Abstract
Telomeres terminate in 3′ overhangs that function in end protection and the formation of t-loops. Determining the steps and factors involved in overhang processing is compromised by the inability to easily and accurately determine overhang size in the presence of many kilobases of double-stranded telomeric DNA. We here describe the use of a double-strand specific nuclease (DSN) that entirely digests double-stranded DNA including telomeres, leaving the overhangs intact so that they can be measured.
INTRODUCTION
Mammalian telomeres consist of repetitive hexamers of TTAGGG terminating in single-stranded overhangs at the 3′ end of the chromosome (1). The overhang is thought to be involved in t-loop formation, structurally masking the chromosome end from being recognized as a double-stranded break (DSB). The overhang also protects telomeres from end-to-end fusions, abnormal recombination and degradation (2). Thus, it is important to investigate and identify the processes and factors involved in production of the single-stranded telomeric overhang. However, technical limitations in the past have made it difficult to study overhang behavior.
Despite several methods developed to determine overhang length, an accurate measurement has been challenging. Both the non-denaturing hybridization assay (3) and the newly developed HPA method (4) only determine the relative strength of overhang signals with respect to total DNA. The telomeric-oligonucleotide ligation assay (T-OLA) (5) gives an imperfect representation of sizes and primarily estimates their maximum length. The primer extension-nick translation method (PENT) (6) can demonstrate the presence of an overhang but gives little information about short overhangs. Electron microscopy requires large amounts of purified telomeres and is unable to measure lengths shorter than ∼75 nt (7). The gp32 overhang protection assay previously developed by our lab can determine longer overhang lengths, but overhangs shorter than <45 nt cannot be assayed (8). The inability of these methods to analyze short overhangs may produce misleading results. For example, our previous study found the overhang length in telomeres produced by leading strand synthesis (∼60 nt) was about twice the size predicted from the rate of telomere shortening (9).
In this report, we present a novel and simple method for direct measurement of overhang length that utilizes Kamchatka crab duplex specific nuclease (DSN). DSN is a newly characterized endonuclease that is highly specific for double-stranded (ds) DNA and is practically inactive towards RNA and single-stranded (ss) DNA (10). DSN has been successfully used for cDNA normalization and the detection of single nucleotide polymorphisms (11,12). DSN digestion should preserve the telomeric overhangs while removing all ds DNA, allowing overhang detection on Southern blots (Figure 1). We show that DSN can successfully determine the size of telomeric overhangs in the presence of total genomic DNA, the shortest human telomeres are 12 nt long, and the average size of the overhangs on leading strand daughter telomeres are only 40 nt long, thus 2–3 times shorter than those on lagging strand daughter telomeric overhangs.
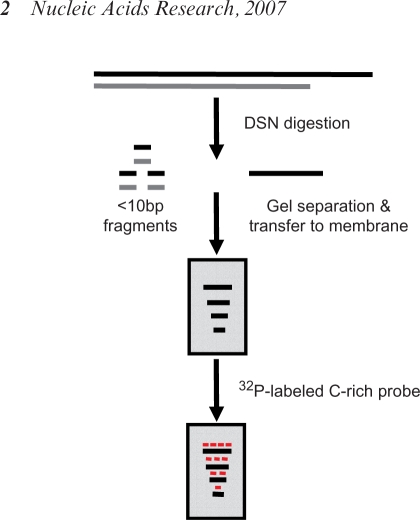
Figure 1.
Strategy of the DSN base overhang assay. Complete and specific digestion of all double-stranded DNA should leave the single-stranded TTAGGG sequences present in the telomeric 3′ overhangs available for analysis on denaturing gels.
MATERIALS AND METHODS
Biochemicals
Duplex-specific Nuclease was obtained from Evrogen Joint Stock Company (Russia); Exonuclease I from Epicentre Biotechnologies; and T7 Exonuclease and all restriction enzymes from New England Biolabs, Inc. Oligonucleotides and RNA were synthesized by IDT (Integrated DNA Technologies).
Cell culture
Cells were cultured at 37°C under 5% CO2 in a 4:1 mixture of Dulbecco's modified Eagle's medium/Medium 199 supplemented with 10% cosmic calf serum (HyClone, Logan, UT).
Genomic DNA purification
Genomic DNA was isolated using the DNAeasy kit (QIAGEN). Since DSN is inhibited by salt, the eluted DNA was re-precipitated by adding two volume of 100% ethanol, washed twice with 70% ethanol and then suspended in distilled water (Millipore) at a concentration of <1 µg/µl. Overnight incubation at 37°C was used to ensure that all the DNAs were completely solubilized.
DSN assay for the determination of telomeric overhang length
Typical reactions were performed in 20 μl. Five microgram of genomic DNA in 1× DSN buffer (provided by the manufacturer) was digested with 0.2 U DSN at 37°C for 2 h. As a control, 10 U ExoI was added to genomic DNA prior to DSN treatment and incubated at 37°C for 1 h to digest 3′ overhangs. The digestion was stopped by adding 0.5 μl of 0.5 M EDTA. After the same volume of formamide (deionized) was added, samples were heated at 65°C for 5 min and loaded on 6% denaturing polyacrylamide gels containing 8 M urea. Electrophoresis was performed at 15 V/cm in 0.5× TBE until the Bromphenol Blue tracking dye migrated two-thirds the length of the gel. DNA was transferred to a presoaked Hybond-N+ membrane (Amersham) using the Bio-Rad electronic transfer system in 0.5× TBE buffer at 4°C at 30 V (6 v/cm, constant voltage). After 90 min the membrane was air-dried, UV cross-linked and hybridized to a high specific activity C-rich probe [an 18-mer C-rich probe containing six 32P-dC, prepared as described (13)] at 42°C for at least 4 h in Rapid-hybrid buffer (Amersham Biosciences). After 2 × 15 min washes with 0.1× SSC + 0.5% SDS at 42°C, membranes were exposed to PhosphoImager screens and quantitated using ImageQuant (Amersham Biosciences). Mean sizes of overhangs were calculated using the program Telorun as described (8). Briefly, a column of 100 boxes was overlaid on each lane and the signal intensity (volume) of each box was determined. The background signal from each box in the ExoI-treated lane was then subtracted from the signal in the untreated lane, and then normalized by dividing the signal by the MW of the box. The mean average length was then calculated using the formula ∑ (ODi)/∑ (ODi/Li), where ODi is the PhosphorImager output (signal intensity) and Li is the length of the DNA at position i. Six nucleotides were then subtracted from the calculated average since DSN fails to digest the final 5–6 ds bp at the ds–ss junction (Figure 3). Sizes above 400 nt were excluded from the analysis due to the presence of variable amounts of ExoI insensitive material in this size range.
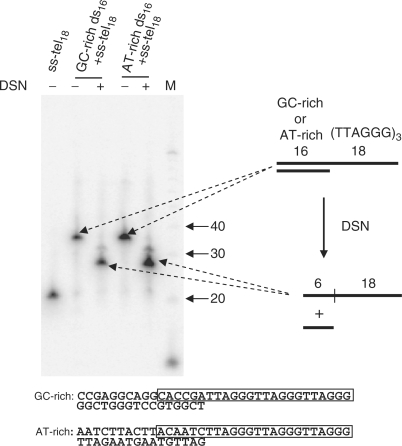
Figure 3.
Five to six ds junctional nucleotides are resistant to DSN treatment. Here, 34 nt oligonucleotides containing either 16 bp of AT-rich or GC-rich sequence followed by 18 nt of ss telomeric sequence were incubated with or without DSN. After incubation, the DNA was loaded onto a 10% polyacrylamide gel with 8 M urea and DNA was then transferred to a membrane and hybridized with a hot C-rich telomeric probe. The sequences of the annealed oligonucleotides are shown, and the regions surviving DSN digestion are boxed. Five to six nucleotides of ds DNA remain undigested regardless of the specific ds–ss junctional sequence.
Separation of leading and lagging daughter telomeres
The procedure was performed as described (9) with minor modifications. Briefly, exponentially growing BJ cells (Population doubling = 35–40, doubling time ∼48 h) were grown in 100 M BrdU for 48 h and then genomic DNA was isolated as described above. All the steps were performed in the dark. Typically, 30 μg genomic DNA was then digested with a mixture of six restriction enzymes (HinfI, RsaI, MspI, HaeIII, AluI, CfoI from NEB) in NEB buffer 2. Digestion was stopped by adding EDTA to 20 mM and the sample was mixed into a CsCl solution (density of 1.75 g/ml containing 5 mM Tris pH 8.0 and 2 mM EDTA). Samples were ultracentrifuged at 55 000 r.p.m. for 20 h at 25°C in a VTI80 vertical rotor (Beckman). Fractions were then collected and aliquots from each fraction were analyzed for density (refractive index) and telomeres (slot blots) as described (9). DNA containing the leading strand daughter DNA was located at a density of ∼1.785 g/ml, the lagging strand daughter DNA was at a density of ∼1.755 g/ml, while the lightest DNA contained the unreplicated DNA (density ∼1.735 g/ml). Three fractions located at each peak were pooled and the DNA was ethanol precipitated after reducing the CsCl concentration by surface dialysis (rocking ∼0.3 ml of solution on a slanted layer of 2% agarose in a 50 ml tube for 30 min at room temperature).
RESULTS
Characterization of DSN
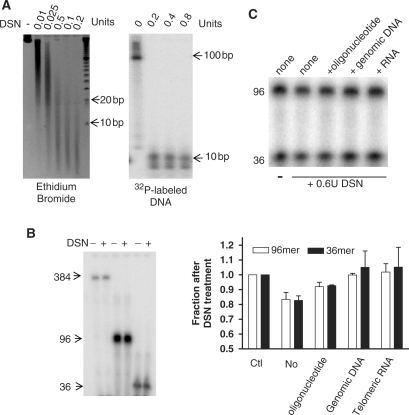
An essential requirement for this approach is that DSN must leave ss overhangs intact while digesting ds telomeric DNA to sizes too small to interfere. DSN (0.2 U) is sufficient to digest 5 µg genomic DNA to <10 bp fragments (Figure 2A, left). In order to follow the behavior of telomeric DNA, continuously labeled radioactive ds TTAGGG repeats (96 bp) were added to the genomic DNA. DSN reduced ds telomeric sequences to ∼10 bp and excess enzyme failed to further reduce their size (Figure 2A, right). Both the size and signal intensity of nanomolar amounts of single-stranded (TTAGGG)n oligonucleotide standards (36–384 nt) remained unaltered after DSN digestion (Figure 2B), indicating that under these condition there is no detectable degradation of ss telomeric DNA. A very small amounts of single-stranded nuclease activity is described by the manufacturer (11) and we were able to detect this minimal nonspecific nuclease activity against ss DNA only if high concentrations of both DSN enzyme (0.6 U) and substrate (µM) were used (Figure 2C). However, competitive ds (genomic) DNA and RNA all quantitatively prevented this potential single-strand degradation (Figure 2C). The 5 µg of total genomic DNA used per reaction will thus eliminate any potential minor degradation of the ∼0.15 fmol of overhangs that are present.
Figure 2.
Characterization of DSN in telomere overhang assays. (A) DSN digests ds telomeres to <10 bp. Left: Five microgram of Hela genomic DNA was digested with DSN and analyzed on 6% polyacrylamide gel with ethidium bromide staining for total DNA. Right: One nanogram of ds telomeric DNA (96 bp, continuously 32P-labeled by primer extension) was added to 5 μg genomic DNA as above. (B) Single-stranded telomeric DNA is resistant to DSN digestion. Approximately two nanomolar of 32P-end-labeled oligonucleotides were digested with 0.2 U of DSN at 37°C for 2 h. (C) Minor ss activity of DSN is eliminated by excess competitor. Top: End-labeled telomeric oligonucleotides (0.5 μM each of 16 and 6 repeats in a 20 μl reaction) were digested with 0.6 U of DSN at 37°C for 1 h. Two microgram of different competitors (a 33-mer random oligonucleotide, genomic DNA from HT1080 cells or a UUAGGGUUAGGGUUAGGGUU RNA) were added. Bottom: The fraction remaining after DSN digestion in the presence of various competitors was quantitated in two experiments, ±SD.
Limits of DSN digestion
In order to test the possibility that DSN might not be able to remove all of the ds nucleotides immediately adjacent to a ss overhang we examined its activity on synthetic model telomeres (16 bp of ds DNA followed by 18 nt of ss TTAGGG sequence). The length after DSN digestion was 24 nt, 6 nt longer than the actual model overhang (Figure 3). Five to six nucleotides of the ds region remained undigested regardless of whether the 16 nt ds segment contained a 75% or a 25% GC content, showing that this limit of digestion was sequence independent. The unexpected property of DSN to leave a relatively precise rather than variable addition at ds–ss telomeric junctions allows one to accurately calculate overhang length by subtracting 6 nt from the measured length.
DSN digestion detects human telomeric overhangs
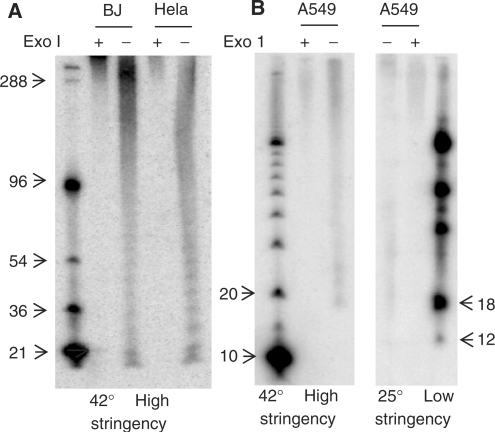
To further validate this method, we compared telomeric overhangs in BJ foreskin fibroblasts and Hela cervical carcinoma cells. The overhangs varied from 20 to 400 nt (Figure 4A), which is consistent with previous results obtained using the gp32 protection assay (8) and electron microscopy (7). The signals below 400 nt are sensitive to the 3′→5′ activity of Exonuclease I, indicating that they are 3′ overhangs and not coming from internal ss DNA released by DSN. No signals were observed using a G-rich telomeric probe (data not shown), confirming that there are no detectable C-rich telomeric overhangs as reported using the T-OLA technique (14) and that no signal arises from incomplete digestion of telomere double-strand DNA. The periodicity in the banding pattern was not always present and remains unexplained. Resection of genomic DNA with T7 exonuclease prior to DSN digestion resulted in elongation of the overhangs as expected (data not shown).
Figure 4.
DSN detects human telomeres as short as 12 nt. (A) Representative DSN overhang assays on BJ fibroblast and Hela adenocarcinoma DNA. Results are quantitated in Table 1. ExoI pretreatment removes the 3′ overhang. (B) A sharp signal loss below 18 nt under normal high stringency hybridization condition is also seen when the membrane was hybridized at low stringency. The signal at 18 nt corresponds to an overhang length of 12 nt.
There is a very sharp disappearance of signal below 18 nt in human telomeres (Figure 4A and B, left). When lower stringency hybridization conditions were used (25°C hybridization and 2× SSC washes instead of 42°C hybridization and 0.1× SSC washes), no additional signal was observed below 18 nt (Figure 4B, right), suggesting that the lack of signal below 18 nt was not due to the inability to stably hybridize to shorter sequences. Therefore, our results imply that the minimum overhang length is 12 nt (18 nt minus 6 junctional bp), and that overhangs shorter than this may not be present.
Table 1 compares the average overhang length of several cultured cells with that obtained by the gp32 protection method (8). Averages are similar but somewhat smaller using DSN because of its ability to include smaller overhangs. As previously observed, we found no reduction of overhang length during cell senescence. Furthermore, our data confirmed that relatively longer overhangs are present in cells lacking telomerase activity, consistent with results obtained by T-OLA analysis of both cultured cells (5) and tissue samples (15).
Table 1.
Comparison of telomere overhang lengths (nt) obtained using the DSN and gp32 protection methods
Leading strand daughter overhangs are much shorter than lagging overhangs
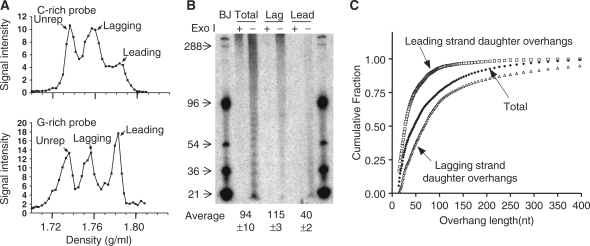
We used CsCl gradients of DNA hemisubstituted with 5-bromodeoxyuridine (9) to separate leading and lagging strand daughter telomeres from BJ fibroblasts and determined their overhang lengths using DSN. Figure 5A shows the profile of separation of unreplicated, lagging and leading telomeres based upon the fraction of BrdU present in each strand. The apparently unequal fraction of lagging versus leading daughters obtained when hybridizing to a C-rich telomeric probe is not seen when a G-rich probe is used. One possible explanation is that BrdU incorporation sensitizes DNA to UV damage. The UV cross-linking used to fix the DNA to positively charged membranes might produce greater amounts of damage that renders the DNA less accessible to hybridization on strands that can incorporate two BrdU per six residues (the TTAGGG sequence) compared to those incorporating only one per six nucleotides (the CCCTAA sequence). As a result, a G-rich probe hybridizing to the better preserved C-strands would give a better representation of the DNA actually present in each fraction. Consistent with this interpretation, C-rich probes gave approximately equal intensities for the leading and lagging peaks when baking rather than UV cross-linking was used to fix the DNA to the membranes (data not shown). Figure 4B shows a typical pattern of overhang distribution, where leading strand overhangs (∼40 nt) are almost three times shorter than those in lagging strands (∼115 nt), which is consistent with our previous calculations (9). However, the fact that >70% of leading overhang are shorter than 50 nt (Figure 2C) demonstrates the large fraction that could not be measured by the prior gp32 overhang protection technique. Our previous speculation (9) that leading overhangs were likely to be ∼35 nt in BJ cells rather than the measured value of 60 nt has thus been confirmed by the DSN method.
Figure 5.
Human leading strand daughter telomeres have very short overhangs. (A) Separation of telomeric leading and lagging strands from BJ cells grown in BrdU for 48 h using CsCl gradients (slot blot results). The signal from leading strand daughter telomeres appeared to be less than that from lagging telomeres only when a C-rich probe was used. (B) Overhangs at lagging and leading strand daughter telomeres in normal fibroblasts. Averages are mean ± SD, n = 2. (C) Distribution of overhang sizes. The cumulative fraction of overhangs shorter than a given size is plotted after quantitating the signals in (B) to illustrate the distribution of lengths.
DISCUSSION
The measurement of very short overhangs is extremely challenging. Electron microscopy analysis demonstrated that normal cells had a long overhang at one end, but the limits of detection did not exclude the possibility that the other end had very short overhangs that were undetectable by EM (7). Using PENT, Makarov et al. claimed that >80% of telomeres had long overhangs in the range of 130–210 (6). The DSN method extends overhang detection limits to 12 nt, which allowed us to reevaluate the distribution of short overhangs. We found that the daughters of leading strand synthesis have overhangs that are as short as 12 nt, that average ∼40 nt, and that are 2–3 times shorter than those on the products of lagging strand synthesis.
Telomeric overhangs are critical for the recruitment of telomerase and proper end-protection mechanisms, but the molecules and steps involved in generating overhangs are largely unknown. Several factors such as Mre11 (16) and Pot1 (17) have been shown to effect overhang size, but their precise role remains obscure and identifying the nucleases and mechanisms producing 3′ overhangs remains a major unsolved issue. This method can determine overhangs as short as 12 nt, is simple and can be done in one day without any unusual reagents other than DSN. This should now enable much more rapid progress in discovering and characterizing the processing events that occur at the ends of human chromosomes.
ACKNOWLEDGEMENTS
This work was funded by National Institute on Aging [AG01228]. Funding to pay the Open Access publication charges for this article was provided by AG01228.
Conflict of interest statement. None declared.
REFERENCES
- 1.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 2.de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- 3.McElligott R, Wellinger RJ. The terminal DNA structure of mammalian chromosomes. EMBO J. 1997;16:3705–3714. doi: 10.1093/emboj/16.12.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tahara H, Kusunoki M, Yamanaka Y, Matsumura S, Ide T. G-tail telomere HPA: simple measurement of human single-stranded telomeric overhangs. Nat. Methods. 2005;2:829–831. doi: 10.1038/nmeth797. [DOI] [PubMed] [Google Scholar]
- 5.Cimino-Reale G, Pascale E, Battiloro E, Starace G, Verna R, D'Ambrosio E. The length of telomeric G-rich strand 3′-overhang measured by oligonucleotide ligation assay. Nucleic Acids Res. 2001;29:E35. doi: 10.1093/nar/29.7.e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makarov VL, Hirose Y, Langmore JP. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- 7.Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chai W, Shay JW, Wright WE. Human telomeres maintain their overhang length at senescence. Mol. Cell. Biol. 2005;25:2158–2168. doi: 10.1128/MCB.25.6.2158-2168.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chai W, Du Q, Shay JW, Wright WE. Human telomeres have different overhang sizes at leading versus lagging strands. Mol. Cell. 2006;21:427–435. doi: 10.1016/j.molcel.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Anisimova VE, Rebrikov DV, Zhulidov PA, Staroverov DB, Lukyanov SA, Shcheglov AS. Renaturation, activation, and practical use of recombinant duplex-specific nuclease from Kamchatka crab. Biochemistry. 2006;71:513–519. doi: 10.1134/s0006297906050075. [DOI] [PubMed] [Google Scholar]
- 11.Shagin DA, Rebrikov DV, Kozhemyako VB, Altshuler IM, Shcheglov AS, Zhulidov PA, Bogdanova EA, Staroverov DB, Rasskazov VA, et al. A novel method for SNP detection using a new duplex-specific nuclease from crab hepatopancreas. Genome Res. 2002;12:1935–1942. doi: 10.1101/gr.547002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhulidov PA, Bogdanova EA, Shcheglov AS, Vagner LL, Khaspekov GL, Kozhemyako VB, Matz MV, Meleshkevitch E, Moroz LL, et al. Simple cDNA normalization using kamchatka crab duplex-specific nuclease. Nucleic Acids Res. 2004;32:e37. doi: 10.1093/nar/gnh031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herbert B, Shay JW, Wright WE. Analysis of telomeres and telomerase. In: Bonifacino JS, Dasso M, Lippincott-Schwartz J, Harford JB, Yamada KM, editors. Current Protocols in Cell Biology (online). Hoboken, NJ: Wiley and Sons; 2003. pp. 18.6.1–18.6.20. [DOI] [PubMed] [Google Scholar]
- 14.Stewart SA, Ben-Porath I, Carey VJ, O’Connor BF, Hahn WC, Weinberg RA. Erosion of the telomeric single-strand overhang at replicative senescence. Nat. Genet. 2003;33:492–496. doi: 10.1038/ng1127. [DOI] [PubMed] [Google Scholar]
- 15.Hashimoto M, Kyo S, Masutomi K, Maida Y, Sakaguchi J, Mizumoto Y, Nakamura M, Hahn WC, Inoue M. Analysis of telomeric single-strand overhang length in human endometrial cancers. FEBS Lett. 2005;579:2959–2964. doi: 10.1016/j.febslet.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 16.Chai W, Sfeir AJ, Hoshiyama H, Shay JW, Wright WE. The involvement of the Mre11/Rad50/Nbs1 complex in the generation of G-overhangs at human telomeres. EMBO Rep. 2006;7:225–230. doi: 10.1038/sj.embor.7400600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hockemeyer D, Sfeir AJ, Shay JW, Wright WE, de Lange T. POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J. 2005;24:2667–2678. doi: 10.1038/sj.emboj.7600733. [DOI] [PMC free article] [PubMed] [Google Scholar]