Abstract
The Escherichia coli phnD gene is hypothesized to code for the periplasmic binding component of a phosphonate uptake system. Here we report the characterization of the phosphonate-binding properties of the phnD protein product. We find that PhnD exhibits high affinity for 2-aminoethylphosphonate (5 nM), the most commonly occurring natural phosphonate produced by lower eukaryotes, but also binds several other phosphonates with micromolar affinities. A significant number of man-made phosphonates, such as insecticides and chemical warfare agents, are chemical threats and environmental pollutants. Consequently, there is an interest in developing methods for the detection and bioremediation of phosphonates. Bacterial periplasmic-binding proteins have been utilized for developing reagentless biosensors that report analytes by coupling ligand-binding events to changes in the emission properties of a covalently conjugated environmentally-sensitive fluorophore. Several PhnD conjugates described here show large changes in fluorescence upon binding to methylphosphonate (MP), with two conjugates exhibiting up to 50% decrease in emission intensity. Since MP is the final degradation product of many nerve agents, these PhnD conjugates can function as components in a biosensor system for chemical warfare agents.
Keywords: fluorescent biosensor, Escherichia coli phnD, methyl phosphonate, nerve agent degradation product, periplasmic-binding protein, 2-aminoethylphosphonate
Phosphonates are characterized by a stable carbon–phosphorus bond, and include a wide range of naturally occurring and synthetic molecules. Natural phosphonates have been discovered in flagellates, protozoa (Horiguchi and Kandatsu 1959), mollusks (Kittredge and Roberts 1969), and some fungi (Wassef and Hendrix 1976). The most common natural phosphonate is 2-aminoethylphosphonate (2-AEP), a precursor in the biosynthesis of phosphonolipids (Baer and Stanacey 1964), phosphonoproteins (Kittredge and Roberts 1969), and phosphonoglycans (Korn et al. 1973). While only few strains of bacteria are able to synthesize phosphonates (Hendlin et al. 1969), numerous Gram-negative species can scavenge carbon–phosphorus containing compounds as a source of phosphorus (Wackett et al. 1987; Schowanek and Verstraete 1990; Dick and Quinn 1995). The Escherichia coli 12.6-kb phn operon (previously known as the psiD locus) codes for 17 genes involved in binding, uptake, and metabolism of phosphonates, and is induced at low phosphate concentrations (Metcalf et al. 1990; Metcalf and Wanner 1991). Based on sequence homology and mutagenesis studies, the gene products of phnC, phnD, and phnE are postulated to code for a phosphonate ABC-transport system (Surin et al. 1985; Metcalf and Wanner 1993). The sequence of phnD is homologous to sulfate- and phosphate-binding proteins (Magota et al. 1984; Sirko et al. 1990), and is therefore postulated to code for a soluble periplasmic-binding protein (PBP) component of the phosphonate uptake system. Furthermore, the Rhizobium meliloti PhnD analog was found to localize in the periplasm (Bardin et al. 1996). Here we report the identification of likely cognate ligands for this protein, and use this information to construct a reagentless fluorescent biosensor for phosphonates.
Synthetic phosphonates include a wide variety of molecules that range from the herbicide glyphosate, to antibiotics (Hendlin et al. 1969), household detergents (Kononova and Nesmeyanova 2002), insecticides (Racke 1993), and chemical warfare agents such as sarin, soman, or VX (Munro et al. 1999). Methods for detection of these molecules are therefore of significant interest (Nowack 2003; Gonzalez-Martinez et al. 2005), and include biosensors (Allert et al. 2004). Fruitful approaches to constructing new biosensors include the exploitation of biodiversity of suitable proteins as well as engineering techniques to alter ligand-binding specificity (Marvin and Hellinga 2001; Dwyer et al. 2003; Looger et al. 2003; Allert et al. 2004). The bacterial PBP superfamily, of which PhnD is a member, is well suited for the development of biosensors (Dwyer and Hellinga 2004). First, these proteins exhibit a high diversity of cognate ligands (de Lorimier et al. 2002). Second, the ligand-binding event is associated with a large conformational change (Quiocho and Ledvina 1996) that can be coupled to a change in the local environment of a covalently coupled reporter group (Dwyer and Hellinga 2004). Third, their binding specificity can be drastically altered to produce novel biosensors using computational design techniques (Marvin and Hellinga 2001; Dwyer et al. 2003; Looger et al. 2003; Allert et al. 2004). Engineered PBPs can function as reagentless biosensors that link a ligand-binding event with a physical signal without change in composition, in contrast to competitive displacement or enzyme-based assays, which require a depletable label or substrate (Hellinga and Marvin 1998; de Lorimier et al. 2002; Dwyer and Hellinga 2004). Here we report the application of these protein engineering techniques to construct a reagentless fluorescent biosensor for phosphonates based on PhnD.
Results
Cloning and purification of PhnD
The phnD gene was cloned by PCR from E. coli genomic DNA (XL1-Blue strain). Flanking primers were designed to introduce a 5′ NdeI site and a 3′ EcoRI site for cloning into a pET21a protein expression vector. The primers also introduced a hexa-histidine fusion at the C terminus. The native periplasmic signal sequence (residues 1–25) was omitted to allow for cytoplasmic expression, and was replaced with a methionine, followed by Glu26 (amino acid numbering system reported here uses the convention that residue number 1 is equivalent to Glu26 of the full-length open reading frame). The DNA sequence of the cloned gene corresponds to the published phnD sequence (Chen et al. 1990). The gene product of the cloned region of phnD, PhnD, was expressed and purified using immobilized metal affinity chromatography as described (de Lorimier et al. 2002) to >95% purity as determined by SDS PAGE, and its mass confirmed by MALDI-TOF.
Identification of cognate ligands for PhnD
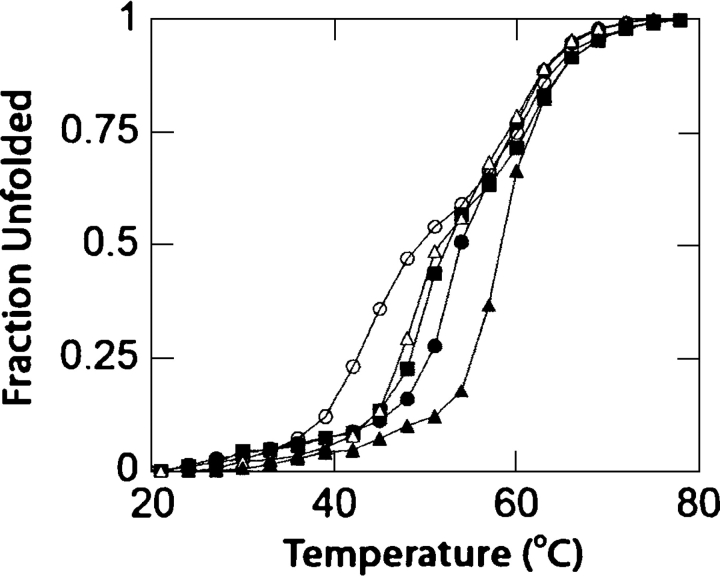
To identify cognate ligands, we took advantage of the increase in protein stability associated with ligand binding (Schellman 1975) to screen several likely ligands for binding to PhnD. Binding of candidate ligands was determined by measuring the ligand-dependent change in thermal stability of PhnD as monitored by circular dichroism to follow protein denaturation (Greenfield 2004). E. coli can utilize methylphosphonate (MP) and ethylphosphonate (EP) as sources of phosphorus under phosphate starvation conditions (Schowanek and Verstraete 1990). Additionally, 2-aminoethylphosphonate (2-AEP) was chosen as a likely candidate for binding to PhnD, as it is the most common naturally occurring phosphonate, produced by many lower eukaryotes (Horiguchi and Kandatsu 1959; Kononova and Nesmeyanova 2002). A large increase in stability (12°C) was observed in the presence of 1 mM 2-AEP (Fig. 1), while smaller shifts in the stability of the protein were observed for EP, MP, and aminomethylphosphonate (AMP) (Fig. 1). A small shift in stability was observed in the presence of 1 mM inorganic phosphate, while no shift in stability was observed in the presence of 10 mM sodium sulfate.
Figure 1.
Thermal denaturation of wild-type PhnD determined by circular dichroism in the absence (open circles) and presence of 1 mM of AMP (open triangles), MP (solid squares), EP (solid circles), or 2-AEP (solid triangles). The apoprotein exhibits a three-state denaturation behavior, presumably due to independent unfolding of the two domains.
Determination of ligand dissociation constants by intrinsic tryptophan fluorescence
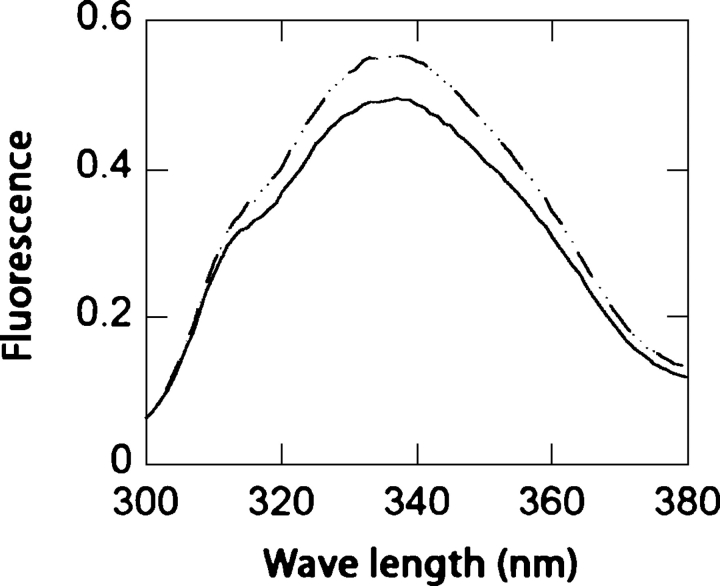
Ligand-mediated changes in intrinsic tryptophan fluorescence were used to determine the dissociation constants of PhnD for each ligand. PhnD was found to exhibit an increase in fluorescence upon addition of the cognate ligands identified by the thermostability assay (Fig. 2). Binding isotherms obtained by fluorescence titration show that PhnD binds preferentially to 2-AEP followed by EP, MP, and phosphate (Table 1), consistent with the observed ligand-mediated changes in stability.
Figure 2.
Tryptophan fluorescence emission spectra of wild type determined in the absence (solid line) and presence or 100 μM 2-AEP (dashed line).
Table 1.
Dissociation constants for selected phosphonates based on tryptophan fluorescence
Construction and analysis of reagentless fluorescent biosensors
Previous work has shown that unique cysteine mutations engineered into PBPs can serve as sites for covalent attachment of single thiol-reactive, environmentally-sensitive reporters that exhibit a change in physical signal in response to a ligand-binding event (de Lorimier et al. 2002). Such signal changes can be monitored as a function of ligand concentration, allowing detection and quantification of an analyte of interest. Construction of reagentless fluorescent biosensors based on PBPs requires engineering of a linkage relationship between a single covalently-attached fluorophore and the ligand-induced conformational transition from the apo (open) form to the ligand-bound (closed) form (Dwyer and Hellinga 2004). The fluorophore is attached at a site that undergoes a significant structural change upon ligand binding. Such positions are either located in the vicinity of the ligand-binding site (peristeric), or near the hinge region away from the binding site (allosteric) (de Lorimier et al. 2002). In the absence of an experimentally determined structure for PhnD, its protein sequence was “threaded” over the crystal structure of sulfate-binding protein, which shares a 37% sequence similarity with PhnD (Fig. 3). Three allosteric positions (A48C, N125C, A155C) and three peristeric positions (Q267C, N272C, E277C) were selected based on the threaded structure (Fig. 3B).
Figure 3.
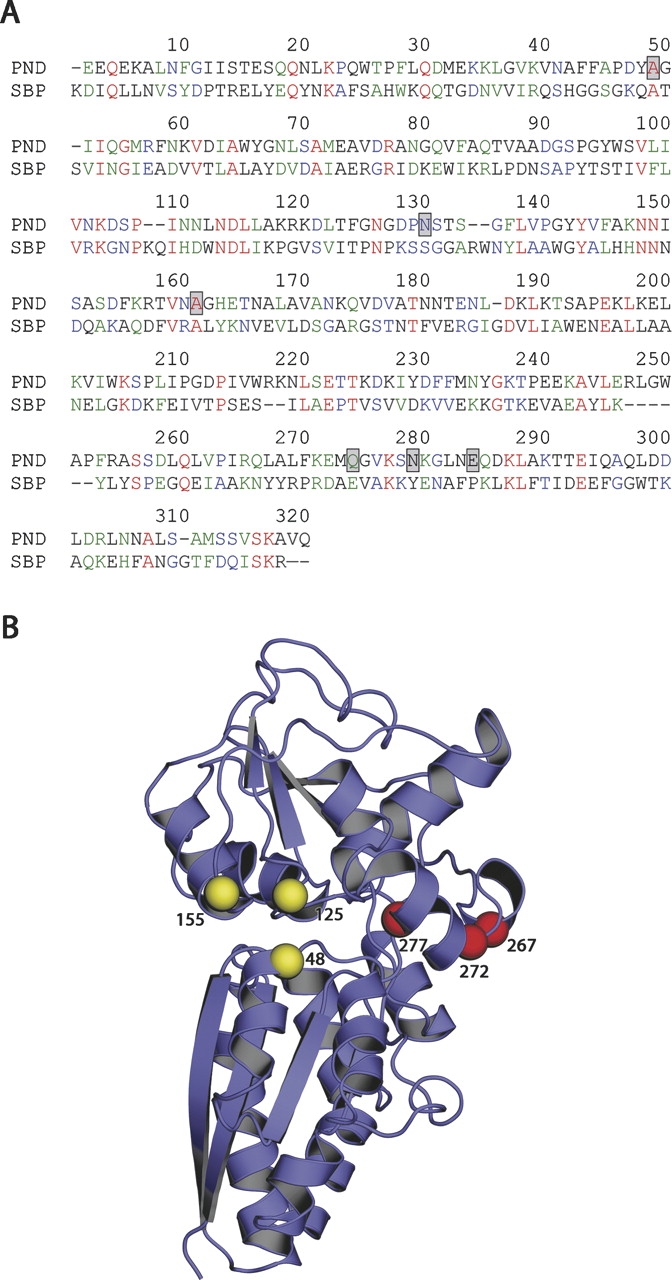
(A) Sequence alignment of PhnD and E. coli sulfate-binding protein (SBP), positions of engineered cysteines are indicated by gray boxes. (B) Location of cysteine mutations mapped onto the crystal structure of SBP (PDB code 1SBP) using the sequence alignment to establish the correct register. Spheres indicate peristeric (yellow) and allosteric (red) positions where cysteine mutations were introduced. Note that PhnD contains several insertions relative to SBP; these amino acids are located in surface loops and are not included in the model shown here.
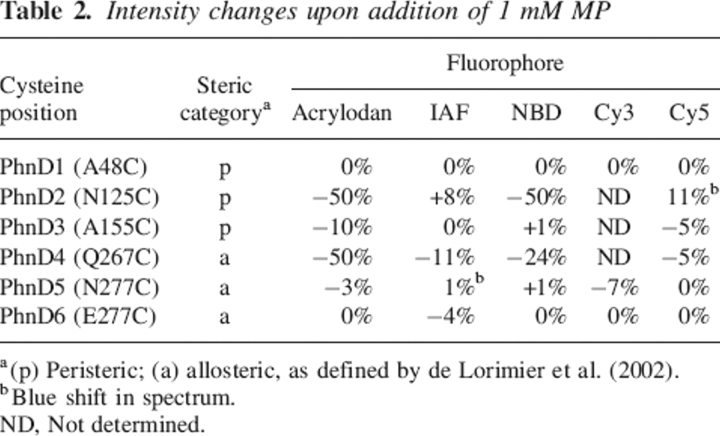
All six cysteine point mutants (PhnD1–6) were constructed using PCR-mediated mutagenesis, expressed, and purified. Each cysteine mutant was singly labeled with one of several thiol-reacive fluorophores. For each protein–fluorophore conjugate fluorescence emission spectra were collected in the absence and presence of a saturating concentration of MP (Table 2). None of the PhnD1 conjugates show significant changes in fluorescence emission upon addition of EP or MP. The fluorescence changes of the PhnD2–6 conjugates range between 1% and 50% in the presence of MP (Table 2). Several conjugates exhibit a decrease in fluorescence in response to MP, some exhibit an increase in emission, while two conjugates, PhnD2–Cy5 and PhnD5–IAF, show a blue shift in the maximum wavelength in the presence of ligand (Table 2).
Table 2.
Intensity changes upon addition of 1 mM MP
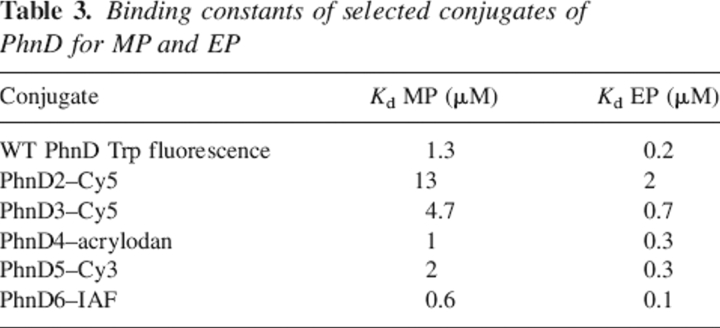
Of each cysteine mutant, one conjugate was chosen for fluorescence titration with EP and MP to determine the effect of the cysteine mutation on affinity (Table 3). With the exception of PhnD4–acrylodan, all conjugates tested retain the ratio of Kd values between EP and MP observed with the cysteine-free PhnD using tryptophan fluorescence. PhnD2 labeled with Cy5 exhibits a 10-fold decrease in affinity for both EP and MP. Other positions tested show smaller decreases in affinity for both ligands. In contrast, PhnD6–IAF shows a twofold increase in affinity for both MP and EP (Table 3), as is often observed with allosteric conjugates (de Lorimier et al. 2002).
Table 3.
Binding constants of selected conjugates of PhnD for MP and EP
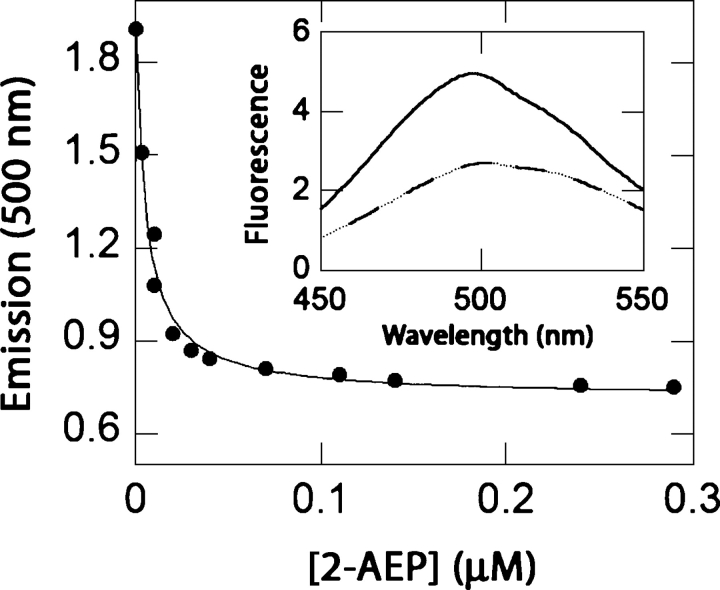
The binding specificity of PhnD was tested in the PhnD4–acrylodan conjugate because its Kd values are closest to those of the cysteine-free protein for MP and EP (Table 3), and it exhibits a large change in emission upon addition of ligand (Fig. 4). The conjugate was found to bind to a variety of natural and synthetic phosphonates, with its highest affinity for 2-AEP (Kd = 5 nM), the most common naturally occurring phosphonate (Fig. 4). The protein also binds other biologically available phosphorus-containing compounds such as phosphonoacetate (Kd = 0.9 μM) and phosphate (Kd = 50 μM) (Table 4). Furthermore, the conjugate responds to phenylphosphonate or glyphosate (Table 4), which are synthetic phosphonates that are not metabolized by E. coli (Schowanek and Verstraete 1990).
Figure 4.
Titration of the PhnD4-acrylodan conjugate with 2-AEP. (Inset) Fluorescence emission spectra of PhnD4-acrylodan in the apoprotein (solid line) and in the presence of 100 μM 2-AEP (dashed line).
Table 4.
Specificity of PhnD4-acrylodan for phosphorus-containing compounds
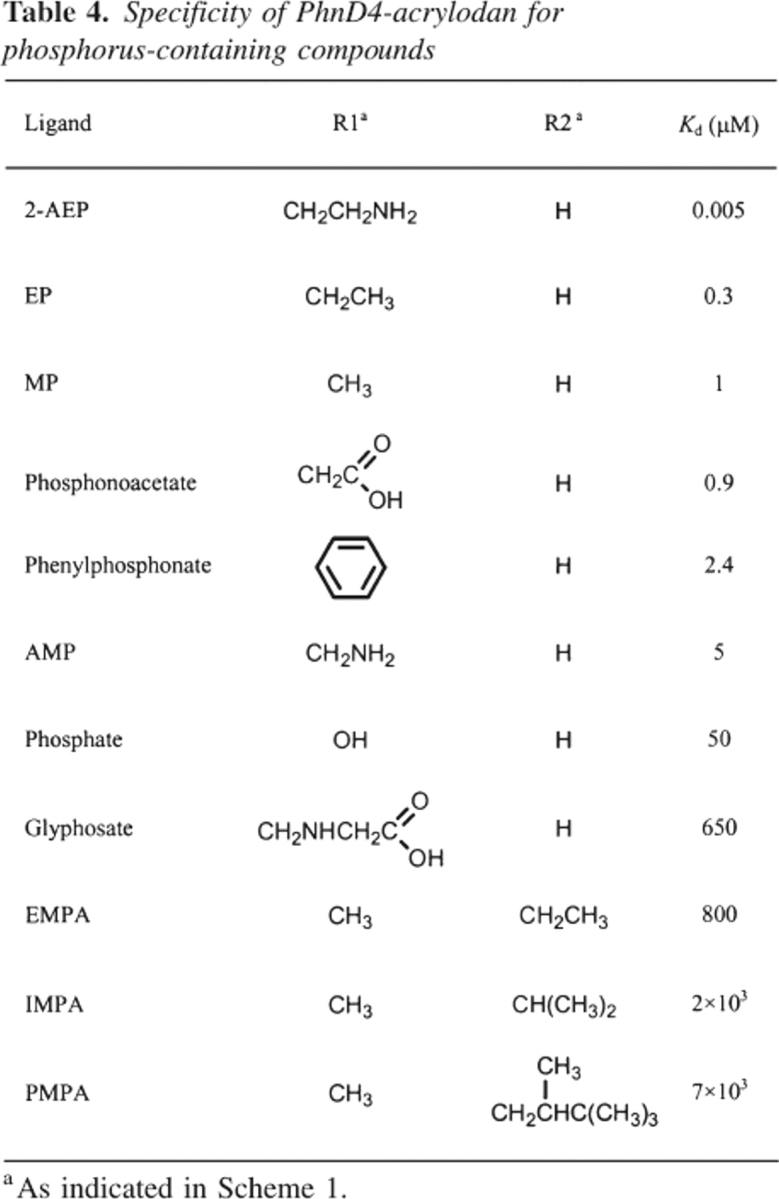
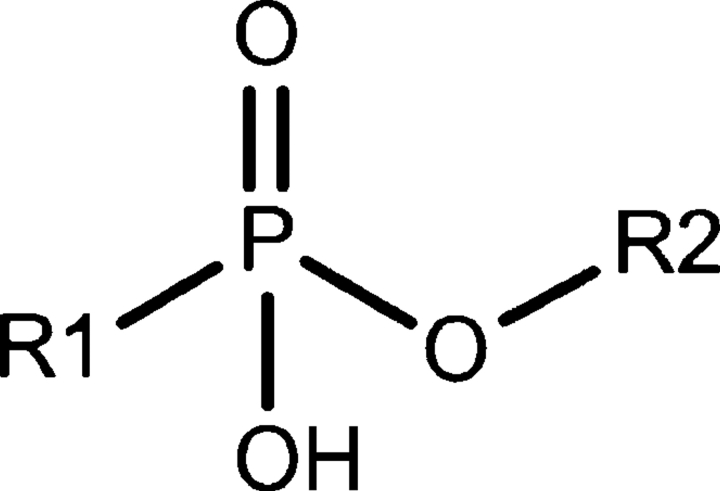
In addition to alkylphosphonates, PhnD4–acrylodan was tested for binding the nerve agent degradation products ethylmethylphosphonic acid (EMPA), isopropylmethylphosphonic acid (IMPA), and pinacolylmethyl phosphonic acid (PMPA). These compounds are methylphosphonates with extensions at one of the phosphonoester oxygens (R2 in Scheme 1). All three compounds show high Kd values (>800 μM) that increase with the size of the alkyl substituent. For example, the protein binds EMPA with a Kd of 800 μM, 2.5-fold more tightly than IMPA, which binds 3.5-fold tighter than PMPA (Table 4).
Scheme 1.
Discussion
Most bacteria are unable to synthesize phosphonates, but many have evolved elaborate mechanisms to scavenge organic phosphorus produced by lower eukaryotes (Kononova and Nesmeyanova 2002). The E. coli phnD gene has been postulated to code for a periplasmic phosphonate-binding protein (Surin et al. 1985; Metcalf and Wanner 1993). However, the expression and characterization of its protein product, PhnD, have not been previously reported. In this study, we report binding interactions of PhnD for a panel of phosphonates (Table 4) that were chosen as representatives of natural sources of phosphorus under phosphate starvation conditions (MP, EP, 2-AEP, and phosphonoacetate), pesticides, or pollutants (glyphosate and phenylphosphonate), and degradation products of nerve agents (EMP, IMPA, and PMPA). Tryptophan fluorescence titrations show that binding to 2-AEP (the most common naturally occurring phosphonate) is most favored, followed by EP and MP (Table 1).
In the absence of structural data for PhnD, comparing dissociation constants of the tested phosphonates serves as a crude indication of the structural specificity of the ligand-binding site. PhnD binds phosphonates with groups attached to the phosphonate carbon, with affinities correlated with the deviation of the R-group from that of 2-AEP. Additionally, the binding pocket does not accommodate molecules with modifications at the phosphonoester oxygen, evident by the relatively low affinities exhibited for EMPA, IMPA, and PMPA (Table 4). Although the binding pocket of PhnD is promiscuous for several phosphonates, it exhibits an extremely high affinity (5 nM) for 2-AEP, a precursor in the biosynthesis of phosphonolipids, phosphonoproteins, and phosphonoglycans (Kononova and Nesmeyanova 2002). The ability to utilize 2-AEP as a source of phosphorus, carbon, and nitrogen enables bacteria to thrive in hostile environments (Chen et al. 2002) and to survive in the absence of inorganic phosphate (Metcalf et al. 1990). We therefore conclude that 2-AEP is the natural cognate ligand for PhnD, the periplasmic binding component of the E. coli phosphorus scavenging system.
In addition to identifying the likely natural cognate ligand for PhnD, we report that PhnD can be used to develop a reagentless biosensor for MP, a degradation product of many nerve agents (Munro et al. 1999). Previously, we have reported reagentless sensors for the nerve agent surrogate PMPA, developed by engineering the binding specificity of ribose- and glucose-binding proteins (Allert et al. 2004). Although these sensors have a high affinity for their target ligand, like most biomolecules, they do not exhibit absolute specificity and also bind MP, a further degradation product of soman (Munro et al. 1999), albeit at significantly lower affinities (Allert et al. 2004). To develop reliable sensor systems, it is therefore necessary to accurately detect not only the target ligand, but also a panel of likely decoys (Albert et al. 2000). The reagentless phosphonate biosensor reported here describes a first step toward that goal.
Cysteine scanning mutagenesis for suitable fluorophore conjugation sites was carried out based on a structure-guided sequence alignment with sulfate-binding protein, which shares a 37% sequence similarity with PhnD. This approach has previously been successful in developing biosensors for the neurotransmitter glutamate, taking advantage of sequence similarity with glutamine- and histidine-binding proteins (de Lorimier et al. 2002). Five of the six PhnD cysteine mutants reported here show change in fluorescence upon addition of MP when conjugated with at least one of five fluorophores tested. Although conjugation affected the overall affinity of the protein for both molecules compared to the cysteine-free construct, in almost all conjugates tested the ratio of binding constants, as determined independently by tryptophan fluorescence, between EP and MP was retained. This suggests that conjugation does not affect the binding mechanism of PhnD, but instead shifts the equilibrium between the unbound and the bound forms of the protein. Based on previously established criteria (de Lorimier et al. 2002), the PhnD2–acrylodan, PhnD3–acrylodan, PhnD2–NBD, PhnD4–NBD, and PhnD2–Cy5 conjugates are acceptable reagentless biosensors. The biosensors described here therefore represent a first step toward the development of a biosensor system for nerve agents.
Materials and methods
Sequences of PhnD and SBP were aligned using ClustalX (Higgins and Sharp 1988). The amino acid sequence of PhnD was threaded over the crystal structure of sulfate-binding protein (PDB code 1SBP) using the program LOOPP (Meller and Elber 2001). The signal peptide of PhnD was predicted using the SignalP algorithm (Nielsen et al. 1997). Acrylodan, IAF (iodoacetamidofluorescein), and NBD (N-((2-(iodoacetoxy)ethyl)-N-methyl)amino-7-nitrobenz-2-oxa-1,3-diazole) were purchased from Molecular Probes (Invitrogen); Cy3 and Cy5 were purchased from Amersham. IMPA was purchased from Cerilliant; 2-AEP and phosphonoacetate from ICN; all other ligands, salts, and buffers from Sigma-Aldrich. PhnD was cloned from XL1-Blue E. coli genomic DNA into a pET21a vector (Novagen) using a 5′ NdeI site and a 3′ EcoRI site with a C-terminal six-histidine tag. The periplasmic signal sequence (predicted to be residues 1–26 by the expasy signalP 3.0 prediction algorithm) (Nielsen et al. 1997) was omitted for cytoplasmic expression and replaced with an N-terminal methionine, where Glu27 was chosen as the first residue of the PhnD open reading frame. Sequences of the wild-type gene and its cysteine mutants were confirmed by DNA sequencing. Proteins were expressed using BL-21 DE3 cells in 0.5 L 2XYT media, induced at 0.6 OD600 with 1 mM IPTG (RPI Corp.) for 2 h at 37°C. Cells were centrifuged for 10 min at 5000g, resuspended in 15 mL of 10 mM imidazole, 20 mM MOPS, 500 mM NaCl (pH 7.8) (resuspension buffer), sonicated on ice for 2 min, centrifuged at 30,000g for 30 min. The supernatant was passed through a 0.45-μm syringe filter and loaded in a 3-mL chelating sepharose column charged with 25 mL of 1 mM NiSO4 and equilibrated with 25 mL of resuspension buffer. The column was washed with 10 mL of resuspension buffer followed by 6 mL of resuspension buffer containing 60 mM imidazole and 3 mL of resuspension buffer containing 400 mM imidazole. The protein was eluted using 3 mL of 400 mM imidazole resuspension buffer and exchanged into 10 mM Tris, 50 mM NaCl (pH 7.9) using an Econo-Pac 10 DG gel filtration column (Bio-Rad). Cysteine mutants were exchanged into 20 mM MOPS, 100 mM NaCl (pH 6.9) for fluorescence labeling with thiol reactive fluorophores for 1 h at room temperature, then exchanged into 10 mM Tris, 50 mM NaCl (pH 7.9). All fluorescence measurements were collected on an Aminco SLM fluorescence spectrometer at 25°C (acrylodan, λex. 390 nm; IAF, λex. 490; NBD, λex. 475 nm; Cy3, λex. 550 nm, Cy5, λex. 650 nm; slit widths: excitation, 4 nm; emission, 8 nm). Ligand-binding isotherms were fitted as described (de Lorimier et al. 2002). Circular dichroism data were obtained on an Aviv CD spectrometer using 15 μM PhnD at 222 nm with a 1-cm path length in the absence or presence of 1 mM ligand.
Acknowledgments
We thank Robert deLorimier for helpful discussions and G. Shirman for assistance with mutagenesis and protein chemistry. This work was supported by grants from the U.S. Department of Homeland Security (W81XWH-05-C-0161) and the NIH Director's pioneer award (5 DP1 OD000122).
Footnotes
Reprint requests to: Homme W. Hellinga, Duke University Medical Center, Department of Biochemistry, Box 3711, Research Drive, 415 Nanaline Duke Building, Durham, NC 27710, USA; e-mail: hwh@biochem.duke.edu; fax: (919) 684-8885.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062135206.
Abbreviations: PhnD, phosphonate-binding protein; PBP, periplasmic-binding protein; MP, methylphosphonate; 2-AEP, 2-aminoethylphosphonate; AMP, aminomethylphosphonate; IAF, iodoacetamidofluorescein; NBD, N-((2-(iodoacetoxy)ethyl)-N-methyl)amino-7-nitrobenz-2-oxa-1,3-diazole; EMPA, ethylmethylphosphonic acid; IMPA, isopropylmethylphosphonic acid; PMPA, pinacolylmethylphosphonic acid.
References
- Albert K.J., Lewis N.S., Schauer C.L., Sotzing G.A., Stitzel S.E., Vaid T.P., Walt D.R. 2000. Cross-reactive chemical sensor arrays. Chem. Rev. 100: 2595–2626. [DOI] [PubMed] [Google Scholar]
- Allert M., Rizk S.S., Looger L.L., Hellinga H.W. 2004. Computational design of receptors for an organophosphate surrogate of the nerve agent soman. Proc. Natl. Acad. Sci. 101: 7907–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer E. and Stanacey N.Z. 1964. Phosphonolipids. I. Synthesis of a phosphonic acid analogue of cephalin. J. Biol. Chem. 239: 3209–3214. [PubMed] [Google Scholar]
- Bardin S., Dan S., Osteras M., Finan T.M. 1996. A phosphate transport system is required for symbiotic nitrogen fixation by Rhizobium meliloti. J. Bacteriol. 178: 4540–4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.M., Ye Q.Z., Zhu Z.M., Wanner B.L., Walsh C.T. 1990. Molecular biology of carbon–phosphorus bond cleavage. Cloning and sequencing of the phn (psiD) genes involved in alkylphosphonate uptake and C-P lyase activity in Escherichia coli B. J. Biol. Chem. 265: 4461–4471. [PubMed] [Google Scholar]
- Chen C.C., Zhang H., Kim A.D., Howard A., Sheldrick G.M., Mariano-Dunaway D., Herzberg O. 2002. Degradation pathway of the phosphonate ciliatine: Crystal structure of 2-aminoethylphosphonate transaminase. Biochemistry 41: 13162–13169. [DOI] [PubMed] [Google Scholar]
- de Lorimier R.M., Smith J.J., Dwyer M.A., Looger L.L., Sali K.M., Paavola C.D., Rizk S.S., Sadigov S., Conrad D.W., Loew L.et al. 2002. Construction of a fluorescent biosensor family. Protein Sci. 11: 2655–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick R.E. and Quinn J.P. 1995. Glyphosate-degrading isolates from environmental samples: Occurrence and pathways of degradation. Appl. Microbiol. Biotechnol. 43: 545–550. [DOI] [PubMed] [Google Scholar]
- Dwyer M.A. and Hellinga H.W. 2004. Periplasmic binding proteins: A versatile superfamily for protein engineering. Curr. Opin. Struct. Biol. 14: 495–504. [DOI] [PubMed] [Google Scholar]
- Dwyer M.A., Looger L.L., Hellinga H.W. 2003. Computational design of a Zn2+ receptor that controls bacterial gene expression. Proc. Natl. Acad. Sci. 100: 11255–11260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Martinez M.A., Brun E.M., Puchades R., Maquieira A., Ramsey K., Rubio F. 2005. Glyphosate immunosensor. Application for water and soil analysis. Anal. Chem. 77: 4219–4227. [DOI] [PubMed] [Google Scholar]
- Greenfield N.J. 2004. Analysis of circular dichroism data. Methods Enzymol. 383: 282–317. [DOI] [PubMed] [Google Scholar]
- Hellinga H.W. and Marvin J.S. 1998. Protein engineering and the development of generic biosensors. Trends Biotechnol. 16: 183–189. [DOI] [PubMed] [Google Scholar]
- Hendlin D., Stapley E.O., Jackson M., Wallick H., Miller A.K., Wolf F.J., Miller T.W., Chaiet L., Kahan F.M., Foltz E.L.et al. 1969. Phosphonomycin, a new antibiotic produced by strains of streptomyces. Science 166: 122–123. [DOI] [PubMed] [Google Scholar]
- Higgins D.G. and Sharp P.M. 1988. CLUSTAL: A package for performing multiple sequence alignment on a microcomputer. Gene 73: 237–244. [DOI] [PubMed] [Google Scholar]
- Horiguchi M. and Kandatsu M. 1959. Isolation of 2-aminoethane phosphonic acid from rumen protozoa. Nature 184: 901–902. [DOI] [PubMed] [Google Scholar]
- Kittredge J.S. and Roberts E. 1969. A carbon–phosphorus bond in nature. Science 164: 37–42. [DOI] [PubMed] [Google Scholar]
- Kononova S.V. and Nesmeyanova M.A. 2002. Phosphonates and their degradation by microorganisms. Biochemistry (Mosc.) 67: 184–195. [DOI] [PubMed] [Google Scholar]
- Korn E.D., Dearborn D.G., Fales H.M., Sokoloski E.A. 1973. Phosphonoglycan. A major polysaccharide constituent of the amoeba plasma membrane contains 2-aminoethylphosphonic acid and 1-hydroxy-2-aminoethylphosphonic acid. J. Biol. Chem. 248: 2257–2259. [PubMed] [Google Scholar]
- Looger L.L., Dwyer M.A., Smith J.J., Hellinga H.W. 2003. Computational design of receptor and sensor proteins with novel functions. Nature 423: 185–190. [DOI] [PubMed] [Google Scholar]
- Magota K., Otsuji N., Miki T., Horiuchi T., Tsunasawa S., Kondo J., Sakiyama F., Amemura M., Morita T., Shinagawa H.et al. 1984. Nucleotide sequence of the phoS gene, the structural gene for the phosphate-binding protein of Escherichia coli. J. Bacteriol. 157: 909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin J.S. and Hellinga H.W. 2001. Conversion of a maltose receptor into a zinc biosensor by computational design. Proc. Natl. Acad. Sci. 98: 4955–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller J. and Elber R. 2001. Linear programming optimization and a double statistical filter for protein threading protocols. Proteins 45: 241–261. [DOI] [PubMed] [Google Scholar]
- Metcalf W.W. and Wanner B.L. 1991. Involvement of the Escherichia coli phn (psiD) gene cluster in assimilation of phosphorus in the form of phosphonates, phosphite, Pi esters, and Pi. J. Bacteriol. 173: 587–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf W.W. and Wanner B.L. 1993. Mutational analysis of an Escherichia coli fourteen-gene operon for phosphonate degradation, using TnphoA′ elements. J. Bacteriol. 175: 3430–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf W.W., Steed P.M., Wanner B.L. 1990. Identification of phosphate starvation-inducible genes in Escherichia coli K-12 by DNA sequence analysis of psi:lacZ(Mu d1) transcriptional fusions. J. Bacteriol. 172: 3191–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro N.B., Talmage S.S., Griffin G.D., Waters L.C., Watson A.P., King J.F., Hauschild V. 1999. The sources, fate, and toxicity of chemical warfare agent degradation products. Environ. Health Perspect. 107: 933–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H., Engelbrecht J., Brunak S., von Heijne G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10: 1–6. [DOI] [PubMed] [Google Scholar]
- Nowack B. 2003. Environmental chemistry of phosphonates. Water Res. 37: 2533–2546. [DOI] [PubMed] [Google Scholar]
- Quiocho F.A. and Ledvina P.S. 1996. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: Variation of common themes. Mol. Microbiol. 20: 17–25. [DOI] [PubMed] [Google Scholar]
- Racke K.D. 1993. Environmental fate of chlorpyrifos. Rev. Environ. Contam. Toxicol. 131: 1–15. [DOI] [PubMed] [Google Scholar]
- Schellman J.A. 1975. Macromolecular binding. Biopolymers 14: 999–1018. [Google Scholar]
- Schowanek D. and Verstraete W. 1990. Phosphonate utilization by bacterial cultures and enrichments from environmental samples. Appl. Environ. Microbiol. 56: 895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirko A., Hryniewicz M., Hulanicka D., Bock A. 1990. Sulfate and thiosulfate transport in Escherichia coli K-12: Nucleotide sequence and expression of the cysTWAM gene cluster. J. Bacteriol. 172: 3351–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surin B.P., Rosenberg H., Cox G.B. 1985. Phosphate-specific transport system of Escherichia coli: Nucleotide sequence and gene-polypeptide relationships. J. Bacteriol. 161: 189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wackett L.P., Shames S.L., Venditti C.P., Walsh C.T. 1987. Bacterial carbon-phosphorus lyase: Products, rates, and regulation of phosphonic and phosphinic acid metabolism. J. Bacteriol. 169: 710–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassef M.K. and Hendrix J.W. 1976. Ceramide aminoethylphosphonate in the fungus Pythium prolatum. Biochim. Biophys. Acta 486: 172–178. [DOI] [PubMed] [Google Scholar]