Abstract
The creation of novel enzymes capable of catalyzing any desired chemical reaction is a grand challenge for computational protein design. Here we describe two new algorithms for enzyme design that employ hashing techniques to allow searching through large numbers of protein scaffolds for optimal catalytic site placement. We also describe an in silico benchmark, based on the recapitulation of the active sites of native enzymes, that allows rapid evaluation and testing of enzyme design methodologies. In the benchmark test, which consists of designing sites for each of 10 different chemical reactions in backbone scaffolds derived from 10 enzymes catalyzing the reactions, the new methods succeed in identifying the native site in the native scaffold and ranking it within the top five designs for six of the 10 reactions. The new methods can be directly applied to the design of new enzymes, and the benchmark provides a powerful in silico test for guiding improvements in computational enzyme design.
Keywords: enzyme design, protein design, active site recapitulation, protein–ligand interactions, geometric hashing
Enzymes are among the most efficient, specific, and selective catalysts known. The ability to design efficient enzymes for a broad class of different reactions would be of tremendous practical interest for both science and the industry. Furthermore, the rational design of enzymes is a stringent test of our understanding of biological catalysis.
There has been exciting progress in enzyme design. On the experimental side, catalytic antibodies, elicited by immunization with transition state analogs, have been shown to possess catalytic activity (Lerner et al. 1991; Hilvert 2000). More recently, several successful enzyme designs have been reported. Kaplan and DeGrado (2004) designed a de novo O2-dependent phenol oxidase within a designed four-helix bundle fold. Using computational protein design, Bolon and Mayo (2001) created a histidine-bearing catalyst for the hydrolysis of p-nitrophenyl acetate into p-nitrophenol. More recently, Dwyer et al. (2004) created a highly active enzyme by grafting the triose phosphate isomerase (TIM) active site on to the ribose binding protein scaffold.
The computational methods used in enzyme site design to date, such as ORBIT from the Mayo group (Dahiyat and Mayo 1996) and Dezymer from the Hellinga group (Hellinga and Richards 1991), have primarily been used to search for catalytic site placement in one or a small number of scaffolds. In contrast, computational methods for searching for functional sites that employ geometric hashing (Russell 1998) are able to search through thousands of scaffolds rapidly. However, these methods have been used in cases where the positions of the side chain functional groups are known, as in the case of proteins of known structure and thus are not immediately applicable to enzyme active site design.
In general, how to evaluate and optimize computational design methods for the creation of new molecules is a nontrivial problem. For robust conclusions, it is desirable to compare alternative methods and parameter choices by comparing results on a representative set of test systems. In the “protein design cycle” approach described by Dahiyat and Mayo (1997), alternative choices in a design method are tested by producing designs and experimentally characterizing them, and the choice is selected that produces designs with the desired properties. While this is a very powerful approach, experimentally characterizing a large number of designs for a number of different methods is slow and expensive, and therefore, it is desirable to have a faster and cheaper test. A purely in silico test for monomeric protein design developed in our group based on recapitulation of native protein sequences (Kuhlman and Baker 2000) has proven invaluable in guiding improvement of our protein design methodology.
In this study, we describe an in silico benchmark for computational enzyme design based on recapitulation of the locations and structures of native enzyme active sites in a set of naturally occurring enzymatic scaffolds. Given the backbone coordinates of 10 naturally occurring enzymes, and a list of the 10 reactions they catalyze, active sites are designed for each reaction in each scaffold. The designs for each reaction are collectively ranked based on their computed catalytic efficacy (see Materials and Methods). To evaluate and guide the optimization of enzyme design methodology, we make the assumption that the actual native enzyme is likely to be a better catalyst than any of the designed enzymes. With this assumption, alternative design methods can be evaluated based on the ranks of the actual native active site for each reaction among all the designs found and the associated computational cost required for the large number of calculations involved.
We also describe two new methods for computational enzyme design that utilize hashing algorithms to enable active site searches in large numbers of scaffolds. Given a description of a catalytic site consisting of a transition state structure surrounded by protein functional groups in geometrical positions optimal for catalysis and a set of protein scaffolds, the methods first search for sites in the scaffolds where the active site can be recapitulated. In the first method, an “inverse rotamer tree” approach is used with a modified version of the geometric hashing algorithm (Bachar et al. 1993) to find positions in a set of scaffolds that can support the catalytic site. In the second method, based on iterative side chain placement and hashing in six-dimensional space, candidate catalytic sites in scaffolds are detected in linear time. Both methods are followed by the design of the pocket using the standard Rosetta design methodology (Kuhlman and Baker 2000). We describe the performance of the two methods in the in silico enzyme design benchmark.
Results
Summary of the methods
In this work, we have developed general methods for searching for new active sites in a library of protein scaffolds and designing the residues surrounding these potential active sites to further stabilize the transition state. In the first, “inside-out” method, an “inverse rotamer tree” is built up from the active site description, and the backbone coordinates of all the rotamer combinations are compared to backbone coordinates of the set of scaffolds using a geometric-hashing based algorithm. In the second, “outside-in” method, side chain rotamers and the transition state (TS) model are sequentially placed at all scaffold positions, and the position of the TS model is recorded in a hash table. The hash table is then scanned for TS positions that are found when placing each of the catalytic side chains independently. These positions represent sites in the scaffolds where the specified active site can be successfully constructed. The two methods have complementary strengths and weaknesses. The first method can search through large numbers of scaffolds, since the spatial relations between residues are all precomputed, but it requires combinatorial enumeration of catalytic side chain rotamer positions. The second method is comparable for searching through a set of scaffolds for a relatively simple site, but because the catalytic side chains are treated independently rather than combinatorially, it is the method of choice for searching complex active sites with finer side chain rotamer sampling. After putative active sites have been identified by one of the two methods, the remaining residues in the pocket around the docked TS model are redesigned to optimize transition state binding affinity. The resulting designs are ranked based on their catalytic efficacies as estimated based on the fit of the catalytic residues to the active site description and the computed TS binding energy.
Recapitulation of native enzymatic sites
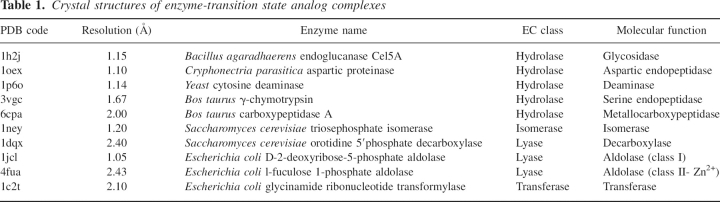
We use two native active site recapitulation tests to benchmark the two new methods. Ten crystal structures of enzyme-transition state analog complexes or enzyme–inhibitor complexes with a resolution of 2.5 Å or better were taken from the Protein Data Bank (PDB) (Berman et al. 2002). The resulting benchmark set includes members of the hydrolase, lyase, isomerase, and transferase enzyme families (Table 1); the only major family missing is the oxidoreductase family, which typically employs non-protein cofactors. The number of catalytic residues at the active sites varies from two to four, and the catalytic amino acids include Asp, Glu, Asn, His, Cys, Ser, Tyr, and Lys (Table 1). The catalytic residues documented as being involved in the catalytic mechanism for each enzyme of our benchmark are used to build the catalytic site descriptions for the corresponding reaction. For each chemical reaction, two benchmark tests were carried out using the complete protocol described in Figure 1, using either the inverse rotamer tree method or the RosettaMatch method. In the first benchmark test, the geometrical parameters relating the TS analog and the functional atoms are taken directly from the crystal structure of the complex. In the second benchmark test, the geometrical parameters are set to optimal values based on the simple rules described in Table 2. The challenge is to recapitulate the native active site by correctly identifying among all designs in all scaffolds the native site in the native scaffold based on the predicted catalytic efficacy.
Table 1.
Crystal structures of enzyme-transition state analog complexes
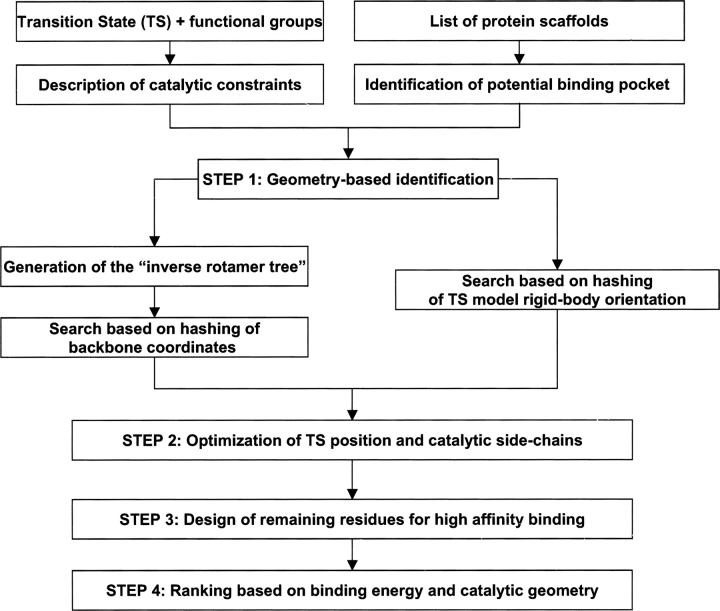
Figure 1.
Diagram of the computational enzyme design procedure.
Table 2.
Catalytic geometry parameters used in benchmark II
Benchmark results starting from native catalytic geometries
For the first test, the TS model and the functional group geometry, but not the conformations of the catalytic side chains, are taken directly from the crystal structure. The results using both match methods are reported at each stage in the design process in Table 3. We expect that a good enzyme design method should identify the naturally occurring site in the correct scaffold and rank it relatively high compared to non-native sites. For all 10 native active sites, both native matches (native catalytic residues at native sequence positions) as well as cross-matches (different positions in the native scaffold or a non-native scaffold) are found. Encouragingly, the rank of matches in the native scaffold in the native positions improves throughout the design process: after minimization (described in step 2 of the Materials and Methods section) and design (step 3), both methods lead to a remarkably good native site recapitulation (Fig. 2). In six out of the 10 benchmark sets, the design predicted to bind the TS model the tightest is in the native scaffold in the native positions. For the remaining benchmark cases, the rank is usually within the first percentile, except for the deoxyribose-phosphate aldolase (DERA) and aspartic proteinase cases with the RosettaMatch method. Both methods not only recapture the native enzymatic site in most cases but also accurately reproduce the TS model position and active site side chain conformations. Two examples of active site recapitulation are shown in Figure 3. The results for the benchmark show that the inverse rotamer tree and RosettaMatch perform equally well on average for the test cases, leading to good discrimination by score between native and non-native matches after minimization and design. The nonidentical ranking of the native matches found using the two methods is due in part to the use of different rotamer libraries (RosettaMatch uses the Dunbrack backbone-dependent rotamer library; the inverse rotamer tree method uses the backbone-independent rotamer library), and hence the reconstructed sites are not identical.
Table 3.
Benchmark I results using inverse rotamer tree approach (A) and the RosettaMatch method (B)
Figure 2.
Energy distribution of native and non-native designed sites. (Left panel) The distribution of virtual energy and LJ repulsive energy between the TS model and the protein scaffold after minimizing the catalytic residues. (Right panel) The distribution of the computed catalytic efficacy (TS binding energy) after designing the binding pocket. The native matches (in the native scaffold in the native positions) are in black; the matches in alternate sites and/or scaffolds are in gray.
Figure 3.

Superposition of native and predicted active sites for fuculose 1-phosphate aldolase (4fua) and DERA aldolase (1jcl). Orange, native TS model position and catalytic side chains; green, designed TS model position and catalytic side chains. The TS model is represented with thick sticks; the catalytic side chains, with thin sticks.
Benchmark results starting from idealized catalytic geometries
In the second benchmark test, the geometrical parameters defining the functional group from the catalytic residues are chosen using the geometrical rules listed in Table 2. Since some of the degrees of freedom are free to adopt a range of discrete values, the number of possible matches is much larger than in the previous test. Because of the combinatorial explosion of possible active sites, the inverse rotamer tree cannot easily handle such a problem since all combinations must be enumerated prior to searching. The RosettaMatch method avoids the combinatorial explosion by treating each catalytic side chain independently; results of this benchmark test for this method are summarized in Table 4. Because the active site descriptions are considerably more general than in the first benchmark, the rank of the native active site is not always high, but in four of the nine test cases reported, the native active site has the highest rank after minimization and design. The complete design process requires one or two CPU days per scaffold on an Intel Xeon at 2.8 Ghz with 2 Gb of RAM with full diversification of the free degrees of freedom for a three-residue active site (type II aldolase). Thus, the computational design strategy allows for rapid identification and evaluation of designed sites on many scaffolds which can be tested experimentally.
Table 4.
Benchmark II results using RosettaMatch
Sensitivity to backbone variation
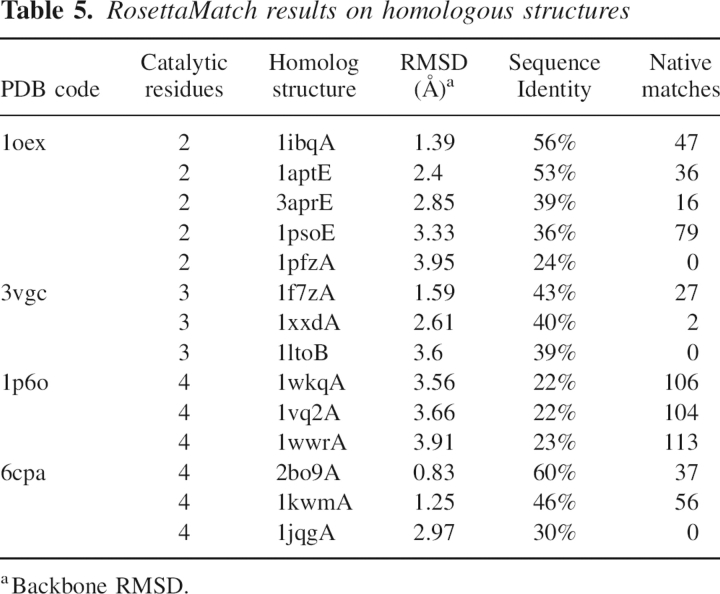
To quantitate the sensitivity of the RosettaMatch algorithm to the precise positions of the backbone atoms, we investigated the performance of the method in recognizing native matches in homologous scaffolds. We used PSIBLAST (Altschul et al. 1997) to identify sequence homologs with known structures for four of the enzymes in our benchmark set: aspartic proteinase, γ-chymotrypsin, cytosine deaminase, and bovine carboxypeptidase A, which contain two, three, four, and four catalytic residues, respectively. The number of homolog structures and their backbone root mean square deviation (RMSD) to the query structure for each enzyme are summarized in Table 5. As indicated in Table 2, our methods are capable of finding the active site for homolog structures of up to ∼4.0 Å backbone RMSD, showing that they are tolerant of variation in backbone coordinates up to this level (the native site can be found multiple times because of the fineness of the rotamer sampling).
Table 5.
RosettaMatch results on homologous structures
Discussion
The enzyme active site recapitulation test presented in this article provides a rapid and comprehensive benchmark to evaluate and guide the improvement of enzyme design methods. However, the experimental characterization of future computationally designed enzymes is, of course, the ultimate proof of the power of a design method.
Although the algorithms we describe are new, the overall approach of starting with a geometric description of an active site, searching through a protein scaffold for positions where it can be placed, and designing the surrounding residues has been used in previous studies (Hellinga and Richards 1991; Bolon and Mayo 2001). The algorithms described here have several advantages over previously described methods. The inverse rotamer tree–based search complexity does not depend on the number of scaffolds searched, whereas previous methods scale at least linearly with the number of positions (and, consequently, scaffolds searched). Dezymer (Hellinga and Richards 1991), for example, places all rotamers for the anchor residue at each position, thereby scaling at least proportionally with the number of positions considered. The approach taken by Bolon and Mayo (2001) also places an extended rotamer (that includes the TS model) on each search position, leading to the same dependence. The computational efficiency of the inverse rotamer tree–based algorithm can be a tremendous advantage if large-scale enzyme site searches are required. The algorithm, however, is limited by its exponential dependence on the number of rotamer combinations considered. In the case of active sites with four or more active site residues, the algorithm performs poorly. Since it is not possible to use large rotamer libraries, the use of this algorithm is limited to a more coarse-grained search.
The RosettaMatch method avoids the combinatorial explosion by treating each catalytic side chain independently in building up the hash table. It thus scales linearly with the number of rotamer combinations considered. Once the hash maps have been built up, the complexity of the look-up step is constant time on average. In the worse-case scenario (i.e., when many TS models placed in different boxes map to the same hash key), the hash look up scales as O(N), where N is the number of entries for the box. Although it is not easy to directly compare the complexity of the algorithm with Hellinga's Dezymer, the RosettaMatch method has the advantage that the algorithm complexity depends only linearly on the number of residues making up the active site and the total number of rotamers used.
The design methods in their current form can be used to design new active sites in existing scaffolds based either on the structures of naturally occurring active sites or on chemical intuition; the speed of the methods makes it possible to search large sets of scaffolds for optimal active site placements. In the benchmark test, a number of the non-native designs have nearly perfect catalytic geometries and transition state binding energies as low or lower than the native match and potentially represent viable enzymes. As an example, Figure 4 shows a design for an aldolase active site built on a decarboxylase scaffold with a calculated binding energy after design comparable to the native enzyme. The experimental evaluation of the activity of such high-ranking designs in non-native scaffolds will test our understanding of the mechanisms of enzyme catalysis. To extend to new reactions for which natural enzymes provide less guidance, it should be very advantageous to use quantum chemistry methods to compute transition states and ideal active site geometries. In particular, the “theozyme” concept developed by Houk and coworkers (Tantillo et al. 1998) fits very nicely into our approach as the coordinates of the theozyme can be used directly as input for the matching process.
Figure 4.

Grafting an aldolase active site onto a decarboxylase scaffold. The top panel is an overall view of the designed protein; the bottom panel is a closer view of the active site. The protein backbone is shown in cartoon mode and colored according to its secondary structure. The TS model is shown in ball-and-stick mode, and the TS analog carbon atoms are colored in orange. The designed catalytic residues (Lys, Asp, Lys) are shown as sticks, and their carbon atoms are colored in green.
Materials and methods
Overview of enzyme design methodology
Starting from an active site description consisting of a TS model surrounded by appropriately placed protein functional groups (geometrical parameters are specified in Fig. 5 and Table 2), a set of protein scaffold candidates is searched to construct a catalytic site that binds tightly to the TS and retains the desired functional group geometry. The design process consists of four steps (Fig. 1). In step 1, a list of scaffolds is searched for positions that can hold the TS model and catalytic residues in the correct orientation. We describe two different methods for step 1: an inside-out method, based on the inverse rotamer tree technique, and an outside-in method called RosettaMatch. In step 2, the TS model and the catalytic side chains placed in step 1 are refined to eliminate clashes and optimize the catalytic geometry. In step 3, the identity and conformations of amino acid residues located near the active site are optimized using the RosettaDesign method. Finally, in step 4, the designs in step 3 are ranked based on the computed TS binding energy, considering only designs where the catalytic constraints are satisfied. We refer to this combination of transition state stabilization with catalytic residues geometry as the predicted catalytic efficacy throughout the text, but we emphasize that determination of the catalytic efficacy of a design requires experimental characterization.
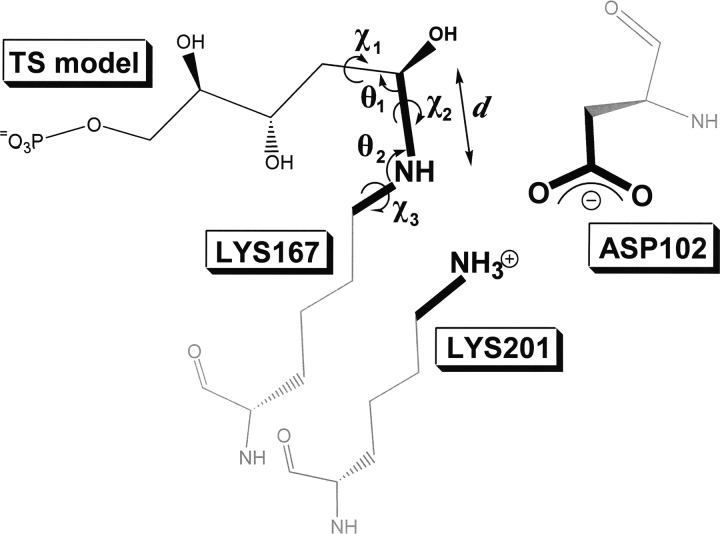
Figure 5.
Illustration of geometric parameters used in active site description for deoxyribose-phosphate aldolase (DERA). Six geometrical parameters (d indicates distance; θ1 and θ2 indicate bond angles; χ1, χ2, and χ3 indicate torsional angles) are specified to describe the spatial positions of the functional groups relative to the TS.
Step 1: Geometry-based site identification
The “inverse rotamer tree” approach
The idea of the inverse rotamer tree is to convert the description of the active site in terms of functional groups into a description in terms of protein backbone coordinates that can then be used to search a set of protein scaffolds or to guide de novo scaffold design. This is the inverse of the standard side chain packing problem in which the positions of the backbone coordinates are known. We use a standard rotameric description of the side chains to solve the problem (Dunbrack and Cohen 1997); but rather than building outward from the backbone coordinates, we grow side chains backward from the functional group positions that are placed around the TS model in positions optimal for catalysis. This generates an inverse rotamer tree specifying the possible placements of the protein backbone around the TS model that are compatible with the specified active site in the sense that the relevant amino acids can be placed to achieve the desired active site geometry. Figure 6 shows the inverse rotamer tree generated for the DERA active site.
Figure 6.
Inverse rotamer tree for deoxyribose-phosphate aldolase (DERA) active site. The transition state is colored in yellow, and the key functional groups of the catalytic residues are in gold. The remainder of the side chains in the rotamer trees are shown using thinner lines in CPK coloring.
Once the inverse rotamer tree has been built, each combination of backbone coordinates for the catalytic residues is searched against the set of scaffolds (a step subsequently referred to as matching) using a geometric hashing based approach. Given the set of scaffolds to be searched, the algorithm begins by building a multiple key hash table: The backbone coordinates (N,Cα,C) for each pair of residues for each scaffold are mapped onto a unique key that is computed from the Cα–Cβ distance and the [Cα,Cβ] vector orientations. For speed, all the scaffolds are mapped into a single hash in memory at the beginning of the program. Each combination of backbone atom coordinates from the inverse rotamer tree is matched against the backbone distances and orientations stored in the hash table using a subgraph isomorphism algorithm similar to that described by Russell (1998). Matches are ranked based on their structural similarity (in RMSD) to the specified active site geometry, and the absence of atomic clashes between the TS model, the placed catalytic side chains, and the protein backbone.
The RosettaMatch approach
The idea of this approach is to build forward from the protein backbone to the TS model for each catalytic side chain independently, and then to identify TS placements compatible with placement of each catalytic residue. The method may be viewed as an extension of the MetalSearch algorithm (Clarke and Yuan 1995) to include ligand orientation as well as center of mass coordinates. We describe first the storage of the position of the TS model for each catalytic side chain rotamer placed at each position using a hash table and, second, the processing of the hash table to extract sets of positions compatible with the specified active site geometry. Finally, we describe performance enhancements to the method using precomputed grids to restrict TS placement to clefts and pockets in the scaffolds, and to speed up the evaluation of atomic clashes with the protein backbone.
For each protein scaffold, a set of potential active site positions is predefined, either all positions in the protein or positions lining cavities or small molecule binding sites. For each amino acid residue in the catalytic site description, all rotamers form the Dunbrack backbone dependent library (Dunbrack and Cohen 1997) are placed at each position. If there is no clash with protein backbone, the TS model for the reaction is positioned as specified in the catalytic site definition. For catalytic side chain–TS interactions such as hydrogen bonds, where there are many chemically equivalent interaction geometries, a large set of TS model placements are considered; the fineness of the sampling around the varying degrees of freedom (the side chain–TS dihedral in the hydrogen bonding case) is specified in Table 2. Each TS rigid body placement is represented by [v,q], where v is the vector of the coordinates of the center of mass (x,y,z) of the TS model, and q is the unit quaternion (q1,q2,q3,q4) (Latombe 1991) associated with the rotation that moves the TS model from a reference frame to its current placement. TS model placements are recorded in a hash table if there are no clashes with the protein backbone or the catalytic side chain using the key K computed as follows:
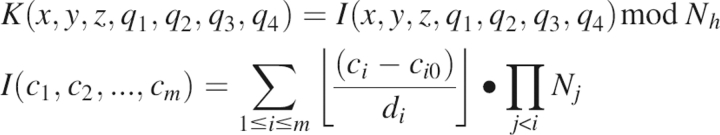 |
where the bracket is the integer part, N h is the expected size of the hash, c i is the coordinate in direction i, c i0 is the origin for the direction i, d i is grid spacing for each direction, and N j is the total number of grid points in direction j.
For each placement of the TS model, the following information is stored in the hash table at the position identified by the key K: the box coordinates (c 1,…,c 7) in which the TS model falls, the position in the protein sequence, the residue type (e.g., His, Asp, etc.), the index of the rotamer in the backbone-dependent library, and the rigid body orientation of the TS model [v,q]. The position in the hash does not suffice to specify the TS position because the hash operator cannot be inverted. For each key K, one list per catalytic residue is kept that records all the information described above for each TS model that hashed with the key K.
Each key of the hash table (corresponding to each discrete box of the six-dimensional space) thus contains N lists, where N is the number of residues making up the catalytic site. If at least one of the N lists is empty, a catalytic site with the specified geometry does not exist for the corresponding TS model location. If the N lists are all not empty, a complete active site can be generated, and every combination of catalytic residues for which there are no significant atomic clashes between the catalytic side chains and no two residues originate in the same backbone position are selected for subsequent minimization and design as described below.
Finding the active site matches requires on the order of 15 min per scaffold on an Intel Xeon machine at 2.8 Ghz with 2 Gb of RAM, with no diversification for the three-residue active site for type II aldolase. The runs take ∼2 h on the same machine with full diversification of the free degrees of freedom for the same active site. In addition, the RosettaMatch method is easily amenable to parallelization by splitting the pocket into different spatial regions and distributing the building of the hash table among different processors.
To focus the design calculations on promising regions of the scaffold, the center of mass of the TS model may be restricted to clefts or pockets that are likely to be large enough to comprise a viable active site. A square grid box is first constructed that covers the regions targeted for active site design. This grid is then trimmed to remove all the grid points that are <2.25 Å from any protein backbone atom. Any residue on the protein backbone that has a Cα–Cβ vector pointing toward one of those grid points and a Cα <3.5 Å from any grid point is then included in the set of active site positions. In practice, the use of the grid does not substantially reduce the number of matches found, but it considerably speeds up the search process by eliminating regions unlikely to contribute high ranking active site designs.
To speed up the evaluation of clashes between the TS model and the protein backbone, a “backbone” grid is constructed that contains points that are <2.25 Å from any backbone atom. TS model placements for which atoms overlap the backbone grid are not included in the hash.
Step 2: Optimization of catalytic site placement in scaffold
For each match found with the inverse rotamer tree or the RosettaMatch method, residues around the TS model, other than the catalytic residues, are truncated to glycines. The initial placements of the TS model and catalytic side chain conformations are optimized by rigid body minimization followed by side chain minimization using Rosetta (Gray et al. 2003a,b; Wang et al. 2005). The potential used for minimization consists of the repulsive part of a standard Lennard Jones 6–12 potential (Kuhlman and Baker 2004), a side chain torsional statistical potential (Dunbrack and Cohen 1997) complemented by a “virtual energy” term that describes the extent to which the functional groups on the catalytic side chains satisfy the ideal geometry described in the active site. The virtual energy term is a quadratic penalty function of the geometrical parameters that relate the functional groups of the catalytic residues to the TS (Fig. 5). Minimization is carried out multiple times using Powell's method (Flannery et al. 2002), gradually increasing the weight on the repulsive interactions between iterations. A very low value is used initially to avoid repulsion of the TS model from the active site.
Step 3: Sequence optimization around the TS model
The minimization step leads to pockets in which a non-clashing TS model is placed with catalytic side chains positioned with functional atoms close to the optimal geometry required for catalysis. It is then necessary to design the surrounding, noncatalytic protein residues to maximally stabilize the transition state. The conformations and identities of residues surrounding the TS model are optimized using Monte Carlo simulated annealing as described previously (Kuhlman and Baker 2000). The potential consists of (1) a 12-6 Lennard-Jones potential with an attenuated repulsive component (Kuhlman and Baker 2004), (2) an implicit solvation model (Lazaridis and Karplus 1999), (3) an orientation-dependent hydrogen bonding term (Kortemme and Baker 2002; Kortemme et al. 2003, 2004; Jiang et al. 2005), (4) a Coulomb model with a distance dependent dielectric constant, (5) a pair potential derived from the Protein Data Bank (Simons et al. 1999) that captures features of side chain side chain electrostatics, and (6) a backbone dependent side chain torsional potential derived from known structures (Dunbrack and Cohen 1997). This potential has performed very well in protein–small molecule docking calculations (Meiler and Baker 2006).
Acknowledgments
We thank the Baker laboratory members for helpful discussions and comments. This work was supported by DARPA (Grant no. HR0011-05-1-0044) and HHMI.
Footnotes
Reprint requests to: David Baker, Department of Biochemistry, University of Washington, Seattle, WA 98195, USA; e-mail: dabaker@u.washington.edu; fax: (206) 685-1792.
Abbreviations: TS, transition state; RMSD, root mean square deviation; DERA, deoxyribose-phosphate aldolase.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062353106.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachar, O., Fischer, D., Nussinov, R., and Wolfson, H. 1993. A computer vision based technique for 3-D sequence-independent structural comparison of proteins. Protein Eng. 6: 279–288. [DOI] [PubMed] [Google Scholar]
- Berman, H.M., Battistuz, T., Bhat, T.N., Bluhm, W.F., Bourne, P.E., Burkhardt, K., Feng, Z., Gilliland, G.L., Iype, L., and Jain, S., et al. 2002. The Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr. 58: 899–907. [DOI] [PubMed] [Google Scholar]
- Bolon, D.N. and Mayo, S.L. 2001. Enzyme-like proteins by computational design. Proc. Natl. Acad. Sci. 98: 14274–14279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, N.D. and Yuan, S.M. 1995. Metal search: A computer program that helps design tetrahedral metal-binding sites. Proteins 23: 256–263. [DOI] [PubMed] [Google Scholar]
- Dahiyat, B.I. and Mayo, S.L. 1996. Protein design automation. Protein Sci. 5: 895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiyat, B.I. and Mayo, S.L. 1997. De novo protein design: Fully automated sequence selection. Science 278: 82–87. [DOI] [PubMed] [Google Scholar]
- Dunbrack Jr., R.L. and Cohen, F.E. 1997. Bayesian statistical analysis of protein side-chain rotamer preferences. Protein Sci. 6: 1661–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer, M.A., Looger, L.L., and Hellinga, H.W. 2004. Computational design of a biologically active enzyme. Science. 304: 1967–1971. [DOI] [PubMed] [Google Scholar]
- Flannery, B., Vetterling, W.T., Teukolsky, S.A., and Press, W.H. 2002. Numerical recipes in C++, 2nd ed. Cambridge University Press, Cambridge, UK.
- Gray, J.J., Moughon, S., Wang, C., Schueler-Furman, O., Kuhlman, B., Rohl, C.A., and Baker, D. 2003a. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J. Mol. Biol. 331: 281–299. [DOI] [PubMed] [Google Scholar]
- Gray, J.J., Moughon, S.E., Kortemme, T., Schueler-Furman, O., Misura, K.M., Morozov, A.V., and Baker, D. 2003b. Protein-protein docking predictions for the CAPRI experiment. Proteins 52: 118–122. [DOI] [PubMed] [Google Scholar]
- Hellinga, H.W. and Richards, F.M. 1991. Construction of new ligand binding sites in proteins of known structure. I. Computer-aided modeling of sites with pre-defined geometry. J. Mol. Biol. 222: 763–785. [DOI] [PubMed] [Google Scholar]
- Hilvert, D. 2000. Critical analysis of antibody catalysis. Annu. Rev. Biochem. 69: 751–793. [DOI] [PubMed] [Google Scholar]
- Jiang, L., Kuhlman, B., Kortemme, T., and Baker, D. 2005. A “solvated rotamer” approach to modeling water-mediated hydrogen bonds at protein-protein interfaces. Proteins 58: 893–904. [DOI] [PubMed] [Google Scholar]
- Kaplan, J. and DeGrado, W.F. 2004. De novo design of catalytic proteins. Proc. Natl. Acad. Sci. 101: 11566–11570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortemme, T. and Baker, D. 2002. A simple physical model for binding energy hot spots in protein-protein complexes. Proc. Natl. Acad. Sci. 99: 14116–14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortemme, T., Morozov, A.V., and Baker, D. 2003. An orientation-dependent hydrogen bonding potential improves prediction of specificity and structure for proteins and protein-protein complexes. J. Mol. Biol. 326: 1239–1259. [DOI] [PubMed] [Google Scholar]
- Kortemme, T., Joachimiak, L.A., Bullock, A.N., Schuler, A.D., Stoddard, B.L., and Baker, D. 2004. Computational redesign of protein-protein interaction specificity. Nat. Struct. Mol. Biol. 11: 371–379. [DOI] [PubMed] [Google Scholar]
- Kuhlman, B. and Baker, D. 2000. Native protein sequences are close to optimal for their structures. Proc. Natl. Acad. Sci. 97: 10383–10388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman, B. and Baker, D. 2004. Exploring folding free energy landscapes using computational protein design. Curr. Opin. Struct. Biol. 14: 89–95. [DOI] [PubMed] [Google Scholar]
- Latombe, J. 1991. Quaternions. In Robot motion planning,, pp. 72–74. Kluwer Academic Publishers, Boston.
- Lazaridis, T. and Karplus, M. 1999. Effective energy function for proteins in solution. Proteins 35: 133–152. [DOI] [PubMed] [Google Scholar]
- Lerner, R.A., Benkovic, S.J., and Schultz, P.G. 1991. At the crossroads of chemistry and immunology: Catalytic antibodies. Science 252: 659–667. [DOI] [PubMed] [Google Scholar]
- Meiler, J. and Baker, D. 2006. ROSETTALIGAND: Protein-small molecule docking with full side-chain flexibility. Proteins 65: 538–548. [DOI] [PubMed] [Google Scholar]
- Russell, R.B. 1998. Detection of protein three-dimensional side-chain patterns: New examples of convergent evolution. J. Mol. Biol. 279: 1211–1227. [DOI] [PubMed] [Google Scholar]
- Simons, K.T., Ruczinski, I., Kooperberg, C., Fox, B.A., Bystroff, C., and Baker, D. 1999. Improved recognition of native-like protein structures using a combination of sequence-dependent and sequence-independent features of proteins. Proteins 34: 82–95. [DOI] [PubMed] [Google Scholar]
- Tantillo, D.J., Chen, J., and Houk, K.N. 1998. Theozymes and compuzymes: Theoretical models for biological catalysis. Curr. Opin. Chem. Biol. 2: 743–750. [DOI] [PubMed] [Google Scholar]
- Wang, C., Schueler-Furman, O., and Baker, D. 2005. Improved side-chain modeling for protein-protein docking. Protein Sci. 14: 1328–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]