Val162 of the EH domain. The Trp309 indole proton is within hydrogen-bonding distance to the backbone of Gly166. One function of this Trp residue might be to stabilize β-turn formation as it is in close proximity to Leu316 and might position the amide proton of Leu316 for hydrogen bonding with Asn313. Mutation of Trp309 to Ala reduces the binding affinity between Eps15–EH2 and stonin2 by about sixfold (Kd value of 0.9 μM) (ITC no. 11; Table I).
The second NPF motif of stonin2 inserts into a hydrophobic pocket on EH2 that is not conserved among human EH domains. Furthermore, the interaction between the second binding site and stonin2 is characterized by a large number of contacts not limited to those made with the NPF2 tripeptide, but including several residues outside the core motif. Pro325 (−4 position relative to NPF2) packs against a hydrophobic patch in the helix-α2 of EH2 and contacts Pro153 and Leu156. Strong NOEs with Pro153 and Val154 position Ser326 between these two residues. Pro327 is largely solvent-exposed, whereas Ile328 is located in a hydrophobic pocket in close proximity to Val154 and Ile135 (Figure 5B). Mutation of Pro327 and Ile328 preceding NPF2 to Val and Thr (the epsin1 counterparts) reduced binding sevenfold, emphasizing the importance of these hydrophobic contacts (ITC no. 12; Table I). The molecular determinants for this interaction are thus in contrast to NPF1 binding, where the residues at positions −1 and −2 are preferentially small and polar, and NMR structures of binding site 1 complexes show that these residues do not contribute to complex formation (de Beer et al, 2000).
The second NPF motif of stonin2 exhibits a slightly different conformation when compared with NPF1 (r.m.s.d. of 0.9 Å for the lowest energy structure). Whereas the backbone conformation is very similar to NPF1, the side chains superimpose less well. In particular, the orientation of Asn329 makes hydrogen bonding to the backbone of Ser332 (equivalent to Leu316 in NPF1) unlikely. Pro330 makes important hydrophobic contacts with Ile135, Val154, Met189 and Tyr193. Phe331 is positioned in a hydrophobic pocket flanked by Met189, Val192, Tyr193 and Leu196, while the main interactions occur via π-stacking with Tyr193. Whereas the residues following NPF2, Ala332 and Ser333, are completely solvent exposed, the two Phe residues at position +3 and +4 are significantly involved in binding. This arrangement is achieved by the formation of a helical turn of the peptide backbone, which enables the three Phe residues 331, 334 and 335 to cluster around Leu196 on the EH domain. Additionally Phe334 interacts with Glu197 and Tyr193, and is positioned on top of Tyr193 and Phe331. Phe335 is situated near the loop connecting helices α2 and α3 of the EH domain, and interacts with Leu196 as well as the aliphatic side chain of Lys159. The important role of the two Phe residues following NPF2 in complex formation was probed by mutational analysis; the four residues SAFF were exchanged for their epsin1 counterparts, QPAP (see Figure 1). This mutation results in an almost 20-fold decrease in affinity (Kd of 2.8 μM) (ITC no. 13; Table I). The remarkable influence of the flanking regions of NPF2 is further emphasized by an NPF2-AAA mutant: this mutant is still able to occupy both sites on the EH domain, as observed in NMR titrations (Supplementary Figure 4), even though the second core motif is missing.
On the other hand, EH2 exhibits chemical-shift perturbations only in the first binding pocket when titrated with epsin1 (amino acids 491–526), shown in the 1H, 15N HSQC overlay (Supplementary Figure 5). Despite its very similar motif pattern, epsin1 only binds to the conserved site, which is consistent with the comparably low affinity of the interaction. The presence of two NPF motifs alone is neither sufficient to bind both sites on EH2, nor to form a high-affinity complex.
Role of the second binding site in specific ligand recognition
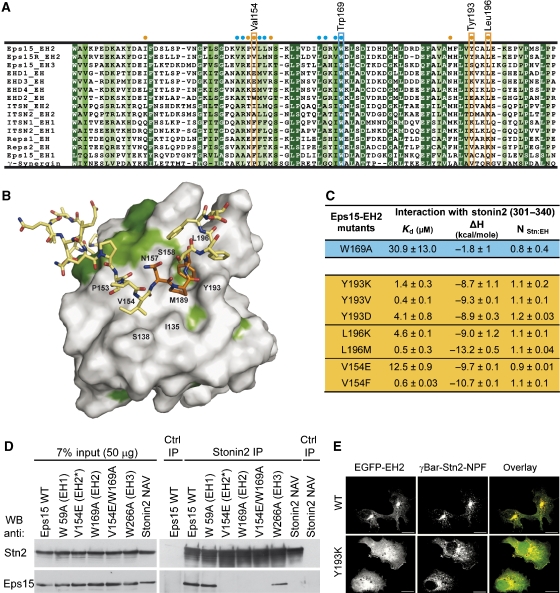
Figure 6B shows the EH domain in a surface representation, which highlights the binding of NPF2 into the previously uncharacterized hydrophobic groove on the domain. The surface is coloured according to a sequence alignment (Figure 6A) of all human EH domains. The second binding site lies 90° rotated to the first site and is much less conserved (Figure 6B). It comprises a more shallow and extended interaction surface than the first binding pocket, thus allowing more contacts with the residues flanking NPF2 (e.g., with Ile328, Phe334, Phe335). However, the residues specifically recognizing the flanking regions are least conserved within the alignment of EH domains (Figure 6A). Leu196 is conserved in more than half of the human EH domains, but in other cases it is replaced by Lys (mostly), Met, Arg or Gln. Another important amino acid that interacts with stonin2 is Val154, which is often exchanged for bulkier, hydrophobic or highly charged residues (Phe≫Glu/Ile>Lys/Trp/Ala). Tyr193, one of the key mediators of interaction with NPF2 and unique to Eps15–EH2 and Eps15R-EH2, corresponds to charged, small residues in other EH domains (Lys≫Val>Asp>Ser/Glu/Ala).
Figure 6.
Mutations in the second binding pocket on the EH domain affect the Eps15–stonin2 interaction. (A) Alignment of all human EH domains where conserved residues are highlighted in green depending on their level of conservation (dark green=identity, green=homology, light green=similarity). Cyan and orange dots indicate the position of the most important residues contacting NPF1 and NPF2, respectively. Key residues in the first and second binding pocket, which were subjected to mutagenesis, are highlighted by a cyan or orange box. (B) The second NPF motif binds to a novel site, which is much less conserved compared with other EH domains. Surface representation of the novel binding site on EH2 according to the colour scheme described in panel A. EH-domain residues that are involved in ligand binding are labeled. The second NPF motif of stonin2 (orange) and flanking regions (yellow) are depicted as sticks. (C) Mutations within the second binding pocket of EH2. Affinity values for EH domain mutants of the second binding site and stonin2. Amino-acid exchanges that occur in other EH domains lead to significant reduction of affinity for stonin2. (D) Mutating crucial residues in the Eps15–EH2 domain or the stonin2 NPF motifs abolishes the interaction between Eps15 and stonin2. Inducibly stonin2-HA-expressing cells were transfected with Flag-tagged Eps15 WT and mutant constructs. An HA-stonin2 NPF to NAV mutant was co-transfected with Flag-tagged WT Eps15 into HEK293 cells. IP was performed using antibodies directed against stonin2 or preimmune serum as control. Immunoprecipitated proteins were detected by immunoblotting for stonin2 and Flag (for Eps15). A total of 7% of the input was loaded as standard. (E) Reduced colocalization of an EGFP–EH2 second-site mutant (Y193K) with γBAR–stonin2. Localization of EGFP–Eps15–EH2 WT and Y193K-mutant proteins in Cos7 fibroblasts overexpressing c-myc-tagged γBAR–stonin2 (red) was analysed by spinning-disc confocal imaging. The Y193K mutation reduces the colocalization of both proteins significantly (overlay). Scale bar, 10 μm.
To underline the importance of the second site and to determine the residues conferring specificity, we created mutants in the second site of EH2 (Figure 6C). The conserved Leu196, which is surrounded by the three Phe residues of stonin2, was mutated to Lys or Met, resulting in a 30-fold and threefold respective reduction of binding affinity (Kd of 4.6 and 0.5 μM) when compared with the wild type. Incorporation of the basic lysine residue disrupts the interaction with the aromatic residues notably, whereas a methionine at this position does not disturb the hydrophobic contacts. Mutation of Val154 to Glu had the strongest effect on binding, with a dissociation constant of 11.8 μM, whereas a Phe mutant only altered the affinity marginally (0.5 μM). Tyr193, which is involved in π-stacking interactions with Phe331 and Phe334, was exchanged with Lys, Asp or Val. If the hydrophobic nature of the amino acid is retained (Tyr193Val), the affinity is reduced only slightly (0.36 μM) as seen for the other two hydrophobic key residues. Introduction of a positively charged residue (lysine) however, has a significant effect on binding (1.2 μM). Mutation of Tyr193 to Lys is less dramatic than expected from the structure. An explanation for this could be that upon mutation the long aliphatic portion of lysine might still be able to contact the aromatic stonin2 residues. A more pronounced effect is observed in the Tyr193Asp mutant that binds stonin2 with a Kd of 4.1 μM. The lack of high-affinity binding of stonin2 to other EH domains may be attributed to the sometimes drastic exchanges of second-site key residues in EH domains from EHD1, Reps1 or ITSN. Binding could be observed for some of these proteins, but the affinity constants measured were in the micromolar range (Supplementary Figure 6). Eps15R–EH2, which shares 77% sequence identity with Eps15–EH2, displays all the critical second-site residues and, consequently, binds stonin2 with sub-micromolar affinity (ITC no. 14; Table I). The less conserved second binding site could thus represent a specificity pocket, which allows differential recognition among EH domains.
The importance of the second binding pocket in EH2 was probed by mutation in the context of full-length Eps15 by co-IP assays. Full-length Eps15 mutated within the NPF2-binding site of EH2 (V154E) failed to co-precipitate with stonin2. Similar results were seen if Eps15 mutated in the conserved NPF1 binding pocket within EH2 (W169A) was analysed. By contrast and in agreement with the biophysical data, no change in binding to stonin2 was observed when the conserved tryptophan residues within EH1 (W59A) or EH3 (W266A) were replaced by alanines. Finally, another stonin2–NPF binding-defective Eps15–EH2 mutant (Y193K) also failed to be recruited to TGN area in fibroblasts overexpressing γBAR–stonin2 (Figure 6E). We conclude that both NPF motifs and their flanking regions within stonin2 are required for high-affinity interaction with Eps15–EH2 in vitro and in living cells.
Discussion
In order to understand the molecular basis of ligand recognition and binding specificity of the Eps15–EH2 domain in molecular detail, we characterized its association with stonin2 by NMR spectroscopy and calorimetric titrations.
Our data demonstrate the high-affinity EH domain–NPF complex between the second EH domain of Eps15 and the NPF region of stonin2. In addition, this complex is the first example of a two-site binding mode for EH domains and thereby constitutes a novel mechanism by which specific ligand recognition is achieved. The first binding site represents the conserved binding pocket and superposes well with the previously described complex structures of EH2 (de Beer et al, 2000). The exceptionally high affinity of the complex, compared with other EH–NPF interactions, is achieved by insertion of the second NPF motif of stonin2 into a novel binding pocket on the domain. The second site differs from conventional EH–NPF interactions, as its binding surface is considerably extended to include flanking regions that contribute appreciably to complex formation. These flanking regions, that are unique to stonin2, mediate specificity. Accordingly, we see dramatic differences (almost 1000-fold difference in Kd) when comparing two different ligands, epsin1 and stonin2, each containing two NPF motifs 13 residues apart, with different flanking regions: stonin2 binds at two sites, whereas epsin1 only occupies the first binding pocket. These results imply that EH domains can specifically recognize their targets and discriminate between several NPF-containing binding partners.
Sequence alignments illustrate that EH domains differ substantially in the region that constitutes the second binding groove in Eps15–EH2. A hydrophobic environment is required to accommodate the proline and phenylalanine residues of NPF motifs, whereas the region surrounding the NPF motif may vary, thereby defining the specificity of the ligand for its cognate EH-recognition domain.
Some of the critical EH-domain residues, which specifically interact with stonin2–NPF2 are almost exclusively found in Eps15–EH2. Not surprisingly, one exception among the human EH domains is Eps15R, which is highly homologous to Eps15 and shares overlapping binding partners and biochemical properties (Coda et al, 1998). However, when comparing sequences from Eps15–EH2 homologues in other organisms, these residues are conserved from worms and flies to humans.
Another example for a possible two-site complex between an EH domain and an NPF ligand has been published previously. Qualmann and co-workers have shown that syndapins bind specifically to EHD proteins and that mutation of one of three NPF motifs within syndapin II either completely abrogates or reduces the interaction (Braun et al, 2005). Two structures of EHD–EH domains were released recently: the solution structure of EHD1–EH (Kieken et al, 2007) and the crystal structure of full-length EHD2 (Daumke et al, 2007). The binding pocket of the EHD–EH domains in both structures exhibit a particular surface potential, which is characterized by a high amount of basic charges. When comparing the sequences of EH domains from Eps15 and EHD proteins, it becomes evident that key residues particularly of the second binding site in Eps15–EH2 correspond to charged or polar residues in EHD1, thus complementing the acidic residues surrounding the NPF motifs within their cognate ligands (syndapins). Change of surface charge might reflect ligand-binding preference. In order to understand EH-domain ligand specificity and identify high-affinity complexes, it will be necessary to study larger fragments, including flanking regions and potential additional binding motifs.
It has generally been assumed that small modular domains, like EH and SH3 domains, bind their cognate peptide motifs with moderate affinity, using a one-to-one stoichiometry. For SH3 domains, several studies have shown that other binding modes may be employed. Some ligands appear to bind at two distinct sites within one domain and thereby increase specificity and affinity considerably (Kami et al, 2002; Dutta et al, 2004). Other SH3 domain-containing proteins use two domains to interact with a single ligand (Groemping et al, 2003).
Our results indicate that different binding modes also exist for EH domains and thereby could be employed to modulate binding affinities. Ligands can bind at one or two sites within the EH domain or possibly interact with separate EH domains within the same molecule or a protein oligomer.
The EH2 domain of Eps15 is not the only example for a two-site interaction domain in endocytosis. The adaptor protein AP-2, which constitutes a major hub in the assembly of multi-protein complexes at the site of CCP formation, binds its ligands via multiple binding sites (Mishra et al, 2004; Praefcke et al, 2004; Edeling et al, 2006; Motley et al, 2006; Schmid et al, 2006). Ligands that interact with the so-called top and side site simultaneously display a very high affinity; an example for a tight binding ligand is Eps15, for which a dissociation constant of 20 nM was determined (Praefcke et al, 2004).
High-affinity interactions might ensure a sufficient degree of complex formation in solution, for example, at early stages of endocytosis. Low-affinity interactions, on the other hand, might be enhanced when endocytic partner proteins are concentrated during CCP maturation (Schmid et al, 2006). McMahon and co-workers proposed a model for CCP maturation, according to which protein–protein interactions are defined on the basis of the affinity of their components in solution during early stages of CCP formation. In a second phase, after recruitment of endocytic proteins, the interactions are dominated by avidity effects arising from multiple interactions and high local protein concentrations. Finally, upon clathrin polymerization, the interactions are enforced by the matrix-like state (Schmid et al, 2006). With respect to this model, the broad range of affinities determined for EH (or other protein interaction) domains might correlate with different stages of endocytosis and hence network assembly. A high-affinity complex as that observed for Eps15 and stonin2 would allow complex formation at early, solution-like stages. To assess such a hierarchical grade of binding, it is necessary to conduct further in vivo and in vitro studies, taking into account additional binding sites that could modulate specificity and affinity further.
Materials and methods
Cloning, expression and purification
Proteins were expressed and purified according to standard methods; a detailed description is given in the Supplementary data.
Cell culture, antibodies and imaging experiments
Primary astrocytes isolated from newborn rats (P1, 8 DIV) were transfected using Lipofectamine 2000 (0.5 μg DNA and 1 μl Lipofectamine 2000 per well in 12-well plate). Astrocytes were fixed in 4% PFA at 12 DIV and processed for immunostaining. Cos7 cells were transfected using Lipofectamine 2000 (see above) and analysed by 24 h post-transfection by immunostaining following PFA (4%) fixation. The following antibodies were used: anti-HA (mouse monoclonal; Babco Inc.), anti-c-myc (clone 9E10; Haucke lab), anti-stonin2 (polyclonal antisera) and anti-Eps15 (a kind gift from PP Di Fiore). Images were acquired with a motorized Carl Zeiss Axiovert M200 inverted microscope under the control of the Stallion system (Intelligent Imaging Innovations), and processed by nearest neighbour deconvolution.
For the γBAR–stonin2 recruitment assay, Cos7 cells were cotransfected with a chimaeric construct comprising amino acids 1–100 of γBAR, an myc tag, and the stonin2 NPF region (amino acids 308–351) and EGFP-fused EH domains of Eps15 (EH1, EH2, EH3 and EH1–3), using Lipofectamine 2000 (Invitrogen). Twenty-four hours post-transfection, cells were fixed in 4% paraformaldehyde for 30 min at room temperature (RT). After removing the fixative and washing with PBS, cells were permeabilized and blocked with blocking buffer (15 mM sodium phosphate buffer, pH 7.4, 385 mM NaCl, 0.23% (w/v) Triton X-100, 30% (v/v) goat serum) for 10 min at RT before incubation with the primary anti-myc antibody (clone 9E10) in blocking buffer for 2 h at RT. Cells were washed three times for 10 min with high-salt PBS (20 mM sodium phosphate buffer, pH 7.4, 500 mM NaCl, 0.3% (w/v) Triton X-100) before the secondary antibody (goat anti-mouse-Alexa594) was applied in blocking buffer for 1 h at RT. Cells were washed for 5 min with HSPBS and twice for 5 min with 120 mM sodium phosphate buffer, pH 7.4, before mounting with Immumount mounting reagent (from Thermo Electron) supplemented with 4′,6-diamidino-2-phenylindole (DAPI). Recruitment of Eps15 EH domain to stonin2-containing structures was analysed by spinning-disc confocal microscopy, using the Perkin Elmer Ultra View ERS system, followed by image analysis using Improvision Volocity software.
IP experiments were performed according to standard methods; a detailed description of this experiment and a membrane recruitment assay are given in the Supplementary data.
Isothermal titration calorimetry
Complex formation between EH constructs and stonin2-derived fragments was measured using a VP-ITC microcalorimeter (MicroCal) at 15°C. Proteins were buffer-exchanged into 50 mM Tris–HCl, pH 7.5, 100 mM NaCl, 2 mM CaCl2 and 0.5 mM TCEP. Proteins at concentrations of 10–50 μM were titrated with a 100–500 μM solution of binding partner in 29 steps of 10 μl. The heat of diluting ligand into buffer was subtracted from the raw data. The data were analysed with a one-binding site model (two binding sites in the case of Eps15 EH1–3) using MicroCal Origin for ITC (version 7).
NMR spectroscopy
All complex spectra were recorded on differentially labelled samples of stonin2 301–340 and Eps15–EH2 in 10 mM perdeuterated Tris/HCl, pH 7 (CDN Isotopes, Pointe-Claire, Canada), 100 mM NaCl, 2 mM CaCl2, 1 mM DTT and 0.02% NaN3.
Spectra were processed with NMRPipe (Delaglio et al, 1995) and analysed using NMRView (Johnson and Blevins, 1994). The 1H, 13C and 15N chemical shifts were assigned by standard methods (Sattler et al, 1999). Distance restraints were derived from 15N- or 13C-resolved three-dimensional and 1H homonuclear two-dimensional NOESY. The procedure of extracting additional restraints (RDC, dihedrals and H-bonds) as well as description of titration and relaxation experiments is given in the Supplementary data.
Structure calculation
Only manually assigned NOEs were used to derive distance restraints. The experimentally determined distance, dihedral and dipolar coupling restraints (Table II) were applied in a simulated annealing protocol using ARIA (Linge et al, 2001) and CNS (Brunger et al, 1998). The final ensemble of NMR structures was refined in a shell of water molecules (Linge et al, 2003). Structural quality was analysed using PROCHECK (Laskowski et al, 1996). Analysis of the structured regions of the complex (Eps15–EH2, residues 122–215 and stonin2, residues 309–317 and 326–336) yielded 85.8% of all residues in the core region, 13.1% in the allowed and 0.9% in the disallowed region of the Ramachandran plot. Ribbon and surface representations were prepared with PYMOL (http://www.pymol.org).
Table 2.
NMR and refinement statistics for the Eps15–EH2·Stonin2 complex structure
| EH2–stonin2 complex | |
|---|---|
| NMR distance and dihedral constraints | |
| Distance constraints | |
| Total NOE | 4320 |
| Intra-residue | 1718 |
| Inter-residue | |
| Sequential (∣i−j∣=1) | 820 |
| Medium-range (∣i−j∣<4) | 694 |
| Long-range (∣i−j∣>5) | 688 |
| Intermolecular | 400 |
| Hydrogen bonds | 32 |
| Total dihedral angle restraints | 140 |
| φ | 68 |
| ψ | 68 |
| χ1 | 4 |
| Total RDCs | 118 |
| Q-factor (phage) | 0.13±0.01 |
| Q-factor (PEG) | 0.15±0.01 |
| Structure statistics | |
| Violations (mean and s.d.) | |
| Distance constraints (Å) | 0.03±0.00 |
| Dihedral angle constraints (deg) | 0.66±0.09 |
| Max. dihedral angle violation (deg) | 4.58 |
| Max. distance constraint violation (Å) | 0.44 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.01±0.00 |
| Bond angles (deg) | 0.68±0.02 |
| Impropers (deg) | 1.90±0.05 |
| Average pairwise r.m.s.d.(Å)a | |
| Heavy | 1.03±0.16 |
| Backbone | 0.53±0.13 |
| Bad contacts per 100 amino acids | 5.5±1.8 |
| EH, Eps15 homology; NMR, nuclear magnetic resonance; NOE, nuclear overhauser effect. | |
| aPairwise r.m.s. deviation was calculated among 10 lowest energy structures out of 100 refined structures (residues 122–215 of Eps15 and 309–317, 326–336 of the stonin2 peptide). | |
Accession codes
Coordinates for the Eps15–EH2–stonin2 complex structure have been deposited in the Protein Data Bank (accession code 2jxc). All assignments have been deposited in the BioMagResBank (http://www.bmrb.wisc.edu) under accession number 15554.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Information
Acknowledgments
We are indebted to Theresia Niese and Melanie Mueller for excellent technical support and to Robert Shoeman for mass spectrometry analysis. We are grateful to Dagmar Ringe and Arne Rufer for critical reading of the paper, and to Anton Meinhart for helpful discussions. We thank Ilme Schlichting for continuous support. The plasmids for human Eps15, Eps15R and rat epsin1 were kind gifts from Harvey McMahon, Alexandre Benmerah and Pietro De Camilli, respectively. This work was supported by the Max-Planck-Society and an Emmy-Noether Fellowship from the Deutsche Forschungsgemeinschaft to YG (DFG, GR1985/2-3). MS thanks the Deutsche Forschungsgemeinschaft and the EU (3D repertoire; LSHG-CT-2005-512028) for support, and the Center for Biomolecular Magnetic Resonance in Frankfurt for high-field NMR measurement time. VH acknowledges support by the German Ministry of Science (BMBF, BioDisc2: RENTRAFF) and the Deutsche Forschungsgemeinschaft (DFG, HA2686/1-2 and HA2686/4-1).
References
- Benmerah A, Gagnon J, Begue B, Megarbane B, Dautry-Varsat A, Cerf-Bensussan N (1995) The tyrosine kinase substrate eps15 is constitutively associated with the plasma membrane adaptor AP-2. J Cell Biol 131: 1831–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun A, Pinyol R, Dahlhaus R, Koch D, Fonarev P, Grant BD, Kessels MM, Qualmann B (2005) EHD proteins associate with syndapin I and II and such interactions play a crucial role in endosomal recycling. Mol Biol Cell 16: 3642–3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Chen H, Fre S, Slepnev VI, Capua MR, Takei K, Butler MH, Di Fiore PP, De Camilli P (1998) Epsin is an EH-domain-binding protein implicated in clathrin-mediated endocytosis. Nature 394: 793–797 [DOI] [PubMed] [Google Scholar]
- Coda L, Salcini AE, Confalonieri S, Pelicci G, Sorkina T, Sorkin A, Pelicci PG, Di Fiore PP (1998) Eps15R is a tyrosine kinase substrate with characteristics of a docking protein possibly involved in coated pits-mediated internalization. J Biol Chem 273: 3003–3012 [DOI] [PubMed] [Google Scholar]
- Confalonieri S, Di Fiore PP (2002) The Eps15 homology (EH) domain. FEBS Lett 513: 24–29 [DOI] [PubMed] [Google Scholar]
- Daumke O, Lundmark R, Vallis Y, Martens S, Butler PJ, McMahon HT (2007) Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling. Nature 449: 923–927 [DOI] [PubMed] [Google Scholar]
- de Beer T, Carter RE, Lobel-Rice KE, Sorkin A, Overduin M (1998) Structure and Asn–Pro–Phe binding pocket of the Eps15 homology domain. Science 281: 1357–1360 [DOI] [PubMed] [Google Scholar]
- de Beer T, Hoofnagle AN, Enmon JL, Bowers RC, Yamabhai M, Kay BK, Overduin M (2000) Molecular mechanism of NPF recognition by EH domains. Nat Struct Biol 7: 1018–1022 [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6: 277–293 [DOI] [PubMed] [Google Scholar]
- Di Fiore PP, Pelicci PG, Sorkin A (1997) EH: a novel protein–protein interaction domain potentially involved in intracellular sorting. Trends Biochem Sci 22: 411–413 [DOI] [PubMed] [Google Scholar]
- Diril MK, Wienisch M, Jung N, Klingauf J, Haucke V (2006) Stonin 2 is an AP-2-dependent endocytic sorting adaptor for synaptotagmin internalization and recycling. Dev Cell 10: 233–244 [DOI] [PubMed] [Google Scholar]
- Dutta K, Shi H, Cruz-Chu ER, Kami K, Ghose R (2004) Dynamic influences on a high-affinity, high-specificity interaction involving the C-terminal SH3 domain of p67phox. Biochemistry 43: 8094–8106 [DOI] [PubMed] [Google Scholar]
- Edeling MA, Mishra SK, Keyel PA, Steinhauser AL, Collins BM, Roth R, Heuser JE, Owen DJ, Traub LM (2006) Molecular switches involving the AP-2 beta2 appendage regulate endocytic cargo selection and clathrin coat assembly. Dev Cell 10: 329–342 [DOI] [PubMed] [Google Scholar]
- Enmon JL, de Beer T, Overduin M (2000) Solution structure of Eps15's third EH domain reveals coincident Phe–Trp and Asn–Pro–Phe binding sites. Biochemistry 39: 4309–4319 [DOI] [PubMed] [Google Scholar]
- Groemping Y, Lapouge K, Smerdon SJ, Rittinger K (2003) Molecular basis of phosphorylation-induced activation of the NADPH oxidase. Cell 113: 343–355 [DOI] [PubMed] [Google Scholar]
- Haffner C, Takei K, Chen H, Ringstad N, Hudson A, Butler MH, Salcini AE, Di Fiore PP, De Camilli P (1997) Synaptojanin 1: localization on coated endocytic intermediates in nerve terminals and interaction of its 170 kDa isoform with Eps15. FEBS Lett 419: 175–180 [DOI] [PubMed] [Google Scholar]
- Heuser J (1989) The role of coated vesicles in recycling of synaptic vesicle membrane. Cell Biol Int Rep 13: 1063–1076 [DOI] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA (1994) NMR VIEW—a computer-program for the visualization and analysis of NMR data. J Biomol NMR 4: 603–614 [DOI] [PubMed] [Google Scholar]
- Kami K, Takeya R, Sumimoto H, Kohda D (2002) Diverse recognition of non-PxxP peptide ligands by the SH3 domains from p67(phox), Grb2 and Pex13p. EMBO J 21: 4268–4276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieken F, Jovic M, Naslavsky N, Caplan S, Sorgen PL (2007) EH domain of EHD1. J Biomol NMR 39: 323–329 [DOI] [PubMed] [Google Scholar]
- Kim S, Cullis DN, Feig LA, Baleja JD (2001) Solution structure of the Reps1 EH domain and characterization of its binding to NPF target sequences. Biochemistry 40: 6776–6785 [DOI] [PubMed] [Google Scholar]
- Koshiba S, Kigawa T, Iwahara J, Kikuchi A, Yokoyama S (1999) Solution structure of the Eps15 homology domain of a human POB1 (partner of RalBP1). FEBS Lett 442: 138–142 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8: 477–486 [DOI] [PubMed] [Google Scholar]
- Linge JP, O'Donoghue SI, Nilges M (2001) Automated assignment of ambiguous nuclear overhauser effects with ARIA. Methods Enzymol 339: 71–90 [DOI] [PubMed] [Google Scholar]
- Linge JP, Williams MA, Spronk CA, Bonvin AM, Nilges M (2003) Refinement of protein structures in explicit solvent. Proteins 50: 496–506 [DOI] [PubMed] [Google Scholar]
- Martina JA, Bonangelino CJ, Aguilar RC, Bonifacino JS (2001) Stonin 2: an adaptor-like protein that interacts with components of the endocytic machinery. J Cell Biol 153: 1111–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer BJ (2001) SH3 domains: complexity in moderation. J Cell Sci 114: 1253–1263 [DOI] [PubMed] [Google Scholar]
- Mayer BJ, Eck MJ (1995) SH3 domains. Minding your p's and q's. Curr Biol 5: 364–367 [DOI] [PubMed] [Google Scholar]
- Mishra SK, Hawryluk MJ, Brett TJ, Keyel PA, Dupin AL, Jha A, Heuser JE, Fremont DH, Traub LM (2004) Dual engagement regulation of protein interactions with the AP-2 adaptor alpha appendage. J Biol Chem 279: 46191–46203 [DOI] [PubMed] [Google Scholar]
- Montesinos ML, Castellano-Munoz M, Garcia-Junco-Clemente P, Fernandez-Chacon R (2005) Recycling and EH domain proteins at the synapse. Brain Res Brain Res Rev 49: 416–428 [DOI] [PubMed] [Google Scholar]
- Morgan JR, Prasad K, Jin S, Augustine GJ, Lafer EM (2003) Eps15 homology domain–NPF motif interactions regulate clathrin coat assembly during synaptic vesicle recycling. J Biol Chem 278: 33583–33592 [DOI] [PubMed] [Google Scholar]
- Motley AM, Berg N, Taylor MJ, Sahlender DA, Hirst J, Owen DJ, Robinson MS (2006) Functional analysis of AP-2 alpha and mu2 subunits. Mol Biol Cell 17: 5298–5308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naslavsky N, Caplan S (2005) C-terminal EH-domain-containing proteins: consensus for a role in endocytic trafficking, EH? J Cell Sci 118: 4093–4101 [DOI] [PubMed] [Google Scholar]
- Naslavsky N, Rahajeng J, Chenavas S, Sorgen PL, Caplan S (2007) EHD1 and Eps15 interact with phosphatidylinositols via their Eps15 homology domains. J Biol Chem 282: 16612–16622 [DOI] [PubMed] [Google Scholar]
- Neubrand VE, Will RD, Mobius W, Poustka A, Wiemann S, Schu P, Dotti CG, Pepperkok R, Simpson JC (2005) Gamma-BAR, a novel AP-1-interacting protein involved in post-Golgi trafficking. EMBO J 24: 1122–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoluzi S, Castagnoli L, Lauro I, Salcini AE, Coda L, Fre' S, Confalonieri S, Pelicci PG, Di Fiore PP, Cesareni G (1998) Recognition specificity of individual EH domains of mammals and yeast. EMBO J 17: 6541–6550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke GJ, Ford MG, Schmid EM, Olesen LE, Gallop JL, Peak-Chew SY, Vallis Y, Babu MM, Mills IG, McMahon HT (2004) Evolving nature of the AP2 alpha-appendage hub during clathrin-coated vesicle endocytosis. EMBO J 23: 4371–4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcini AE, Confalonieri S, Doria M, Santolini E, Tassi E, Minenkova O, Cesareni G, Pelicci PG, Di Fiore PP (1997) Binding specificity and in vivo targets of the EH domain, a novel protein–protein interaction module. Genes Dev 11: 2239–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler M, Schleucher J, Griesinger C (1999) Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog NMR Spectrosc 34: 93–158 [Google Scholar]
- Schmid EM, Ford MG, Burtey A, Praefcke GJ, Peak-Chew SY, Mills IG, Benmerah A, McMahon HT (2006) Role of the AP2 beta-appendage hub in recruiting partners for clathrin-coated vesicle assembly. PLoS Biol 4: e262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M (1989) SPXX, a frequent sequence motif in gene regulatory proteins. J Mol Biol 207: 61–84 [DOI] [PubMed] [Google Scholar]
- Tebar F, Confalonieri S, Carter RE, Di Fiore PP, Sorkin A (1997) Eps15 is constitutively oligomerized due to homophilic interaction of its coiled-coil region. J Biol Chem 272: 15413–15418 [DOI] [PubMed] [Google Scholar]
- van Delft S, Schumacher C, Hage W, Verkleij AJ, van Bergen en Henegouwen PM (1997) Association and colocalization of Eps15 with adaptor protein-2 and clathrin. J Cell Biol 136: 811–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther K, Diril MK, Jung N, Haucke V (2004) Functional dissection of the interactions of stonin 2 with the adaptor complex AP-2 and synaptotagmin. Proc Natl Acad Sci USA 101: 964–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman H, Yarden Y (2001) Molecular mechanisms underlying endocytosis and sorting of ErbB receptor tyrosine kinases. FEBS Lett 490: 142–152 [DOI] [PubMed] [Google Scholar]
- Yamabhai M, Hoffman NG, Hardison NL, McPherson PS, Castagnoli L, Cesareni G, Kay BK (1998) Intersectin, a novel adaptor protein with two Eps15 homology and five Src homology 3 domains. J Biol Chem 273: 31401–31407 [DOI] [PubMed] [Google Scholar]
- Zarrinpar A, Bhattacharyya RP, Lim WA (2003) The structure and function of proline recognition domains. Sci STKE 2003: RE8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Information