Abstract
Many multifunctional tumor suppressor proteins have low stability, a property linked to cancer development. The von Hippel–Lindau tumor suppressor protein (pVHL) is one of these proteins. pVHL forms part of the E3 ubiquitin ligase complex that regulates the degradation of the hypoxia-inducible factor (HIF). Under native conditions, free pVHL is a molten globule, but it is stabilized in the E3 complex. By using molecular dynamics simulations, we observed that the interface between the two pVHL domains is the least stable region in unbound pVHL. We designed five stable mutants: one with a mutation at the interdomain interface and the others in the α- or β-domains. Experimentally, type 2B pVHL disease mutant Y98N at the HIF binding site was shown to destabilize pVHL and decrease its binding affinity to HIF. Our simulations showed that the decrease in pVHL stability and binding affinity are allosterically regulated. The mutations designed to stabilize unbound wild-type pVHL, which are away from the elongin C and HIF binding sites, successfully stabilized the Y98N pVHL–elongin C complex and lowered the binding free energy of pVHL with HIF. Our results indicated both the enthalpic and dynamic allosteric components between the elongin C and HIF binding sites in pVHL, in the α- and β-domains, respectively, mediated by the interdomain interface and linker. Drugs mimicking the allosteric effects of these mutants may rescue pVHL function in von Hippel–Lindau disease.
Keywords: allosteric site prediction, drug design, E3 ubiquitin ligase, HIF, molecular dynamics simulations
The von Hippel–Lindau (VHL) tumor suppressor gene is mutated in the von Hippel–Lindau cancer predisposition syndrome and in sporadic, clear-cell renal carcinomas (RCCs) (1–5). Germ-line mutations in this gene can cause the VHL hereditary cancer syndrome, which is characterized by the development of tumors of the central nervous system, kidney, retina, pancreas, and adrenal glands (6, 7). Somatic mutations of the VHL gene also play an important role in the development of sporadic hemangioblastomas and RCC, the most common form of adult kidney cancer (8, 9).
The function of the VHL tumor suppressor protein (pVHL) has been explored extensively. pVHL binds to elongin C and elongin B (10–13) and functions as the substrate-recognition component of the E3 ubiquitin protein ligase complex that negatively regulates the hypoxia-inducible factor (HIF) under normoxic conditions (9, 14–18). HIF is composed of two nonidentical subunits, HIF-α and HIF-β (19, 20). HIF-β is stable but HIF-α is degraded by pVHL during normoxia (17, 20–23). Loss of pVHL or hypoxic conditions results in HIF-α accumulation and, when bound to HIF-β, leads to the formation of the HIF heterodimer (5). The heterodimer binds to specific DNA sequences and activates the transcription of some hypoxia-inducible genes, such as vascular endothelial growth factor (VEGF), which promotes tumor growth and vascularization (24–27).
pVHL has two domains, α and β. The α-domain and the interdomain linker bind to elongin C (13), whereas the β-domain interacts with HIF (15, 28, 29). The carcinogenic mutations are mostly in the HIF and elongin C binding sites, affecting the major function of pVHL. Sutovsky and Gazit (30) reported that free pVHL is a molten globule with marginal stability under native conditions. These authors analyzed the stability of pVHL in solution under physiologically relevant conditions by using several biophysical techniques, including dynamic light-scattering and gel-filtration chromatography, and observed a much larger Stokes radius of purified pVHL than in the crystal structure. Their near-UV circular dichroism experiments further indicated the absence of a tertiary structure. pVHL stability is greatly improved after binding to elongin C and elongin B (31, 32). There is evidence that the renal cell carcinoma risk in type 2 von Hippel–Lindau disease correlates with defects in pVHL stability and HIF-1α interactions (31). Type 2B mutants Y98N and Y112N do not affect the binding of pVHL to elongin C to form the CBCVHL binding complex; however, at physiological temperature its stability is lower. Interestingly, mutants that lowered pVHL stability also significantly influenced the ubiquitin ligase activity toward HIF-1α in vitro (31). Recently, Jung et al. reported that E2-EPF UCP destabilizes pVHL and stabilizes HIF (33). These studies provide evidence indicating that pVHL stability correlates with its function of binding to HIF.
pVHL is not the only marginally stable tumor suppressor protein. Other tumor suppressors, such as p53 and p16, also have low intrinsic thermodynamic stabilities (34–37). Involvement of p53 nonfunctional mutations in cancer has been related to low intrinsic stabilities (34) and considerable efforts have been invested in stable p53 design (35, 38). Comparison of the human and worm p53 structures have also suggested a way to stabilize human p53 (36). However, there are few studies on the origin of pVHL instability, correlated allosteric effects between its two binding sites in the α- and β-domains and the involvement of the interdomain interface and linker in the effects of cancer-related mutations, and on the design of stable pVHL.
In this study, we first performed molecular dynamics simulations on unbound human pVHL and on pVHL bound to elongin C. We observed that the most unstable motifs were in the interface between pVHL α- and β-domains. Among these interdomain interface motifs, the interdomain linker and its linked helix in the α-domain (H1) were also part of the pVHL-elongin C binding site. We mutated residues whose motions were strongly correlated with these two motifs, and following our p53 strategy, selected corresponding residues from the worm pVHL (36). Five mutations were performed creating five mutants. Similar to the p53, here too the worm-based residue substitutions stabilized the human pVHL.
We proceeded to study the renal cell carcinoma type 2B mutant Y98N, experimentally demonstrated to be less stable and to have lower binding affinity with HIF than wild-type pVHL (31). Consistent with experiment, we observed that this mutation destabilized the pVHL and that there is a correlation between the stability and binding affinity of pVHL and HIF. We next inserted the G123F and D179N stabilizing mutations derived from the worm pVHL sequence into the Y98N cancer-related mutant. We observed that these worm-based substitutions not only stabilized the wild-type pVHL, but also pVHL mutant Y98N and improved the stability of pVHL bound to HIF. That is, the G123F and D179N mutations can rescue mutant pVHL stability and thus apparently its function as a HIF binding module. We further observed that these mutations also stabilized the pVHL–elongin C complex. Our results clearly indicated both the enthalpic and dynamic allosteric components between the elongin C and HIF binding sites in pVHL, in the α- and β-domains, respectively, mediated by the interdomain interface. The methods used here could be general and prove useful in identification of allosteric sites for mutant engineering and drug discovery and design.
Results and Discussion
Comparison of Unbound and Bound pVHL.
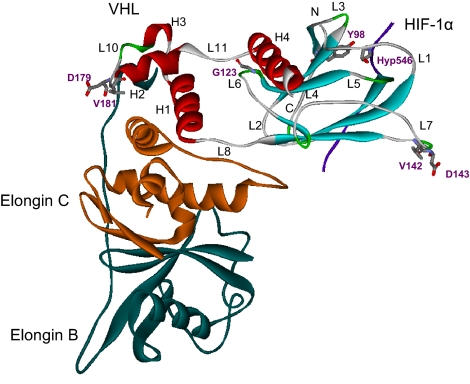
The crystal structure of pVHL bound with elongin C and elongin B (13) had two domains, α and β. The C-terminal α-domain consisted of three α-helices, H1, H2, and H3. The N-terminal β-domain of pVHL consisted of a seven-stranded β-sandwich and an α-helix, H4. The two domains were connected by two linkers, the L8 and L11 loops, and polar interactions between the H1 helix and the L6 loop at the interface. pVHL interacted with elongin C by the α-domain H1, H2, and H3 helices and the L8 loop interdomain linker, as shown in Fig. 1. Sutovsky and Gazit (30) demonstrated that the unbound form of pVHL was a molten globule. However, binding to elongin C and elongin B stabilized pVHL (13). We performed molecular dynamics simulations for both unbound pVHL and pVHL bound to elongin C. As expected, unbound pVHL was much less stable, in agreement with experiment (Fig. 2a). To understand the origin for the unbound pVHL instability, we analyzed the pVHL domains α and β and their interface. The β-domain was the most stable segment in both unbound and bound pVHL with the lowest rmsd fluctuation (Fig. 2b). As to the α domain, even though both the unbound and bound pVHL had increased fluctuations (Fig. 2c) and change in the radius of gyration [supporting information (SI) Fig. 7], the differences between the unbound and bound forms were not significant. However, the interface between the α- and β-domains had increased rmsd (Fig. 2d) and radius of gyration (SI Fig. 7) for the unbound pVHL but not for the bound form. These results suggested that both α- and β-domains were relatively stable in the unbound pVHL; however, the region largely responsible for the structural fluctuation was the interface between these two domains. Binding to elongin C stabilized the interface and thus the whole pVHL. The details of the rmsd values for the bound and unbound pVHL are presented in SI Table 1.
Fig. 1.
Crystal structure of HIF (purple)–VHL (red and cyan)–elongin C (orange)–elongin B (green) complex (PDB ID code 1lm8). The mutated positions are noted as well as the cancer-related mutation site (Y98). The relevant loops and helices that are discussed in the text are marked.
Fig. 2.
C rmsds from MD simulations for unbound (black) and bound (red) pVHL. The bound pVHL was complexed with elongin C in the simulations. (PDB ID code 1vcb). (a) rmsd of the entire pVHL. (b) rmsd of the pVHL β-domain. (c) rmsd of pVHL α-domain. (d) rmsd of pVHL interdomain interface.
Design of Mutants to Stabilize Unbound pVHL.
Because the most unstable part of pVHL was the interface between the α- and β-domains, to stabilize pVHL we needed to stabilize the interface. However, the interface also included regions that were bound to elongin C. To choose mutation sites that stabilized pVHL without interrupting the binding of pVHL to elongin C, covariance matrix maps and residue-based fluctuations were examined. Because we suspected that mutations elsewhere in the structure might have allosteric effects, we first identified regions of pVHL whose motions were strongly correlated with the interface residues. These regions should present the most significant change in their correlated motions to the interdomain interface between the unbound pVHL and the elongin C bound state. The covariance map difference between the unbound and bound forms of pVHL is shown in Fig. 3a. Overall, as expected, the correlated motions between the residues were stronger in the unbound pVHL than in the bound form. The region between the two lines, including L8 and H1, is the interdomain interface. As shown in Fig. 3a, the L2, L3, L4, L6, L7, L8, H1, and L10 motifs (positions marked in Fig. 3b) present the most significant change in the correlated motions to L8 and H1 after binding to elongin C. The motifs with the largest correlated motions with L8 and H1 should be potential mutation targets for stabilizing unbound pVHL. Examination of the root mean squared fluctuations (RMSF) plot (Fig. 3b) confirmed that bound pVHL had lower fluctuations than unbound pVHL in these motifs. Among these, L2 and L4 are from the β-domain and fluctuated to a lesser extent. Both L7 and L10 are away from either interface or binding sites. L7 is from the β-domain and L10 is from at the α-domain (see Fig. 1). The L6, H1, and L8 motifs are at the α- and β-interdomain linker or interface that overall showed the largest instability (Fig. 2). L8 and H1 are also in the binding region to elongin C. We examined the residues in the peaks in these areas. We noticed that the peak residues of L2, L4, L8, and H1 were involved in either binding to elongin C or HIF thus could not be mutated. Therefore, residues G123 from L6 at the interface, V142 and D143 from L7 at the β-domain, and D179 and V181 from L10 in the α-domain were selected as the potential mutation sites to stabilize pVHL (circled in Fig. 3b).
Fig. 3.
Covariance map and RMSFs of bound and unbound pVHL. (a) Covariance map difference between the bound and unbound pVHL. The region between the two lines is the interdomain interface. Circles are areas where mutations were made. (b) RMSFs of bound and unbound pVHL. Circles are residues at binding sites either with HIF or elongin C. The arrows point to loop regions where residues were mutated.
Like pVHL, p53 is also an unstable tumor suppressor protein. Molecular dynamics studies suggested that the worm p53 is more stable than human p53 and that mutating human p53 according to the worm p53 sequence can enhance the stability of human p53 (36). We hypothesized that worm pVHL might also be more stable than human pVHL. Because, unlike p53, the worm pVHL crystal structure is unavailable and we encountered difficulties in building a reasonable homology model, we performed only a sequence alignment between human and worm pVHL (SI Fig. 8). The alignment suggested only 25% overall sequence identity; however, most binding site residues were conserved, suggesting functional conservation between the human and worm pVHL. Nonconserved residues were selected as candidates for mutations in the correlated regions discussed above. Five human pVHL mutants, G123F, V142P, D143R, D179N, and V181I, were generated, located in L6, L7, and L10. Molecular dynamics simulations were performed for these mutants. The rmsd plots are shown in Fig. 4. All five mutants are more stable than the wild type. Among these, the G123F (in L6) and D179N (in L10) mutants stabilize unbound pVHL the most. The covariance maps (SI Fig. 9) also show that G123F and D179N mutants have a lesser change in the correlated motions than the unbound wild type when compared with the bound. L6 is in the interdomain interface. L10 is far away (Fig. 1).
Fig. 4.
The rmsd of unbound pVHL designed mutants. (a) V181I and D179N at L10 of the α-domain. (b) V142P and D143R at L7 of the β-domain. (c) G123F at L6 at the interface between the domain.
Rescuing pVHL-HIF Binding Affinity.
Previously it was suggested that cancer-related type 2B mutation Y98N destabilized the pVHL when it was bound to elongin C (31). The same Y98N mutation also decreased the binding affinity of pVHL with HIF. The association rate of the Y98N mutant with HIF only decreased by twofold compared with the wild type; however, the dissociation rate increased >285-fold. This mutation was in the β-domain, at the HIF binding site (Fig. 1). This was not a unique case for a protein–protein binding dissociation rate to change much more dramatically than an association rate. In 1994, Bjork et al. reported such differential changes in the association and dissociation rate constants. They suggested that the contributing factor was the change in stability rather than the binding site conformational change (39). These observations implied that the stability of pVHL when in complex with elongin C and the pVHL-HIF binding affinity might be correlated, suggesting long-range effects. We performed simulations for the pVHL–elongin C complex for the wild-type and Y98N mutants. The rmsd plot in Fig. 5 showed that the structure of the Y98N mutant was less stable than the wild type, which was consistent with the experiments; at the same time the snapshots in SI Fig. 10 suggested that the conformation was unchanged for the Y98N mutant compared with the wild type.
Fig. 5.
The rmsd of pVHL when bound to elongin C for the wild type (black), cancer mutation Y98N (red), and rescuing mutants G123F_Y98N (green), and D179N_Y98N (blue).
To understand whether the stability and binding affinity are correlated, we needed to stabilize pVHL Y98N mutant when bound to elongin C. Because G123F and D179N were found to be able to stabilize the unbound pVHL, simulations were performed with pVHL–elongin C complex for G123F_Y98N and D179N_Y98N double mutants. We observed that the G123F and D179N mutations (derived from the worm sequence) can stabilize the disturbed Y98N structure of pVHL when bound to elongin C, as shown in Fig. 5. We also performed simulations for the pVHL–elongin C–HIF complex to examine how these mutations would affect the binding free energy of pVHL and HIF. Knauth et al. (31) reported that Y98N had a much lower binding affinity than the wild type. Our calculations showed that G123F_Y98N and D179N_Y98N double mutations significantly lowered the binding free energy by −12.6 and −12.8 kcal/mol, respectively. Neither G123 nor D179 are in the binding site of pVHL and HIF: one is in the interdomain interface and the other is in the α-domain. Nevertheless, mutations far away from the HIF binding site significantly changed the binding free energy.
We investigated the binding site of pVHL-HIF. In the crystal structure, HIF Hyp-546 (hydroxyproline) formed multiple van der Waals contacts with pVHL Trp-88, Tyr-98, Ser-111, His-115, and W117, making a major contribution to the binding of pVHL and HIF. The distances between the residue pairs are shown in SI Fig. 11. Only the wild type had all residue contacts during the simulations. Mutants Y98N, G123F_Y98N, and D179N_Y98N all had loose contact between Hyp-546 and the mutated Asn-98, which is understandable; the change of tyrosine 98 to asparagine alters the hydrophobic core formed in the wild type by Trp-88, Tyr-98, and Trp-117. All of the other residue pair contacts of the mutants were as in the wild type. Thus, except for the loss of the Y98–Hyp-546 interaction, no dramatic changes were observed in the binding site of the Y98N mutant compared with the wild type; yet at the same time, the mutations significantly lowered the binding energy of pVHL and HIF.
The whole picture of pVHL–elongin C–HIF complex may provide an answer for this apparent paradox. Comparisons of the snapshots (Fig. 6; pVHL in blue; elongin C, red; HIF, green) clearly showed that the pVHL-HIF binding sites for these four complexes (the wild type and three mutants) did not change significantly, but the binding sites of pVHL interacting with elongin C were different in these four cases. In the wild type (Fig. 6a), the α-helix of elongin C formed a four-helix hydrophobic core with the pVHL three helices. However, these contacts are lost in the Y98N mutant (Fig. 6b). The double mutations G123F_Y98N and D179N_Y98N (Fig. 6 c and d), in part, restored these contacts. Therefore, even though the double mutants did not change the binding site of pVHL-HIF, they stabilized the binding of pVHL with elongin C compared with Y98N. The allosteric effect of the Y98N mutant at the HIF binding site caused a small conformational change at the pVHL–elongin C binding site, thereby leading to a less favorable enthalpy change; however, the favorable enthalpy change was restored by the double mutations. Those mutations were also away from the pVHL–elongin C binding site, yet their motions correlated with the interdomain linker.
Fig. 6.
Snapshots of pVHL–elongin C–HIF simulations. (a) Wild type. (b) Y98N. (c) G123F_Y98N. (d) D179N_Y98N. The circled areas are the elongin C–pVHL binding site. It is bolded in the cancer-mutant-affected site. Elongin C is in red; VHL, blue; HIF, green.
The covariance maps showed that the correlated motions between HIF and elongin C binding sites in the double mutants (SI Fig. 12b and c) are more similar to those of the wild type than those of the Y98N mutant (SI Fig. 12a). The change in the pVHL-correlated motions between the HIF and elongin C binding sites in the Y98N mutant may be the reason behind the loss of binding of elongin C to pVHL. The snapshots (Fig. 6) confirm that the elongin C-pVHL binding was affected by the pVHL–HIF interaction. Unlike the pVHL–elongin C–HIF complex, the pVHL–elongin C complex (SI Fig. 10) has the stable elongin C–pVHL interface for all of four simulations, including the Y98N mutant. This result confirmed that binding with HIF influences the pVHL–elongin C binding for the Y98N mutant.
In 2006, experiments carried out by Popovych et al. suggested that dynamics-driven allostery can be mediated solely by protein motions (40). The pVHL Y98N mutant presented both conformation- and dynamics-driven allostery. The binding to HIF led to a significant decrease in flexibility and entropy penalty. At the same time, the loss of the favorable pVHL–elongin C binding accounted for the less favorable enthalpy change. The combination of these two factors led to the weaker affinity of Y98N mutant binding to HIF. The decrease in flexibility in the double mutants G123F_Y98N and D179N_Y98N is less significant; thus, the entropy penalty is lower. The remaining pVHL–elongin C binding decreases the loss of enthalpy change. Therefore, these double mutants can increase the binding affinity compared with that of the Y98N mutant binding to HIF.
These results also provided an explanation for the differential change of association and dissociation rate constants for pVHL–HIF binding between the wild type and the Y98N mutant. The fact that the association rate constant of the wild type was only two times that of the Y98N mutant may be explained by the observation that the binding of pVHL–HIF is not greatly affected. Most contacts with Hyp-546 still exist except at the mutation. However, as noted above, the dissociation rate constant of the Y98N mutant increases 285-fold compared with the wild type. This can be explained by the binding of Y98N mutant pVHL to elongin C being affected after the pVHL–HIF interaction. Loss of pVHL–elongin C binding by the Y98N mutant reduced the pVHL–HIF dissociation rate constants significantly because the pVHL–elongin C binding loss contributed significantly to less favorable enthalpy change. Although the double mutants G123F_Y98N and D179N_Y98N did not change the binding sites of pVHL–HIF, they restored the binding of pVHL–elongin C. Therefore, we propose that although the double-mutant association rate constants would not change significantly compared with the Y98N mutant, they would significantly decrease the dissociation rate constants of Y98N, thus enhancing the binding of pVHL and HIF to rescue pVHL function.
Allosteric binding sites can be targets for drug discovery. For G protein-coupled receptors (GPCRs) and ligand-gated ion channels (LGICs), allosteric modulators have been extensively investigated in drug discovery (41, 42). We propose that mimicking the allosteric effects of the stabilizing mutants could be a rescue strategy to restore pVHL function. To check the availability of such drug binding sites, we inspected the surface of pVHL mutants, as shown in SI Fig. 13. Interestingly, for mutant G123F (SI Fig. 13a), the mutation F123 is right in a pocket with depth of 17 Å and width of 20 Å, whereas for mutant D179N (SI Fig. 13b), the mutation N179 is on the edge of a 12-Å-deep and 16-Å-wide pocket. Such pockets provide potential drug binding sites that could mimic the mutants. We further suggest that the computational protocol used here, involving identification of allosteric sites by large correlated motions to binding sites and examination of covariance matrix maps, could prove a general strategy for engineering rescue mutants and for drug discovery and design.
Conclusions
Molecular dynamics simulations of unbound pVHL and pVHL bound to elongin C suggested that the interface between the two domains of pVHL is the region largely responsible for the marginal stability of unbound pVHL. We expected that residues showing motions correlated with this region can be mutated to stabilize the unbound pVHL. We mutated such residues to their corresponding worm pVHL residues based on alignment between the human and the worm pVHL sequences. The simulations suggested that all mutants, G123F, V143P, D143R, D179N, and V181I, can stabilize unbound pVHL. Among these, G123F and D179N stabilized the unbound pVHL the most.
Simulations of the wild-type pVHL–elongin C–HIF and of the complex with pVHL cancer-related mutation Y98N revealed a lower affinity to HIF by this type2B pVHL disease mutation than the wild type. This observation can be understood: On the one hand, the fluctuations in the Y98N mutant in the pVHL–elongin C complex were much higher than those in the wild type; yet the structural fluctuations after binding to HIF were very similar to the wild type. This dynamically driven allostery contributed to a higher entropy penalty. However, binding of Y98N to HIF disrupted the pVHL binding to elongin C. These results also explained the differential change in the dissociation and association rate constants of the wild type and the Y98N mutation as observed in experiment. The designed mutants that can stabilize unbound pVHL, such as G123F and D179N, can improve the binding affinity of pVHL by stabilizing the pVHL–elongin C and rescue the pVHL binding to elongin C during the 10-ns simulations. We propose that drugs mimicking these mutants can stabilize pVHL in the von Hippel–Lindau disease and provide a rescue strategy to restore pVHL function. We further propose that the method described here to identify unknown allosteric sites could be a general strategy, potentially useful in engineering mutants, and in drug discovery and design.
Computational Methods
Molecular dynamics (MD) simulations were performed with a CHARMM 22 (43) force field by using CHARMM (44). The starting structure of the unbound and bound pVHL was constructed from the crystal structure solved by Stebbins et al. (13) (PDB ID code 1vcb). For the structure of the pVHL bound to elongin C, only elongin C residues 58–112 were used because coordinates were unavailable for the eight-residue gap between residues 49 and 58, and the solved elongin C structure for residues 17–49 was far away from the binding site of pVHL–elongin C. The starting structure of pVHL–elongin C–HIF complex was generated from the crystal structure solved by Min et al. (29) (PDB ID code 1lm8) with the same elongin C truncation. The mutations were selected from a sequence alignment with the worm pVHL. Mutant structures were built from the wild-type model, with all of the backbone atoms superimposed on the corresponding atoms of the wild-type structure. The side chains were generated by using CHARMM and underwent 500 steps of energy minimization with the rest of the protein fixed to remove any steric conflicts before the MD simulations were performed. All models were solvated in a TIP3P water box with a minimum distance of 10 Å from the edge of the box to any protein atom. The charges of the system were neutralized by adding chloride or sodium ions. To eliminate residual unfavorable interactions between the solvent and the protein, the solvated systems were first minimized for 500 steps with the protein restrained followed by another 500 steps of minimization for the whole system by using the steepest decent algorithm. After 20 ps equilibration with the NVT ensemble, the production simulations were performed for 10 ns with the NPT ensemble at a temperature of 300 K. During the production simulations, the time step was 2 fs, with a SHAKE constraint on all bonds containing hydrogen atoms, and the nonbonded cutoff was 12Å. Structures were saved every 2 ps. The simulation results were analyzed with CHARMM.
The contribution of mutations to the binding energy of pVHL and HIF was evaluated by using the following equation:
The binding free energies were as follows:
G could be described as
where EMM is the molecular mechanical energy of the system consisting of all components in the CHARMM potential energy function, and Gsolv could be written as two terms: Gelec and Gne. The electrostatic contribution to the solvation energy Gelec was calculated with the Generalized Born using the Molecular Volume (GBMV) method (44, 45). The nonelectrostatic term Gne was calculated through solvent-accessible surface area (SASA) calculations. TSMM is the entropy contribution calculated by the harmonic approximation for the temperature 300 K. Energy calculations were performed on 5,000 structures extracted from the simulation trajectories at 2-ps time intervals during the 10-ns simulations. Statistical analysis was performed by obtaining the overall averages from these 5,000 structures.
Supplementary Material
ACKNOWLEDGMENTS.
We thank members of the R.N. group for discussions. This project was funded by National Cancer Institute, National Institutes of Health, Contract no. N01-CO-12400. This work was supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707401105/DC1.
References
- 1.Kaelin WG, Jr, Maher ER. Trends Genet. 1998;14:423–426. doi: 10.1016/s0168-9525(98)01558-3. [DOI] [PubMed] [Google Scholar]
- 2.Maher ER, Kaelin WG., Jr Medicine (Baltimore) 1997;76:381–391. doi: 10.1097/00005792-199711000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Gnarra JR, et al. Biochim Biophys Acta. 1996;1242:201–210. doi: 10.1016/0304-419x(95)00012-5. [DOI] [PubMed] [Google Scholar]
- 4.Linehan WM, Lerman MI, Zbar B. J Am Med Assoc. 1995;273:564–570. [PubMed] [Google Scholar]
- 5.Kim WY, Kaelin WG. J Clin Oncol. 2004;22:4991–5004. doi: 10.1200/JCO.2004.05.061. [DOI] [PubMed] [Google Scholar]
- 6.van der Harst E, et al. Int J Cancer. 1998;77:337–340. doi: 10.1002/(sici)1097-0215(19980729)77:3<337::aid-ijc5>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 7.Zbar B, et al. Hum Mutat. 1996;8:348–357. doi: 10.1002/(SICI)1098-1004(1996)8:4<348::AID-HUMU8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 8.Shuin T, et al. Contrib Nephrol. 1999;128:1–10. doi: 10.1159/000059976. [DOI] [PubMed] [Google Scholar]
- 9.Kaelin WG., Jr Nat Rev Cancer. 2002;2:673–682. doi: 10.1038/nrc885. [DOI] [PubMed] [Google Scholar]
- 10.Duan DR, et al. Science. 1995;269:1402–1406. doi: 10.1126/science.7660122. [DOI] [PubMed] [Google Scholar]
- 11.Duan DR, et al. Proc Natl Acad Sci USA. 1995;92:6459–6463. doi: 10.1073/pnas.92.14.6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kibel A, Iliopoulos O, DeCaprio JA, Kaelin WG., Jr Science. 1995;269:1444–1446. doi: 10.1126/science.7660130. [DOI] [PubMed] [Google Scholar]
- 13.Stebbins CE, Kaelin WG, Jr, Pavletich NP. Science. 1999;284:455–461. doi: 10.1126/science.284.5413.455. [DOI] [PubMed] [Google Scholar]
- 14.Cockman ME, et al. J Biol Chem. 2000;275:25733–25741. doi: 10.1074/jbc.M002740200. [DOI] [PubMed] [Google Scholar]
- 15.Ohh M, et al. Nat Cell Biol. 2000;2:423–427. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 16.Iwai K, et al. Proc Natl Acad Sci USA. 1999;96:12436–12441. doi: 10.1073/pnas.96.22.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maxwell PH, et al. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 18.Kamura T, et al. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 19.Semenza GL. Curr Opin Cell Biol. 2001;13:167–171. doi: 10.1016/s0955-0674(00)00194-0. [DOI] [PubMed] [Google Scholar]
- 20.Wang GL, Jiang BH, Rue EA, Semenza GL. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang LE, Gu J, Schau M, Bunn HF. Proc Natl Acad Sci USA. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salceda S, Caro J. J Biol Chem. 1997;272:22642–22647. doi: 10.1074/jbc.272.36.22642. [DOI] [PubMed] [Google Scholar]
- 23.Jiang BH, Semenza GL, Bauer C, Marti HH. Am J Physiol. 1996;271:C1172–C1180. doi: 10.1152/ajpcell.1996.271.4.C1172. [DOI] [PubMed] [Google Scholar]
- 24.Semenza GL. Trends Mol Med. 2001;7:345–350. doi: 10.1016/s1471-4914(01)02090-1. [DOI] [PubMed] [Google Scholar]
- 25.Pugh CW, Ratcliffe PJ. Nat Med. 2003;9:677–684. doi: 10.1038/nm0603-677. [DOI] [PubMed] [Google Scholar]
- 26.Mani A, Gelmann EP. J Clin Oncol. 2005;23:4776–4789. doi: 10.1200/JCO.2005.05.081. [DOI] [PubMed] [Google Scholar]
- 27.Iliopoulos O, Levy AP, Jiang C, Kaelin WG, Jr, Goldberg MA. Proc Natl Acad Sci USA. 1996;93:10595–10599. doi: 10.1073/pnas.93.20.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hon WC, et al. Nature. 2002;417:975–978. doi: 10.1038/nature00767. [DOI] [PubMed] [Google Scholar]
- 29.Min JH, et al. Science. 2002;296:1886–1889. doi: 10.1126/science.1073440. [DOI] [PubMed] [Google Scholar]
- 30.Sutovsky H, Gazit E. J Biol Chem. 2004;279:17190–17196. doi: 10.1074/jbc.M311225200. [DOI] [PubMed] [Google Scholar]
- 31.Knauth K, Bex C, Jemth P, Buchberger A. Oncogene. 2006;25:370–377. doi: 10.1038/sj.onc.1209062. [DOI] [PubMed] [Google Scholar]
- 32.Schoenfeld AR, Davidowitz EJ, Burk RD. Proc Natl Acad Sci USA. 2000;97:8507–8512. doi: 10.1073/pnas.97.15.8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jung CR, et al. Nat Med. 2006;12:809–816. doi: 10.1038/nm1440. [DOI] [PubMed] [Google Scholar]
- 34.Bullock AN, et al. Proc Natl Acad Sci USA. 1997;94:14338–14342. doi: 10.1073/pnas.94.26.14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joerger AC, Allen MD, Fersht AR. J Biol Chem. 2004;279:1291–1296. doi: 10.1074/jbc.M309732200. [DOI] [PubMed] [Google Scholar]
- 36.Pan Y, Ma B, Levine AJ, Nussinov R. Biochemistry. 2006;45:3925–3933. doi: 10.1021/bi052242n. [DOI] [PubMed] [Google Scholar]
- 37.Tang KS, Guralnick BJ, Wang WK, Fersht AR, Itzhaki LS. J Mol Biol. 1999;285:1869–1886. doi: 10.1006/jmbi.1998.2420. [DOI] [PubMed] [Google Scholar]
- 38.Pan Y, Ma B, Venkataraghavan RB, Levine AJ, Nussinov R. Biochemistry. 2005;44:1423–1432. doi: 10.1021/bi047845y. [DOI] [PubMed] [Google Scholar]
- 39.Bjork I, et al. Biochem J. 1994;299:219–225. doi: 10.1042/bj2990219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Popovych N, Sun S, Ebright RH, Kalodimos CG. Nat Struct Mol Biol. 2006;13:831–838. doi: 10.1038/nsmb1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Christopoulos A. Nat Rev Drug Discov. 2002;1:198–210. doi: 10.1038/nrd746. [DOI] [PubMed] [Google Scholar]
- 42.Gunasekaran K, Ma B, Nussinov R. Proteins. 2004;57:433–443. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- 43.MacKerell AD, Jr, et al. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 44.Lee MS, Salsbury FR, Brooks CLI. J Chem Phys. 2002;116:10606–10614. [Google Scholar]
- 45.Still WC, Tempczyk A, Hawley RC, Hendrickson T. J Am Chem Soc. 1990;112:6127–6129. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.