Abstract
Phytopathogenic oomycetes cause some of the most devastating diseases affecting agricultural crops. Hyaloperonospora parasitica is a native oomycete pathogen of Arabidopsis and is related to other oomycete phytopathogens that include several species of Phytophthora, including the causal agent of potato late blight. Recently, four oomycete effector genes have been isolated, and several oomycete genomes have been sequenced. We have developed an efficient and genetically amenable system to test putative effector genes using the bacterial pathogen Pseudomonas syringae pv. tomato DC3000. The H. parasitica effector protein ATR13 was delivered via P. syringae by fusing the ATR13 gene with the avrRpm1 type three secretion signal peptide, a bacterial sequence that allows transfer of proteins into the host cell through the bacterial type III secretion system. We also inserted ATR13 into the genome of the turnip mosaic virus, a single-stranded RNA virus. Our results show that delivery of ATR13 via the bacterial or viral pathogen triggers defense responses in plants containing the cognate resistance protein RPP13Nd, which restricts proliferation of both pathogens. Hence, recognition of ATR13 by RPP13 initiates defense responses that are effective against oomycete, bacterial and viral pathogens, pointing to a common defense mechanism. We have characterized regions of the RPP13Nd resistance protein that are essential for effector recognition and/or downstream signaling, using transient coexpression in Nicotiana benthamiana.
Keywords: plant disease resistance, effector-triggered plant immunity, plant pathogenesis
Hyaloperonospora parasitica is a native oomycete pathogen of Arabidopsis (1). This plant–pathogen interaction became established as a laboratory model in the 1990s and led to the identification of many Arabidopsis resistance genes (R-genes) (1). However, cloning H. parasitica's effector genes has been much more difficult because of its obligate biotrophic lifestyle and lack of genetic tractability. Recently, four oomycete effector genes have been isolated, and this coincides with the sequencing of several oomycete genomes (including H. parasitica) and has allowed prediction of a large number of effector genes that share signal peptide sequences with the four known effectors (2). The H. parasitica effector Arabidopsis thaliana recognized 13 (ATR13) is recognized by the A. thaliana R-gene Resistance to Peronospora parasitica13 (RPP13) (3). ATR13 contains two predicted signal sequences at its N terminus. The first sequence is believed to allow secretion of the protein across the oomycete plasma membrane; the second sequence contains an RxLR motif, which has been shown to target Phytophthora infestans effector proteins across the host cell plasma membrane (4). The RxLx motif is also conserved in the human malaria parasite Plasmodium falciparum and is responsible for vacuolar export of proteins (4). Once inside the plant cell, ATR13 is detected by the cognate R-protein RPP13, a NBS-LRR protein of the coiled-coil (CC) class (3). Recognition leads to induction of defense pathways and is visible as an HR (hypersensitive cell death response). Allen et al. (3) showed by biolistic transient expression in Arabidopsis that the RPP13 gene product from ecotype Niederzenz (RPP13Nd) recognizes ATR13 from H. parasitica strains Emco5, Maks9, Aswa1, and Goco1, but not from strains Emoy2 and Hind4.
We have developed an efficient and genetically amenable system to test ATR13-RPP13 recognition using the bacterial pathogen Pseudomonas syringae pv. tomato DC3000 (Pst). Pst is a Gram-negative bacterium that delivers effector proteins into host cells via a type III secretion system (TTSS). We delivered ATR13 alleles via Pst by fusing the ATR13 gene with the AvrRpm1 signal peptide, a bacterial sequence that allows transfer of proteins into the host cell through the TTSS (5, 6). Our results show that delivery of ATR13Emco5, but not ATR13Emoy2, via the bacterial pathogen triggers defense responses in the plant expressing the cognate resistance protein RPP13Nd. This leads to restriction of bacterial proliferation and thus validates our delivery system. The system can now be used to test other predicted effector proteins from H. parasitica for their ability to be recognized by a wide range of diverse ecotypes of A. thaliana.
We also inserted ATR13 into the genome of the turnip mosaic virus, a single-stranded RNA virus. Again, expression of ATR13Emco5 (but not ATR13Emoy2) by the viral pathogen triggers defense responses in A. thaliana containing RPP13Nd and stops viral proliferation. A hypersensitive cell death response (HR) is also triggered when RPP13Nd and ATR13 Emco5 are coexpressed in Nicotiana benthamiana by Agrobacterium infiltration, indicating that N. benthamiana contains the signaling components necessary to trigger HR. This has allowed us to characterize regions of the RPP13Nd R-protein that are essential for effector recognition and/or downstream signaling.
Results
Construction of a Surrogate Bacterial TTSS to Deliver H. parasitica Effector Proteins into Plant Cells.
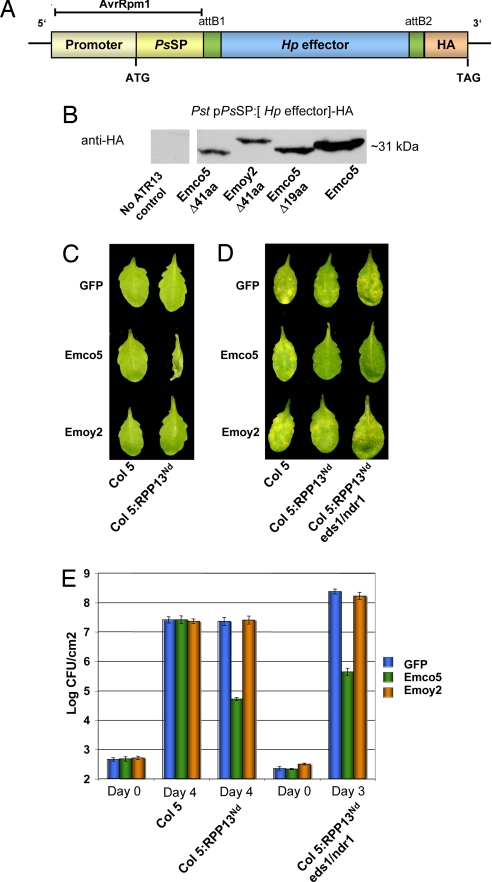
Pst causes disease on A. thaliana, a pathosystem that has been well characterized. To deliver putative H. parasitica (Hp) effectors via Pst DC3000, we used the Pseudomonas avrRpm1 promoter plus secretion-translocation signal of AvrRpm1 reported in refs. 5 and 7). The promoter region plus the Pseudomonas syringae type III secretion signal peptide (PsSP) were cloned into the wide host range vector pVSP61 that can replicate in Pst (Fig. 1A). The addition of the Gateway destination cassette allows rapid cloning of putative effector genes. Pseudomonas constructs were preinduced in Pseudomonas-promoter induction media (8), and HA epitope tagged chimeric effector expression was confirmed by immunoblot detection of the predicted chimeric proteins (Fig. 1B).
Fig. 1.
Surrogate Pst DC3000 bacterial TTSS to deliver H. parasitica effector protein ATR13 to detect RPP13 bacterial resistance. (A) Expression vector pVSP_PsSPdes for delivery of putative effector proteins via the TTSS of Pst DC3000. Expression vectors are gateway-compatible to enable easy effector cloning and epitope tagging. (B) Immunoblot analysis to detect effector expression in Pst DC3000 after preinduction in Pseudomonas-promoter induction media. HA epitope-tagged Hp effector constructs with the ATR13Emco5 full-length (≈32 kDa), ATR13Emco5-Δ19aa (≈31 kDa), ATR13Emco5-41aa (≈28 kDa), and the nonrecognized allele ATR13Emoy2-Δ41aa (≈32 kDa). (C) Leaves were inoculated with high-density suspensions of Pst DC3000 (5 × 107 CFU/ml). Delivery of Δ41ATR13Emco5 triggered an HR within 24 hpi in Col5 plants expressing RPP13Nd, but not in Col5 WT plants. Delivery of GFP or Δ41ATR13Emoy2 does not induce an HR. (D) Leaves were inoculated with low-density suspensions of Pst DC3000 (1 × 104 CFU/ml). Chlorotic disease symptoms do not develop for DC3000 delivering ATR13Emco5 on Col-5 RPP13Nd and Col-5 (ndr1/eds1) RPP13Nd. (E) Bacterial growth levels were assessed 3 and 4 days after inoculation. Delivery of Δ41ATR13Emco5 triggered RPP13-specific resistance, as evident from the absence of chlorotic areas (D) and decreased bacterial growth (E). RPP13Nd signaling is independent of EDS1 and NDR1 because homozygous double mutants still exhibit RPP13-specific resistance to Pst DC3000 delivering Δ41ATR13Emco5. The nonrecognized allele Δ41ATR13Emoy2 does not activate RPP13 resistance. Error bars represent standard deviation for three replicated samples. Growth assay experiments were carried out three times with similar results.
ATR13-Specific Activation of RPP13 Resistance Is Effective Against a Bacterial Pathogen.
We tested Hp effector activation of RPP13 resistance by effector delivery through the PstTTSS of DC3000. We compared alleles of ATR13 from the two Hp strains, one (Emco5) that activates isolate specific resistance on RPP13Nd and one that does not (Emoy2) (9). The first 19 aa constitute a putative Hp signal peptide and cleavage site (3). In addition to full-length ATR13, we made a Δ19 (no Hp signal peptide) version of ATR13Emco5 and ATR13Emoy2. Also Δ41 (no Hp signal peptide or RxLR motif) versions were made to evaluate the role of this RxLR motif. Although Allen et al. (3) reported that a full-length ATR13Emco5 construct with Hp signal peptide delivered biolistically was active, our full-length ATR13Emco5 construct delivered from Pst was not active (data not shown). We detected recognition for both Δ19 (no signal peptide) and Δ41 (no signal peptide or RXLR motif). Detailed disease assays were carried out by using the Δ41 version of ATR13. At high inoculation density, Pst delivering ATR13Emco5 triggered an HR after 24 h on Col-5:RPP13Nd but not on Col-5 WT plants (Fig. 1C). Disease assays were performed 3 or 4 days after low-density inoculation. For Pst delivering ATR13Emco5 on Col-5:RPP13Nd and Col-5(ndr1/eds1):RPP13Nd, there were no chlorotic disease symptoms (Fig. 1D), which was confirmed by in planta bacterial growth assays (Fig. 1E). Bacterial growth was reduced 1,000-fold on Col-5:RPP13Nd for strains delivering Δ41ATR13Emco5, confirming that RPP13 is effective against a bacterial pathogen. Growth was also reduced 1,000-fold on Col-5:RPP13Nd plants carrying the ndr1/eds1 mutations, confirming that the defense response triggered by RPP13 is NDR1- and EDS1-independent (Fig. 1 D and E). The nonactive allele, ATR13Emoy2, also was unable to activate RPP13Nd resistance against the bacterial pathogen. For our AvrRpm1-ATR13 effectors, we observe no enhanced virulence for Pst on Col-5. Resistance to Pst strains delivering the control effectors AvrRpt2 and AvrRps4 was abolished in the double mutant (data not shown), because these signaling pathways require NDR1 and EDS1, respectively (10, 11). We conclude that delivery of the oomycete effector protein ATR13 via the TTSS of Pst triggers a resistance response in Arabidopsis containing the R-gene RPP13 consistent with the Hp isolate-specific resistance response previously reported (9). This resistance is specific to interactions between cognate effector–R-gene pairs, and is functional against the bacterial pathogen Pst DC3000.
Δ41ATR13Emco5 Activates Resistance to a Viral Pathogen in Plants Expressing RPP13Nd.
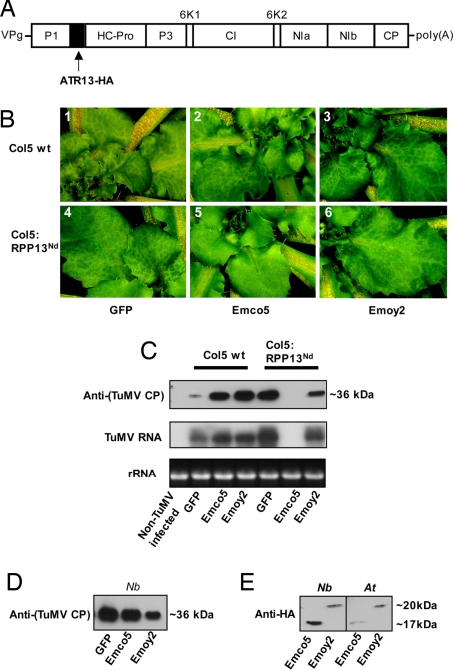
Turnip mosaic virus (TuMV) is able to systemically infect A. thaliana (12, 13). The virus belongs to the class of single-stranded (+) RNA potyviruses, with nonoverlapping ORFs (Fig. 2A). This allows insertion of genes into the viral genome, which are subsequently translated into a polyprotein by the host cell and cleaved into individual proteins by viral proteases. We inserted GFP, Δ41ATR13-HAEmco5 and Δ41ATR13-HAEmoy2 between the P1 and HcPro genes of the TuMV virus. Challenge of WT Col-5 with viral particles led to a systemic infection with visible mottling symptoms on young rosette leaves (Fig. 2B). TuMV-GFP-infected plants exhibited GFP fluorescence under a UV light, which corresponded to mottled areas. This confirms that the symptoms were due to viral spread (data not shown). WT Col5 plants with mottling symptoms contained high levels of coat protein and viral RNA (Fig. 2C). TuMV containing GFP and Δ41ATR13-HAEmoy2 replicated to similar levels on Col-5 plants expressing RPP13Nd, but virus expressing ATR13Emco5 was unable to replicate. No mottling symptoms were observed on leaves, and coat protein and viral RNA were absent (Fig. 2 B and C). Hence, RPP13Nd is able to induce a defense response in the presence of ATR13Emco5 that is effective against the viral pathogen TuMV. This defense response is independent of NDR1 and EDS1, because TuMV containing ATR13Emco5 cannot replicate on Col5 ndr1/eds1 mutants expressing RPP13Nd (data not shown). TuMV containing Δ19ATR13Emco5 also triggered resistance (data not shown). We confirmed that viral particles for all three strains were present at comparable levels in N. benthamiana before harvest of inoculum used to infect A. thaliana (Fig. 2D). To confirm that ATR13-HA protein was processed from the TuMV polyprotein in infected N. benthamiana and Arabidopsis leaves, we analyzed protein expression by immunoblot analysis (Fig. 2E). Δ41ATR13-HAEmco5 (≈17 kDa) and Δ41ATR13-HAEmoy2 (≈20 kDa) were detected in both TuMV-infected N. benthamiana and Arabidopsis and migrated close to their predicted sizes.
Fig. 2.
Delivery of ATR13Emco5 within the turnip mosaic virus (TuMV) genome activates RPP13 resistance against TuMV systemic infection of A. thaliana. (A) TuMV genome. The transgene insertion site (GFP, ATR13Emco5, or ATR13Emoy2) is indicated by an arrow. Upon translation, viral proteases cleave the polyprotein into separate proteins. RPP13Nd recognizes ATR13 from strain Emco5, but not strain Emoy2, when the gene is delivered within the TuMV genome, leading to resistance against the viral pathogen. (B) Leaves were inoculated with infectious viral particles containing GFP, Δ41ATR13Emco5, or Δ41ATR13Emoy2 fused to the HA epitope sequence as described in Materials and Methods. Leaves were inspected for symptoms 8 days pi (after infiltration). Diseased plants showed mottling symptoms on young leaves near the center of the rosette (1–4, 6). Expression of Δ41ATR13Emco5 from within the viral genome triggered resistance in Col5 plants expressing RPP13Nd (5) but not in Col5 WT plants (2), as evident from the absence of mottling symptoms. Resistance is not induced on delivery of GFP or Δ41ATR13Emoy2, with leaves showing clear mottling (4, 6). Contrast and brightness of images was altered in Adobe Photoshop. (C) By using immunoblot analysis and Northern blotting infected plants (1), Col5 WT plants (2) Col-5 RPP13Nd (3) Col-5 (ndr1/eds1) RPP13Nd were tested for levels of viral coat protein and viral RNA 8 days pi. Coat protein (CP) and viral genome were absent in plants expressing RPP13Nd challenged with TuMV containing Δ41ATR13Emco5. Blots were probed with anti-CP antibody and 32P-labeled coat protein DNA, respectively. Ethidium bromide-stained 25S rRNA in the RNA gel is shown as a loading control. A minimum of three independent experiments were carried out for each TuMV strain. (D) Leaf discs were taken for protein extraction from the youngest leaf of TuMV-infected N. benthamiana (Nb) immediately before processing tissue for Arabidopsis infection inoculum. Immunoblot analysis with anticoat protein (CP) detects similar levels for all TuMV strains. (E) Immunoblot analysis of TuMV infected N. benthamiana (Nb) and A. thaliana (At) detects ATR13Emco5-HA and ATR13Emoy2-HA that are processed from the TuMV polypeptide. Protein was extracted from infected leaf discs, and blots were probed with anti-HA antibody.
Coexpression of Cognate RPP13 and ATR13 Proteins Induces a Cell Death Response in N. benthamiana.
Transient coexpression of R-proteins and their corresponding effector proteins in N. benthamiana has been used as a powerful tool to study R-gene–effector gene recognition (14). To determine whether coexpression of RPP13Nd and ATR13Emco5 could trigger a specific cell death response, we transferred each gene into a gateway compatible Agrobacterium binary vector where transient expression is driven by either the 35S promoter or the very strong OCS promoter (15). The resulting plasmids were conjugated into Agrobacterium tumefaciens C58C1. A C58C1 strain carrying 35S-RPP13Nd–HA was coinfiltrated with C58C1 strains carrying either a GUS-control or one of the ATR13 allele binary constructs (Fig. 3A). RPP13Nd-HA and Δ41ATR13Emco5 together induced a clear HR within 22 hours after inoculation (hpi). No HR was detected for coinoculations of 35S-RPP13Nd-HA with GUS control or the nonrecognized allele, Δ41ATR13Emoy2. The coinoculation of Δ41ATR13Emco5 or Δ41ATR13Emoy2 with GUS control was also negative.
Fig. 3.
Agrobacterium-mediated coexpression of ATR13Emco5 and RPP13Nd in N. benthamiana induces an effector-specific hypersensitive cell death response (HR). A. tumefaciens strain (C58C1) was used for transient coexpression, and N. benthamiana leaves were used where coinfiltrated constructs were mixed at a final OD600 of 0.3 for each. (A) tumefaciens delivers the various constructs from three different gateway-compatible binary plasmids, pMD1–35Sdes with the 35S promoter and from pE1776des or pE1776-T7des with the stronger OCS promoter. (A) Coexpression of 35S-RPP13Nd-HA with OCS-Δ41ATR13Emco5 induced an effector-specific HR within 22 hpi. Coexpression of 35S-RPP13Nd-HA with either the nonrecognized ATR13 allele OCS-Δ41ATR13Emoy2 or the OCS-GUS control did not induce an HR. Leaf was photographed at 48 hpi. (B) Coexpression of OCS-RPP13Nd-HA with OCS-Δ41ATR13Emco5 induced an effector-specific (HR) within 18 hpi (data not shown); however, by 48 hpi, coexpression of OCS-RPP13Nd-HA with the OCS–GUS control induced an effector-independent HR. Coexpression of 35S-RPP13Nd-HA and OCS-Δ41ATR13Emco5 gave the same response as in A, and the OCS-RPS2 construct served as a positive HR control. (C) Immunoblot analysis of coexpression constructs at 18 hpi detects elevated levels of RPP13-HA for the strong OCS promoter construct, however, no RPP13 was detected when the effector OCS-Δ41ATR13Emco5 was coexpressed. Loading control (Ponceau S stain of Western blot membrane after probing with anti HA)
A second very strong promoter construct OCS-RPP13Nd–HA triggered an Δ41ATR13Emco5-specific HR at 16 hpi, whereas the 35S-RPP13Nd–HA triggered an HR at 22 hpi (data not shown). By 48 hpi, the OCS-RPP13Nd–HA triggered an Δ41ATR13Emco5-independent HR. This autoactivated HR was not observed for lower expressing 35S-RPP13Nd–HA construct. Immunoblot analysis to detect RPP13 expression at 18 hpi confirms the stronger OCS promoter construct expression (Fig. 3C). In the case of the 35S-RPP13Nd–HA, it appears that the coexpression of the RPP13 construct with Δ41ATR13Emco5 destabilizes the RPP13 protein even before a visible HR is observed.
Characterization of RPP13 Domains Required for ATR13-Specific Induction of the Cell Death Response.
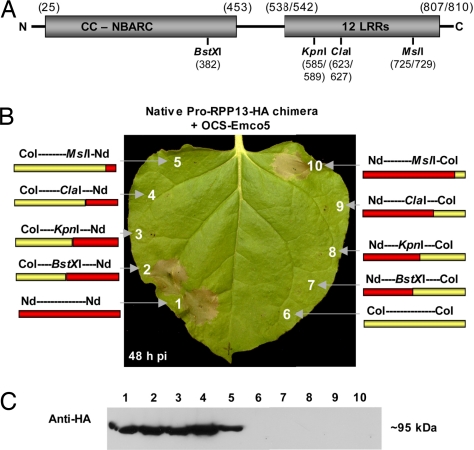
To define regions of the RPP13Nd gene that are necessary for induction of cell death in the N. benthamiana transient expression assay, we constructed chimeras between the active RPP13Nd (Nd) and the inactive RPP13Col (Col) alleles in a native RPP13 promoter Agrobacterium binary vector. We used restriction sites conserved between the two genes to swap 5′ and 3′ ends of the two alleles (Fig. 4A). As expected, Agrobacterium coexpression of the WT RPP13Nd with Δ41ATR13Emco5 induced an HR at 24 hpi (Fig. 4B), whereas coexpression with GUS-control triggered no HR (data not shown). Coexpression of RPP13Col with Δ41ATR13Emco5 induced no HR (Fig. 4B). Only the RPP13 chimera proteins Col-BstXI-Nd and Nd-MslI-Col gave HR at 24 hpi (Fig. 4B). All other chimeras failed to show any responses. The results indicate that amino acids between positions 382–729 of RPP13Nd are required for ATR13Emco5 recognition and cell death induction. This corresponds to the most variable region between these two alleles. Immunoblot analysis with anti-HA for detection of the C-terminal HA epitope tag of all RPP13 constructs revealed that all constructs that have RPP13Nd sequences at the C terminus yield the predicted 95-kDa protein, whereas all constructs with the RPP13Col sequences at the C terminus were not detectable even though Nd-MslI-Col was able to give HR. This suggests that the constructs with the RPP13Col sequences at the C terminus may have been expressed but the HA epitope was not detectable (Fig. 4C). The sequence of the epitope region was reconfirmed for all Agrobacterium ex-conjugates.
Fig. 4.
The LRR domain of RPP13Nd is required for induction of hypersensitive cell death response when ATR13Emco5 is coexpressed with chimeric RPP13 constructs between the active Niederzenz allele and the inactive Columbia allele. All Agrobacterium-transient expression constructs were inoculated at OD600 0.3. (A) Diagram of the RPP13 protein. Amino acid positions are indicated by numbers. Where the amino acid positions differ between the RPP13Nd and RPP13Col alleles, both are indicated in the format RPP13Nd/RPP13Col. KpnI cuts within the third LRR, ClaI cuts between the fourth and fifth LRRs, and MslI cuts between the eighth and ninth LRRs (9). (B) OCS-ATR13Emco5 is coexpressed with chimeric native promoter RPP13 constructs in N. benthamiana leaves at a final OD600 of 0.3 for each strain. Strains carried the binary plasmid pMD1 nP13des containing the indicated chimeric constructs of RPP13Nd and RPP13Col (format: N terminus–restriction site–C terminus). All chimeras were coexpressed with OCS-Δ41ATR13Emco5). Coexpression of RPP13 constructs Nd–Nd, Col-BstXI-Nd, and Nd-MslI-Col with ATR13Emco5 induced an HR by 24 hpi. All other constructs failed to induce an HR. The image was taken 48 hpi. (C) Immunoblot analysis with anti-HA for detection of the C-terminal HA epitope tag of all RPP13 constructs revealed that all constructs that have RPP13Nd at the C terminus yield the predicted 95-kDa protein, whereas all constructs with the RPP13Col at the C terminus were not detectable even though Nd-MslI-Col was able to give a cell death response.
Discussion
The genomes of H. parasitica, P. infestans, Phytophthora ramorum, Phytophthora sojae, and Phytophthora capsici have recently been sequenced (http://genome.jgi-psf.org and http://phytophthora.vbi.vt.edu), enabling us to predict putative effector proteins by computational analysis. In Hp alone, 149 candidate effector proteins were predicted based on the presence of signal peptide and RxLR motifs (2). To date, genetic analysis of oomycete effector proteins has been hampered by the difficulty (or inability) to culture these organisms in vitro and to genetically manipulate strains. To circumvent this problem, we tested whether we could individually deliver oomycete effector proteins into resistant Arabidopsis lines via the TTSS of the phytopathogenic bacterium PstDC3000 and trigger a disease-resistance response. We constructed a broad host range vector that contained the first 89 aa of the bacterial AvrRpm1 effector protein (which encodes a signal for type III secretion), a C-terminal HA epitope tag sequence, and the Gateway ccdB cassette, allowing for rapid cloning of candidate effector genes into our expression vector. The resulting plasmid was then introduced into Pst and inoculated onto Arabidopsis plants and scored for disease resistance.
Plants have developed the capacity to recognize pathogens from diverse classes and induce defenses, which inhibit pathogen growth and multiplication. To date, the majority of identified plant R proteins fall into the category of proteins containing NBS-LRR motifs. These proteins have been shown to confer resistance to all major classes of pathogens, including viruses, bacteria, fungi, oomycetes, nematodes, and insects. In this study, we demonstrate that the resistance response triggered by activation of the Arabidopsis R-gene RPP13 is effective not only against an oomycete but also against bacterial and viral pathogens. The resistance response was specific to the cognate effector and R-protein pair.
Transient coexpression of effector proteins with their cognate R-proteins in N. benthamiana has served as a powerful tool to investigate the specificity of R-gene recognition, protein localization, and protein–protein interactions (16). When we coexpressed RPP13Nd with ATR13Emco5, we observed a rapid cell death response in N. benthamiana (Fig. 3A). This response was specific to the interaction of cognate effector and R-protein, because coexpression with GUS or ATR13Emoy2 failed to induce an HR. When expressed at very high levels, RPP13 alone did induce a delayed HR (Fig. 3B), indicating that the protein is kept in an inactive state, which can be activated if overexpressed.
The RPP13 allele from Arabidopsis ecotype Columbia is unable to recognize ATR13Emco5 (3, 17–19). To identify essential regions for ATR13 recognition and signaling within the RPP13Nd protein, we created chimeras between the active RPP13Nd allele and the inactive RPP13Col allele (Fig. 4B). Our results show that the C-terminal LRR region between amino acids 382 and 725 of RPP13Nd is required for induction of cell death upon coexpression with ATR13Emco5 (Fig. 4B) and is correspondingly the region of highest variability between the two alleles. LRR domains are commonly involved in protein–protein interactions and ligand binding (20) and would therefore be expected to mediate binding to the effector protein. Indeed, genetic analysis of the flax L protein, a TIR-NBS-LRR, has suggested that the LRR is critical for determining the specificity of effector recognition (21), supported by a recent report of direct interactions of this R-protein with its effector in a yeast-two-hybrid assay (22). However, other NBS-LRR proteins, such as the tobacco N protein and Arabidopsis R-proteins RPM1 and RPS5, have been shown to interact with their effectors or other signaling molecules through the N-terminal TIR or CC domains (23–25).
In brief, we have shown that ATR13 recognition by its cognate RPP13 allele confers resistance to pathogens from different kingdoms that employ various infection strategies. The mechanism of this resistance has yet to be determined, but a forward genetic mutagenesis screen of Col5:RPP13Nd seed may shed light on the pathway involved in RPP13-mediated disease resistance. Additionally, the region of RPP13 implicated in ATR13 recognition has been narrowed down to the amino acid residues between 382 and 725 of the LRR. Focusing on this region, a positive selection analysis will be performed to identify critical residues involved in the ATR13 recognition event, followed by site-directed mutagenesis of these residues to determine functionality and possibly discover novel specificities.
Materials and Methods
Strains and Growth.
Escherichia coli strains DH5α, Top10, and DB3.1 and A. tumefaciens strains C58C1 and GV2260 were grown on Luria–Bertani agar containing the appropriate antibiotics, at 37°C and 28°C respectively. Pst was grown on NYGA agar at 28°C. N. benthamiana plants were grown at 24°C, 16-h light/8-h dark cycle. A. thaliana plants were grown at 22°C, 8-h light/16-h dark cycle.
Plasmid Constructs.
For Hp effector delivery via the TTSS of Pst DC3000, a destination vector was constructed, by using the broad host-range vector pVSP61 as a backbone. The promoter (201 bp) and signal peptide leader sequence (267 bp) of AvrRpm1 (5, 26) were amplified by PCR with primers 5′-agtcgaattcGGAGGCCTGCAGTATTCGG-3′ and 5′-aatgggatccGATATCCTCGGTTGCACCATCAGTTTTTCT-3′. This EcoRI-AvrRpm1-EcoRV-BamHI fragment was digested with EcoRI and BamHI and inserted into pVSP61. An HA epitope plus stop codon tag annealed from two primers (5′-GATCCTATCCGTACGACGTACCAGACTACGCATGAA-3′, and 5′-AGCTTTCATGCGTAGTCTGGTACGTCGTACGGATAG-3′) was inserted at the BamHI and HindIII sites. This intermediate plasmid was redigested with BamHI, filled in with Klenow, and religated so that when the destination cassette is cloned in later, the HA epitope is maintained as a translational fusion onto the C terminus of various pENTR clones. This intermediate plasmid was cut at the introduced EcoRV site for cloning a EcoRV fragment containing the ccdB cassette reading frame A (Invitrogen) between the AvrRpm1 TTSS leader sequence and the HA epitope plus stop codon sequence to make the P. syringe TTSS signal peptide (PsSP) destination vector pVSP PsSPdes.
Alleles of ATR13 from H. parasitica isolates Emco5 and Emoy2 were amplified from spore cDNA by PCR and cloned into pENTR by using the Directional TOPO Cloning kit (Invitrogen). The following primers were used: forward Δ19 (no signal peptide) 5′-CACCAATCTGCTCCACGCCCATGCGCT-3′, Δ41 (no RXLR motif) 5′-CACCGCAGCCAGCGAAGTATTTGG-3′; reverse: ATR13Emco5 5′-CTGTCTGTCAAGAGCATCCCGA-3′, ATR13Emoy2 5′-CTGACTGGCAACGGCAGTCTG-3′. The pENTR ATR13 constructs were transferred to pVSP PsSPdes by performing an LR recombination reaction (Invitrogen). For ATR13 delivery by TuMV, Δ19 ATR13Emco5, Δ41ATR13Emco5, and Δ41ATR13Emoy2 fragments were amplified from plasmid DNA with primers adding a 5′ NcoI site and a 3′ HA epitope sequence, followed by a KpnI site. The PCR product was cloned into vector p5PKTuMCS, inserting ATR13 between the viral P1 and HC-Pro genes. This plasmid was digested with SmaI and KpnI, and the fragment containing ATR13 was cloned into p35STuMV. The resulting p35STuMVATR13 plasmid was digested with NcoI and ApaI, and the fragment containing ATR13 was ligated into the binary vector pCBTuMV.
For ATR13 delivery by Agrobacterium-mediated transient expression, the binary vector pE1776 with a MAS promoter plus triple chimera OCS UAS and one MAS UAS (15) was first modified with a T7 epitope plus start codon tag inserted at the XhoI and SpeI sites. This intermediate plasmid was redigested with SpeI and filled in with Klenow for cloning an EcoRV fragment containing the ccdB cassette reading frame B to make pE1776-T7des. ATR13 Emco5Δ19+stop and Emoy2Δ19+stop constructs were PCR amplified with the same forward Δ19 primer and the Emco5 and Emoy2 reverse primers, with an additional (5′-CTA-3′) stop codon added, were cloned directionally in pENTR. The pENTR ATR13 constructs were transferred to pE1776-T7des by performing an LR recombination reaction (Invitrogen).
For RPP13 delivery by Agrobacterium-mediated transient expression, three binary vectors with different strength promoters were used. For the 35S promoter, pMD1 (27) was first modified with a linker annealed from two primers (5′-TCGAGACTAGTTATCCCTACGACGTACCAGACTACGC A T T G C A AGCTCCACCCGCTGAGAGGGCTACACTGTAGG-3′) and (5′-TCGACCTACAGTGTAGCCCTCTCAGCGGGTGGAGCTTGCAATGCGTAGTCTGGTACGTCGTAG G G A TAACTAGTC-3′) and inserted at the XhoI site. This intermediate plasmid was redigested with SmaI for cloning an EcoRV fragment containing the ccdB cassette reading frame B to make pMD1–35Sdes. For the strong OCS promoter, an EcoRV fragment containing the ccdB cassette reading frame A was cloned into T4 filled-in SacI and KpnI sites of pE1776 to make pE1776des. For RPP13 native promoter, the 35S promoter of pMD1–35Sdes vector was replaced with a HindIII and XbaI native promoter fragment for RPP13 generated with primers: forward (5′-AAGCTTCGTGATATCGCAATCTGT-3′), reverse (5′-TCTAGACTTCTCAATATCGATTTGA-3′) to make pMD1-nP13des. RPP13Nd and RPP13Col alleles were PCR amplified from genomic DNA from Arabidopsis ecotypes Niederzenz and Columbia, respectively, and cloned into pENTR by using primers: common forward (5′-CACCATGGTAGATGCGATCACGGAGTTCGT-3′); reverse with a SpeI site before the stop codon: RPP13Nd (5′-CTAACTAGTAACGCTCGCAATCGGTTTG-3′) and RPP13Col (5′-CTAACTAGTGCAGTAGATTTGACCAAACGCT-3′). The pENTR RPP13Nd and pENTR RPP13Col constructs were further modified by adding an HA epitope plus stop codon at the SpeI site. The pENTR RPP13 constructs were transferred to both pE1776-T7des and pMD1–35Sdes by performing an LR recombination reaction.
For pENTR RPP13 Nd and Col chimera constructs, the common BstXI, KpnI, ClaI, and MslI sites of the pENTR RPP13Nd and pENTR RPP13Col were used with the unique AscI pENTR vector site to generate the various combinations. The pENTR RPP13 chimera constructs were transferred to pMD1 nP13des by performing an LR recombination reaction.
Bacterial Disease Assays.
All A. thaliana bacterial disease assays were performed as described (16).
TuMV Infection.
A. tumefaciens strain GV2260 with binary TuMV constructs were first used to infect N. benthamiana and subsequent infectious TuMV extracts from N. benthamiana were used to infect A. thaliana plants as described in ref. 13. Mottling of young A. thaliana leaves was observed after 6–8 days. Viability of viral particles from infected A. thaliana were also tested by reinfecting N. benthamiana. Infected leaves displayed comparable numbers of distinct lesions 5–6 days pi (data not shown).
Agrobacterium-Mediated Transient Expression.
A. tumefaciens strain C58C1 carrying the binary vector with the gene of interest were cultured, preinduced, and infiltrated into leaves of N. benthamiana as described (5).
Northern Blot Analysis.
Total RNA was extracted from individual plants infected with TuMV by using the RNeasy Plant Total RNA kit (Qiagen) according to the manufacturer's instructions. Northern blot analysis was performed as described (28). All probes were prepared by PCR, by using the following primers: coat protein: CP for 5′-TGACAGACGAGCAAAAGCAG-3′; CP rev 5′-ATGTTCCGATTAACGTCCTC-3′ (from pCBTuMVGFP plasmid) tub for 5′-GTGAACTCCATCTCGTCCAT-3′; tub rev 5′-CCTGATAACTTCGTCTTTGG-3′ (from cDNA).
Plant and Bacterial Immunoblot Analysis.
N. benthamiana transiently expressed proteins were extracted and immunoblot analysis performed as described in ref. 5. Pst cultures were washed and cultured for 3 h in the Pseudomonas effector promoter induction media with sucrose (8) before protein extraction and immunoblot analysis. Protein bolts were probed with either Anti-TuMV-CP: rabbit polyclonal antibody PVAS34 or Anti-HA: mouse monoclonal antibody(Covance) and secondary HRP-anti-rabbit or HRP-anti-mouse (Bio-Rad).
ACKNOWLEDGMENTS.
We thank John M. McDowell (Virginia Technical Institute and State University, Blacksburg, VA) for H. parasitica cDNA; Jonathan Jones (Sainsbury Laboratory, John Innes Centre, Norwich, U.K.) for H. parasitica DNA and for strategic discussions concerning the delivery of H. parasitica effectors via the TTSS of Pseudomonas syringae; Jim Carrington (Oregon State University, Corvallis, OR) for the plasmids p5PKTuMCS, p35STuMV, and pCBTuMVGFP; and Jim Beynon (University of Warwick, Coventry, U.K.) for the Col-5 RPP13Nd transgenic seed. We also thank all members of the B.J.S. laboratory for comments and critical reading of the manuscript. This work was supported by National Institutes of Health Grant R01 GM069680-01 (to B.J.S.).
Note Added in Proof.
Sohn et al. (29) have also shown that ATR13 and ATR1 can be delivered to plant cells via the P. syringae TTSS and can also promote disease susceptibility.
Footnotes
The authors declare no conflict of interest.
References
- 1.Koch E, Slusarenko A. Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell. 1990;2:437–445. doi: 10.1105/tpc.2.5.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Win J, et al. Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes. Plant Cell. 2007;19:2349–2369. doi: 10.1105/tpc.107.051037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen RL, et al. Host–parasite coevolutionary conflict between Arabidopsis and downy mildew. Science. 2004;306:1957–1960. doi: 10.1126/science.1104022. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharjee S, et al. The malarial host-targeting signal is conserved in the Irish potato famine pathogen. PLoS Pathog. 2006;2:e50. doi: 10.1371/journal.ppat.0020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leister RT, et al. Molecular genetic evidence for the role of SGT1 in the intramolecular complementation of Bs2 protein activity in Nicotiana benthamiana. Plant Cell. 2005;17:1268–1278. doi: 10.1105/tpc.104.029637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grant SR, Fisher EJ, Chang JH, Mole BM, Dangl JL. Subterfuge and manipulation: Type III effector proteins of phytopathogenic bacteria. Annu Rev Microbiol. 2006;60:425–449. doi: 10.1146/annurev.micro.60.080805.142251. [DOI] [PubMed] [Google Scholar]
- 7.Guttman DS, Greenberg JT. Functional analysis of the type III effectors AvrRpt2 and AvrRpm1 of Pseudomonas syringae with the use of a single-copy genomic integration system. Mol Plant–Microbe Interact. 2001;14:145–155. doi: 10.1094/MPMI.2001.14.2.145. [DOI] [PubMed] [Google Scholar]
- 8.Huynh TV, Dahlbeck D, Staskawicz BJ. Bacterial blight of soybean: Regulation of a pathogen gene determining host cultivar specificity. Science. 1989;245:1374–1377. doi: 10.1126/science.2781284. [DOI] [PubMed] [Google Scholar]
- 9.Bittner-Eddy PD, Crute IR, Holub EB, Beynon JL. RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant J. 2000;21:177–188. doi: 10.1046/j.1365-313x.2000.00664.x. [DOI] [PubMed] [Google Scholar]
- 10.Century KS, Holub EB, Staskawicz BJ. NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc Natl Acad Sci USA. 1995;9:6597–6601. doi: 10.1073/pnas.92.14.6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aarts N, et al. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:10306–10311. doi: 10.1073/pnas.95.17.10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lellis AD, Kasschau KD, Whitham SA, Carrington JC. Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr Biol. 2002;12:1046–1051. doi: 10.1016/s0960-9822(02)00898-9. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez F, Martinez-Herrera D, Aguilar I, Ponz F. Infectivity of turnip mosaic potyvirus cDNA clones and transcripts on the systemic host Arabidopsis thaliana and local lesion hosts. Virus Res. 1998;55:207–219. doi: 10.1016/s0168-1702(98)00049-5. [DOI] [PubMed] [Google Scholar]
- 14.Scofield SR, et al. Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- 15.Ni M, et al. Strength and tissue specificity of chimeric promoters derived from the octopine and mannopine synthase genes. Plant J. 1995;7:661–676. [Google Scholar]
- 16.Day B, Dahlbeck D, Staskawicz BJ. NDR1 interaction with RIN4 mediates the differential activation of multiple disease resistance pathways in Arabidopsis. Plant Cell. 2006;18:2782–2791. doi: 10.1105/tpc.106.044693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding J, et al. Contrasting patterns of evolution between allelic groups at a single locus in Arabidopsis. Genetica. 2007;129:235–242. doi: 10.1007/s10709-006-0002-9. [DOI] [PubMed] [Google Scholar]
- 18.Bakker EG, Toomajian C, Kreitman M, Bergelson J. A genome-wide survey of R gene polymorphisms in Arabidopsis. Plant Cell. 2006;18:1803–1818. doi: 10.1105/tpc.106.042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rose LE, et al. The maintenance of extreme amino acid diversity at the disease resistance gene, RPP13, in Arabidopsis thaliana. Genetics. 2004;166:1517–1527. doi: 10.1534/genetics.166.3.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McHale L, Tan X, Koehl P, Michelmore RW. Plant NBS-LRR proteins: adaptable guards. Genome Biol. 2006;7:212. doi: 10.1186/gb-2006-7-4-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellis J, Dodds P, Pryor T. Structure, function and evolution of plant disease resistance genes. Curr Opin Plant Biol. 2000;3:278–284. doi: 10.1016/s1369-5266(00)00080-7. [DOI] [PubMed] [Google Scholar]
- 22.Dodds PN, et al. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci USA. 2006;103:8888–8893. doi: 10.1073/pnas.0602577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burch-Smith TM, Dinesh-Kumar SP. The functions of plant TIR domains. Sci STKE. 2007;2007:pe46. doi: 10.1126/stke.4012007pe46. [DOI] [PubMed] [Google Scholar]
- 24.Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- 25.Ade J, DeYoung BJ, Golstein C, Innes RW. Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci USA. 2007;104:2531–2536. doi: 10.1073/pnas.0608779104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ritter C, Dangl JL. The avrRPm1 gene of Pseudomonas syringae pv maculicola is required for virulence on Arabidopsis. Mol Plant–Microbe Interact. 1995;8:444–453. doi: 10.1094/mpmi-8-0444. [DOI] [PubMed] [Google Scholar]
- 27.Tai TH, et al. Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc Natl Acad Sci USA. 1999;96:14153–14158. doi: 10.1073/pnas.96.24.14153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown T. In: Current Protocols in Molecular Biology. Ausubel FM, et al., editors. New York: Wiley; 1994. pp. 4.9.14–4.9.16. [Google Scholar]
- 29.Sohn KH, Lei R, Nemri A, Jones JDG. The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana. Plant Cell. 2007 doi: 10.1105/tpc.107.054262. [DOI] [PMC free article] [PubMed] [Google Scholar]