Abstract
The diversity of terpenoid compounds produced by plants plays an important role in mediating various plant–herbivore, plant–pollinator, and plant–pathogen interactions. This diversity has resulted from gene duplication and neofunctionalization of the enzymes that synthesize and subsequently modify terpenes. Two diterpene synthases in Norway spruce (Picea abies), isopimaradiene synthase and levopimaradiene/abietadiene synthase, provide the hydrocarbon precursors for most of the diterpene resin acids found in the defensive oleoresin of conifers. Although these paralogous enzymes are 91% identical at the amino acid level, one is a single-product enzyme, whereas the other is a multiproduct enzyme that forms completely different products. We used a rational approach of homology modeling, protein sequence comparison, domain swapping, and a series of reciprocal site-directed mutagenesis to identify the specific residues that direct the different product outcomes. A one-amino acid mutation switched the levopimaradiene/abietadiene synthase into producing isopimaradiene and sandaracopimaradiene and none of its normal products. Four mutations were sufficient to reciprocally reverse the product profiles for both of these paralogous enzymes while maintaining catalytic efficiencies similar to the wild-type enzymes. This study illustrates how neofunctionalization can result from relatively minor changes in protein sequence, increasing the diversity of secondary metabolites important for conifer defense.
Keywords: biochemical evolution, natural product biosynthesis, plant defense, terpene diversity, site-directed mutagenesis
Conifers produce an oleoresin containing approximately equal amounts of monoterpenoids and diterpene acids with smaller amounts of sesquiterpenoids. The role of this secretion is thought to be largely for defense (1–3). In addition to its roles as a sticky physical barrier to invading pests and pathogens, the myriad of compounds that make up this secretion can have various behavioral and physiological roles in plant–herbivore and plant–pathogen interactions. After exposure to the atmosphere, the more volatile compounds evaporate to leave the diterpene resin acids that can harden to seal wounds (2).
Terpenoids are formed by terpene synthases (TPSs), which, particularly those of secondary metabolism in plants, comprise a large gene family believed to result from multiple gene duplications and subsequent neofunctionalization and subfunctionalization (4–6). TPSs are unique enzymes in that most form multiple products from a single substrate by providing a 3D template and stabilizing various intermediate carbocations during cyclization (7). As such, these enzymes are often suggested to have molecular promiscuity resulting from plasticity residues that slightly change the template or carbocation stabilization when the amino acid is changed and thus direct product outcome. Research in recent years has addressed how specific residues or domains in various mono-, sesqui-, and di-TPSs direct product profiles, whether it is in naturally occurring systems or through directed mutagenesis (8–15).
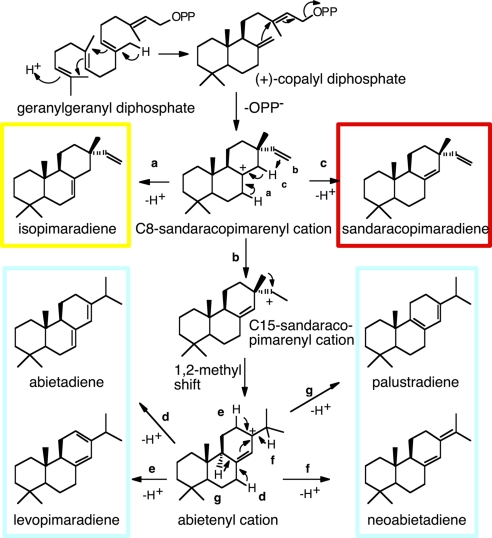
Diterpene resin acids are formed in two major steps. Di-TPSs use geranylgeranyl diphosphate (GGPP) to produce the diterpene olefins (5, 16), which are then oxidized to the corresponding acids by multisubstrate and multifunctional cytochromes P450 (17, 18). To date, only a handful of di-TPSs involved in diterpene resin acid biosynthesis have been characterized, with all of them originating from gymnosperm trees (2). A series of papers from Croteau, Peters, and coworkers (14, 15, 19–23) studied an abietadiene synthase (AS) from Grand fir [Abies grandis (Ag)] to establish the general mechanism and specific features of this type of di-TPS. Two closely related di-TPSs identified in Norway spruce [Picea abies (Pa)] have 91% amino acid identity but nonoverlapping product profiles (5). One, isopimaradiene synthase (Iso), exclusively produces isopimaradiene (isopimara-7,15-diene) (Fig. 1). The other, levopimaradiene/AS (LAS), produces a mixture of four diterpenes: abietadiene (abieta-7,13-diene), levopimaradiene [abieta-8(14),12-diene], neoabietadiene [abieta-8(14),13(15)-diene], and palustradiene (abieta-8,13-diene), similar to the product profile of AgAS (20). Orthologous LAS from Loblolly pine (Pinus taeda) (Pt) (24) and a levopimaradiene synthase (LS) from Ginkgo biloba (Gb) (25) have also been characterized.
Fig. 1.
Proposed reaction mechanisms for the di-TPSs PaIso and PaLAS. Routes a and b are the pathways for PaIso and PaLAS, respectively. Route c occurs with some of the mutants. Deprotonations leading to the multiple products of PaLAS are indicated by routes d–g.
All of these enzymes are bifunctional with two catalytically independent active sites. The first active site (class II) in the N-terminal domain contains a DXDD motif that catalyzes the protonation-initiated cyclization of GGPP to (+)-copalyl diphosphate [(+)-CPP; Fig. 1]. The (+)-CPP then freely diffuses to a second active site (class I) in the C-terminal domain, containing the Mg2+-binding DDXXD motif that catalyzes the diphosphate ionization-initiated cyclization of (+)-CPP to produce the C8-sandaracopimarenyl cation intermediate (21–23). Deprotonation of this carbocation at alternate sites results in isopimaradiene or sandaracopimaradiene [sandaracopimara-8(14),15-diene] (Fig. 1, routes a and c). Alternatively, intramolecular proton transfer and a 1,2-methyl migration result in the abietenyl cation intermediate (Fig. 1, route b) that is subsequently deprotonated at one of four sites to produce abietadiene, levopimaradiene, neoabietadiene, or palustradiene.
The few amino acid differences (Fig. 2) in the second active site between the paralogous PaIso and PaLAS enzymes are sufficient to direct PaIso to deprotonate the C8-sandaracopimarenyl cation while PaLAS proceeds further through an abietenyl cation intermediate, resulting in alternate products (Fig. 1). These paralogous enzymes provide a unique opportunity to examine the domains and residues that determine product diversity of di-TPSs involved in conifer secondary metabolism. The objective of the present work was to identify those residues that are different between PaLAS and PaIso and result in the observed product profile differences. We used reciprocal domain-swapping and mutagenesis strategies directed by homology modeling and sequence comparisons to delineate and identify the specific amino acids that determine the product profiles of these naturally occurring paralogues.
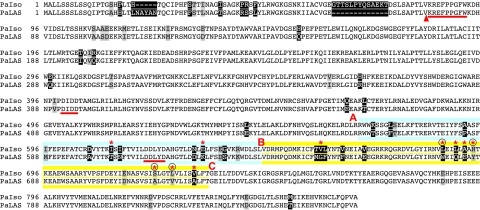
Fig. 2.
Protein alignment of PaIso and PaLAS. Amino acids with white, gray, and black backgrounds indicate identical, similar, and nonsimilar residues, respectively. Asterisks indicate residues mutated in this study. Circles indicate the four residues found in this study that determined product profile differences. Red letters and colored backgrounds indicate AB and BC domain boundaries. A triangle indicates the start of the pm constructs. The conserved DXDD and DDXXD motifs for the first and second active sites, respectively, and the KRX6W motif are underlined.
Results
We developed a homology model of the second active site for both PaIso and PaLAS by using Deep View/Swiss-PDBViewer and SWISS-MODEL (26) based on the structure of 5-epi-aristolochene synthase from Nicotiana tabacum (Protein Data Bank ID code 5EAT) (27). Although 5-epi-aristolochene synthase is an angiosperm sesqui-TPS that shares only 18% identity to gymnosperm PaIso and PaLAS, the tertiary structures of TPSs are well conserved (7, 27), allowing us to model these enzymes when no comparable di-TPS structure has been described.
To delineate the larger regions of PaIso and PaLAS that determine the different product outcomes, we prepared a series of chimeric constructs dividing the second active site into two sections that were reciprocally swapped, alone or in combination, between PaLAS and PaIso (Fig. 2). Most of the residues within the modeled second active site occurred in an area of increased sequence divergence between PaIso and PaLAS (residues 571–730 of PaIso; Fig. 2). We named the portion of these enzymes corresponding to residues 568–640 of PaIso the AB domain and the portion corresponding to residues 651–733 of PaIso the BC domain, as shown in Fig. 2.
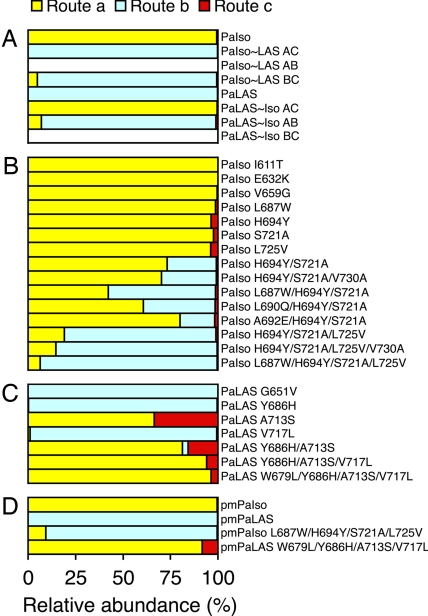
By swapping the full AC domain between PaIso and PaLAS, we completely reversed the product profiles of these two enzymes (Fig. 3A). Thus, the AC domain fully encompassed those residues that determine product profile differences between the two wild-type enzymes. We then characterized chimeras for which the AB and BC domains were individually swapped. PaIso containing the BC domain of PaLAS (PaIso∼LAS BC) and PaLAS∼Iso AB chimeras formed products similar to each other and wild-type PaLAS (Fig. 3) except for additionally producing 6% isopimaradiene (the product of route a in Fig. 1). Expression of both the PaIso∼LAS AB and PaLAS∼Iso BC chimeras failed to produce any His-tagged enzyme after Ni-affinity purification. Analysis of the active chimeras indicated that the BC domain determined almost all of the functional differences between PaIso and PaLAS but that the AB domain also had a minor influence on product profile. Precise quantitative data for the results shown in Fig. 3 appear in supporting information (SI) Table 2.
Fig. 3.
Relative abundance of enzyme assay products from routes a, b, and c (see Fig. 1) for each enzyme tested. (A) Wild-type and chimeric enzymes. (B) Site-directed mutants of PaIso. (C) Site-directed mutants of PaLAS. (D) Pm wild-type and quadruple mutants.
We then examined residues of PaIso and PaLAS in an increasing sphere corresponding to that around the phosphorus atom of the hydroxyphosphonate group of the substrate analogue in the sesqui-TPS structure superimposed on our di-TPS homology models. Table 1 lists the residues divergent between PaIso and PaLAS within a 20-Å sphere. We focused site-directed mutations to those uniquely different in PaIso compared with four other related di-TPSs (PaLAS, AgAS, PtLAS, and GbLS; Table 1 and SI Fig. 6), which all favor formation of the abietenyl cation (Fig. 1, route b). We hypothesized that those residues that were conserved between the four enzymes but divergent in PaIso were most likely to direct differences in product outcomes (Table 1, Fig. 2, and SI Fig. 6).
Table 1.
Divergent residues in PaIso and PaLAS homology models within an increasing radius from the phosphorous atom of the substrate analogue in the structure of 5-epi-aristolochene synthase from N. tabacum and the corresponding residues in other related di-TPSs
| Radius, Å | Residue in: |
Domain | Unique | Mutated | ||||
|---|---|---|---|---|---|---|---|---|
| PaIso | PaLAS | PtLAS | AgAS | GbLS | ||||
| 5 | ||||||||
| 10 | L690 | Q682 | Q | Q | L | BC | PaIso | |
| H694 | Y686 | Y | Y | Y | BC | Yes | Both | |
| S721 | A713 | A | A | A | BC | Yes | Both | |
| 15 | I611 | T603 | T | T | T | AB | Yes | PaIso |
| I613 | N605 | N | N | C | AB | |||
| L687 | W679 | W | W | W | BC | Yes | Both | |
| A692 | E684 | E | E | A | BC | PaIso | ||
| E714 | D706 | G | E | E | BC | |||
| L725 | V717 | V | V | V | BC | Yes | Both | |
| 20 | A592 | P584 | P | P | V | AB | ||
| N630 | D622 | N | D | D | AB | |||
| E632 | K624 | K | K | K | AB | Yes | PaIso | |
| V659 | G651 | G | G | G | BC | Yes | Both | |
| L660 | F652 | F | F | L | BC | |||
| E842 | D834 | D | Q | Y | ||||
| M852 | T844 | M | M | K | ||||
| >20 | V730 | A722 | A | A | I | BC | PaIso | |
The domain in which the residue resides and whether the residue is uniquely divergent in PaIso are indicated. The rightmost column indicates whether mutants in PaIso, both PaIso and PaLAS, or neither were characterized at this residue.
Within a predicted 10-Å sphere, we identified two residues, H694 and S721 in PaIso, and Y686 and A713 in PaLAS, that had previously been speculated to determine product outcome in these enzymes (5). However, reciprocal mutagenesis at these residues was not sufficient to fully reverse product profiles (Fig. 3B). Both PaIso H694Y and PaIso S721A mutations individually did not result in products from route b but did produce 2–4% sandaracopimaradiene, the product from route c. However, the double mutant PaIso H694Y/S721A produced 26% of the products from route b. The reciprocal PaLAS Y686H mutant did not change product outcome from wild-type PaLAS, but the PaLAS A713S mutant completely abolished products from route b and instead produced a 2:1 ratio of products from routes a and c. The double mutant PaLAS Y686H/A713S produced an even greater ratio of product from route a but also produced 3% products from route b. Thus, although reciprocally mutating these two residues was not sufficient to completely reverse product profiles, it had a substantial impact on redirecting catalytic routes and thus appeared necessary for determining product profiles.
We then used site-directed mutagenesis to characterize additional divergent residues within the AC domain as shown in Table 1 that may direct product profile differences. Because the initial mutations of PaIso and PaLAS described above suggested that it was harder to change the function of PaIso rather than that of PaLAS, PaIso was used as the initial template for further mutations. Although none of the individual mutations of PaIso gave products from route b, the quadruple mutant PaIso L687W/H694Y/S721A/L725V almost exclusively formed products from route b (93%; Fig. 3B). All four of these mutated residues occurred within the BC domain. A comparison of the product profile of this mutant with those of PaIso∼LAS BC and PaLAS∼Iso AB suggested that these four mutations account for all of the critical residues in the BC domain necessary for directing product outcome (Fig. 3 A and B). Other mutations characterized within the AC domain did not produce substantial differences in product outcome.
To further validate the role of these four residues in determining product outcome, we made reciprocal mutations with PaLAS as template and determined that the PaLAS W679L/Y686H/A713S/V717L mutant no longer produced any diterpenes from route b but rather 96% from route a and 4% from route c (Fig. 3C). Although the single-residue A713S mutation of PaLAS appeared to have a major impact redirecting product profile, the additional three mutations substantially enhanced the product outcome toward that of PaIso. Thus in both cases, the quadruple reciprocal mutants were effective in nearly completely reversing the product profiles for these two paralogous diTPS enzymes, PaLAS and PaIso (Figs. 3 and 4).
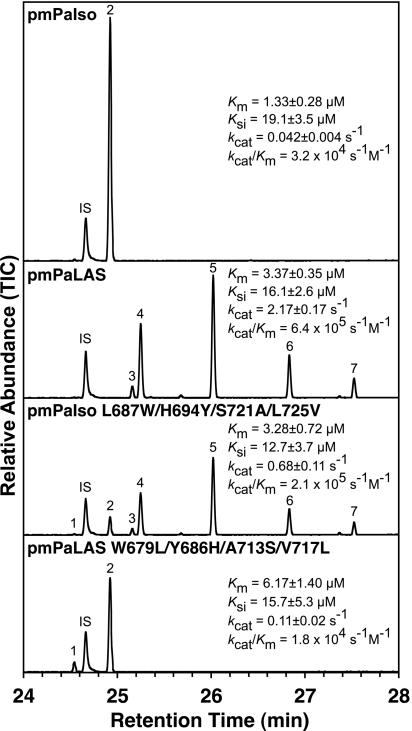
Fig. 4.
GCMS total ion chromatograms on an HP5ms column of products of the pm enzymes with GGPP. TIC, total ion current. Compound identities: 1, sandaracopimaradiene; 2, isopimaradiene; 3, palustradiene; 4, levopimaradiene; 5, abietadiene; 6, neoabietadiene; 7, unknown; IS, internal standard. Kinetic parameters were determined at 22°C with GGPP as substrate. Cf. pmAgAS: Km = 3.0 ± 0.8 μM, Ksi = 5 ± 2 μM, kcat = 0.75 ± 0.15 s−1 and kcat/Km = 2.5 × 105 s−1·M−1 (20).
We prepared pseudomature (pm) constructs of PaIso, PaLAS, PaIso L687W/H694Y/S721A/L725V, and PaLAS W679L/Y686H/A713S/V717L (truncated at residues 84 and 76 of PaIso and PaLAS, respectively) to allow direct comparison of the kinetic parameters with the pm version of AgAS (19) and evaluate the catalytic efficiency of the PaIso and PaLAS mutants. The products from these N-terminally His-tagged pm enzymes did not differ from their full-length counterparts (Fig. 3 A and D) or C-terminally His-tagged pm constructs (data not shown). Kinetic data for the pm enzymes with GGPP as substrate are shown in Fig. 4. We observed substrate inhibition and overall kinetic properties similar to those reported for AgAS (20). PaLAS appeared a more efficient enzyme than PaIso. In addition, the efficiency of the quadruple mutants increased or decreased toward the efficiency of the reciprocal wild-type enzyme, suggesting that these reciprocal mutations did not impair catalytic function. Although the kinetics parameters were not determined for the other intermediate mutants, none appeared to be severely impaired in our assay conditions.
The four pm enzymes were also characterized at different pHs and temperatures. These enzymes were active between pH 6.0 and 9.5 but showed greatly reduced or no activity at lower pH (data not shown). Both pmPaIso and pmPaLAS W679L/Y686H/A713S/V717L did not change product profiles over this pH range, but pmPaLAS and pmPaIso L687W/H694Y/S721A/L725V produced slightly more abietadiene at lower pH. We observed no differences in product profiles over the range of 4°C to 37°C with the enzymes tested except for a slight increase in the amount of sandaracopimaradiene produced with increasing temperature for pmPaLAS W679L/Y686H/A713S/V717L (data not shown).
Discussion
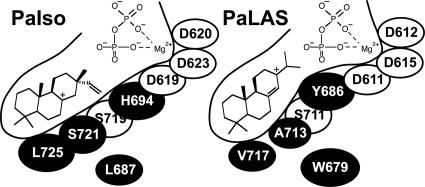
Our rational approach to identifying the specific residues that determine product profile differences between PaIso and PaLAS was successful in identifying four residues that nearly completely reverse the product outcome between these two enzymes. By examining the location of these residues within the homology models, we found that the three residues (H694, S721, and L725) in PaIso, and the corresponding residues in PaLAS, form a line of amino acids running the length of the predicted active-site cavity, with H694 nearest the entrance and L725 furthest into the cavity (Fig. 5). PaIso L687 and PaLAS W679 do not line the active-site cavity, but because of their proximity to the cavity and their relative size differences may have a secondary effect on the size and shape of the active-site cavity that modestly influences product outcome.
Fig. 5.
Schematic of the modeled active-site cavities of PaIso and PaLAS. The positions of the ionized diphosphate and the C8-sandaracopimarenyl and abietenyl cations are shown. The residues identified in this study that direct product outcome differences between PaIso and PaLAS are shown with black backgrounds.
In general, PaIso and PaLAS form different products because the predicted active-site cavities of the two enzymes differ in size, shape, and polarity. Once formed, the C8-sandaracopimarenyl cation seems to be stabilized deeper into the cavity of PaIso and in a slightly different orientation than in PaLAS. The polarity of S721 of PaIso appears positioned to stabilize the C8-sandaracopimarenyl cation. This residue is an alanine in PaLAS and thus cannot stabilize the cation. The dramatic change in product profile from the PaLAS A713S mutant supports this residue's influence in directing product outcome. Near this residue is a conserved serine (PaIso S719/PaLAS S711), which may also play a role in stabilizing both the C8-sandaracopimarenyl and abietenyl cations. In AgAS, mutation of this conserved serine to alanine greatly impairs catalysis (15). Although PaLAS has this conserved serine to stabilize the C8-sandaracopimarenyl cation, Y686 in PaLAS is better positioned to stabilize the abietenyl cation (Fig. 5). The corresponding residue in PaIso is H694, which, owing to its positive charge, may destabilize the abietenyl cation. PaIso S721 and H694, and the corresponding PaLAS A713 and Y686, appear to strongly influence the polarity of the active site and thus determine which of the alternate carbocation intermediates is favorably stabilized. PaIso L725 and the corresponding PaLAS V717 residue influence the shape in the deeper portions of the cavity, with PaIso L725 impinging into the modeled cavity further than does PaLAS V717. As this single mutation has a negligible influence on product outcome, we hypothesize that this difference changes the orientation of the intermediates and thus affects differential stabilization of the carbocations by other residues.
Interestingly, mutation of PaLAS more easily resulted in large changes in product profiles compared with the reciprocal mutations in PaIso. This finding may suggest that the subsequent formation of the abietenyl cation from the C8-sandaracopimarenyl cation (Fig. 1) is particularly sensitive to single amino acid changes in the active site. The single PaLAS A713S mutation dramatically changed product profile, abolishing all products derived from the abietenyl cation and instead producing a 2:1 ratio of isopimaradiene and sandaracopimaradiene. No mutants of PaIso produced as much sandaracopimaradiene as did the mutants of PaLAS.
There is conservation around the DDXXD motif between these two enzymes (Fig. 2), and our focus of study was those residues that influence product profile after formation of the C8-sandaracopimarenyl cation. Thus, it would seem that the differences we observed in the kinetics between enzymes were not caused by differences in the kinetics of initial formation of the C8-sandaracopimardienyl cation but rather in final product formation and subsequent release of the products from the enzyme. Although formation of the abietenyl cation in route b requires an intramolecular proton transfer, passes through a less stable secondary carbocation intermediate, and includes a methyl migration, the kinetic parameters of enzymes that used this pathway appeared more efficient than the enzymes that simply deprotonate the C8-sandaracopimarenyl cation in route a. The difference in kinetic efficiencies between these enzymes cannot be explained with our current understanding of these enzymes.
For those enzymes expressed in both full-length and pm forms, the resulting product profiles did not differ between full-length and pm forms. Interestingly, the relative ratio of products resulting from route b (i.e., abietadiene, levopimaradiene, neoabietadiene, palustradiene, and two minor unknown products not previously identified from these enzymes) did not differ among all of the mutations, even though the relative amounts of products from routes a, b, and c varied widely. However, mutations in this study only targeted those residues that differ between PaLAS and PaIso in a reciprocal fashion. Different amino acid substitutions at these residues, or changes at other residues, may direct changes to the ratio of these products. A previous mutational study with AgAS (15) and the single-product LS from Gb (25) suggest that such changes to the ratios are possible. The AgAS mutations did not target the same residues as we selected in the present study (the residues previously targeted are conserved between PaIso and PaLAS). However, some mutations of AgAS did result in product ratio changes and mutation of one residue directed the production of only sandaracopimaradiene, albeit with greatly impaired catalysis (15). Recently, an Ile to Thr mutation at the equivalent residue of PaIso L725 and PaLAS V717 was shown to change an isokaurene synthase into a pimara-8(14),15-diene synthase in rice (28). This observation shows that these same residues influence product profiles in other di-TPSs, even though this enzyme uses (−)-CPP rather than (+)-CPP as substrate.
In conclusion, we observed large changes in product outcomes by making only a few amino acid changes in the two paralogous di-TPSs that account for most of the diterpene resin acid diversity of the oleoresin defense secretion of Norway spruce and other conifers. This study highlights how TPS gene duplication can easily be followed by neofunctionalization based on a few nonsynonymous nucleotide substitutions, resulting in the diversification of chemical defense profiles that can be a selective advantage to the conifer in plant–herbivore and plant–pathogen interactions. This functional diversification and evolutionary plasticity of enzymes appears particularly apparent in the large gene family of TPSs of plant secondary metabolism.
Materials and Methods
Molecular Modeling.
We used Deep View/Swiss-PDBViewer and SWISS-MODEL (26) to develop a 3D homology model of the second active site of both PaIso and PaLAS based on the structure of 5-epi-aristolochene synthase from N. tabacum containing the substrate analogue farnesyl hydroxyphosphonate (Protein Data Bank ID code 5EAT) (27).
Preparation of Mutants and Chimeras.
Wild-type cDNAs were in pCR BLUNT as described (5). Initial site-directed mutagenesis used these plasmids with the QuikChange II XL site-directed mutagenesis kit (Stratagene). After sequence verification, inserts were subcloned into the pET100 TOPO/D expression vector (Invitrogen), resulting in full-length N-terminally His-tagged constructs. For comparison, wild-type constructs were prepared in a similar manner. Additional mutations were completed on the pET100 constructs (wild type or mutated) with the QuikChange Multi site-directed mutagenesis kit, and sequences were verified.
Reciprocal chimeric constructs PaIso∼LAS AC, PaIso∼LAS AB, PaIso∼LAS BC, PaLAS∼Iso AC, PaLAS∼Iso AB, and PaLAS∼Iso BC were prepared from the full-length wild-type PaIso and PaLAS constructs in pCR BLUNT by using a mega-primer approach (29) with primers perfectly complementary to both sequences and the Quik-Change XL site-directed mutagenesis kit. After sequence verification, inserts were subcloned into pET100 TOPO/D as full-length N-terminally His-tagged constructs, and sequences were verified.
Preparation of Pm Versions of PaIso and PaLAS.
For kinetic analysis and pH and temperature studies, PaIso, PaLAS, PaIso L687W/H694Y/S721A/L725V, and PaLAS W679L/Y686H/A713S/V717L were subcloned by using a sticky-end PCR protocol (30) into the NdeI/HindIII sites of pET28b(+) (Novagen) as N-terminally His-tagged proteins, truncated at residues 84 and 76 for PaIso and PaLAS, respectively. These truncations correspond closely to the pm M84 truncation of AgAS previously characterized (19) and are thus designated with a pm prefix.
Heterologous Expression of Recombinant Protein.
Plasmids were transformed into C41 Escherichia coli (www.overexpress.com) containing the pRARE 2 plasmid isolated from Rosetta 2 cells (Novagen) to negate codon bias. Luria–Bertani containing ampicillin (100 mg/liter) and chloramphenicol (50 mg/liter) was inoculated with three individual colonies and cultured overnight at 37°C and 220 rpm. Terrific Broth containing carbencillin (100 mg/liter) and chloramphenicol (50 mg/liter) was then inoculated with a 1:100 dilution of the overnight culture and grown at 37°C and 220 rpm until OD600 reached ≈0.5. Cultures were then cooled to 16°C, induced with 0.2 mM isopropyl β-D-thiogalactoside, and cultured for ≈16 h at 16°C and 220 rpm before harvesting.
Ni-Affinity Purification of Recombinant Enzymes.
For semipreparative purifications, cell pellets from 50-ml cultures were resuspended, lysed, and sonicated in (1.5 ml/g pellet) ice-cold 20 mM NaPO4, 500 mM NaCl, 30 mM imidazole, 0.04 mg/ml DNase, 1 mM MgCl2, 5 mM PMSF, and 0.5 mg/ml lysozyme, pH 7.4 and then clarified by centrifugation. The cleared lysates were applied to His SpinTrap Ni-affinity columns (GE Healthcare) and eluted with 20 mM NaPO4, 500 mM NaCl, and 500 mM imidazole, pH 7.4 at 4°C following the manufacturer's protocol. Purified enzymes were used immediately for enzyme assays as described below.
For preparative purifications, cell pellets from 1-liter cultures were resuspended in (2 ml/g pellet) ice-cold 20 mM Hepes (pH 7.5), 150 mM NaCl, 20 mM imidazole, 5 mM DTT, 5 mM PMSF, and 0.5 mg/ml lysozyme. After 30 min of gentle mixing at 4°C, the lysates were sonicated, clarified by centrifugation, and applied to a 1-ml HisTrap HP Ni-affinity column by using an ÄKTApurifier 10 fast protein liquid chromatograph (GE Healthcare). The column was washed with 60 ml of 20 mM Hepes, 150 mM NaCl, and 20 mM imidazole, pH 7.5 before eluting the His-tagged proteins with 20 mM Hepes, 150 mM NaCl, and 350 mM imidazole, pH 7.5. Buffer composition of eluate was exchanged to 20 mM Hepes and 150 NaCl, pH 7.5 with an Econo-Pac 10DG desalting column (Bio-Rad). Protein concentration was quantified (7–13 mg/ml) by direct measurement of absorbance at 280 nm with a spectrophotometer (NanoDrop Technologies) using the molar extinction coefficients calculated by Vector NTI software (Invitrogen). Fresh DTT was added to 5 mM final concentration and then the purified enzymes were aliquoted, flash-frozen in liquid N2, and stored at −80°C where they were stable for several months.
Single-Vial Enzyme Assays.
Enzyme assays were completed in 2-ml amber glass GC sample vials as described (31). Buffer (500 μl) consisted of 20 mM Hepes, 20 mM Mes, 20 mM Mops (pH 7.0), 100 mM KCl, 7.5 mM MgCl2, 0.02 mM MnCl2, 5 mM fresh DTT, 5% glycerol, and 40 μM GGPP (Sigma–Aldrich). Assays were started by adding 50–100 μg of purified enzyme, gently mixed, overlaid with 500 μl of pentane containing 1.6 μM 1-eicosene as an internal standard, and then placed in a 30°C water bath for 1 h. The vials were then vigorously vortexed for 30 s to stop the reaction (by denaturing the enzyme) and to extract the products from the aqueous layer. Phases were separated by centrifugation at 1,000 × g for 30 min at 4°C.
GCMS Analysis.
Assay products were analyzed on Agilent HP5ms (5% phenyl methyl siloxane, 30 m × 250 μm i.d., 0.25-μm film) and DB-WAX (polyethylene glycol, 30 m × 250 μm i.d., 0.25-μm film) columns at 1 ml/min He on an Agilent 6890N GC, 7683B series autosampler (vertical syringe position of 8), and a 5975 Inert XL MS Detector at 70 eV. GC temperature programs were as follows: HP5ms, 40°C for 1 min, 7.5°C/min to 250°C, hold 2 min, pulsed splitless injector held at 250°C; DB-WAX, 40°C for 3 min, 10°C/min to 240°C, hold 15 min, pulsed splitless injector held at 240°C. Compounds were identified by comparison to authentic standards, retention indices, and mass spectra from Adams (32) and National Institute of Standards and Technology 2005 MS library searches. The relative contributions of the different pathway products of mutant and wild-type enzymes were calculated by using total ion currents.
Analysis of Enzyme Activity and Product Profiles at Various pHs and Temperatures.
To assess the effect of pH on enzyme activity and product profile, single-vial assays were performed as above on preparatively Ni-affinity-purified pmPaLAS, pmPaIso, pmPaIso L687W/H694Y/S721A/L725V, and pmPaLAS W679L/Y686H/A713S/V717L in 20 mM Hepes, 20 mM Mes, 20 mM Mops, 100 mM KCl, 7.5 mM MgCl2, 0.02 mM MnCl2, 5 mM fresh DTT, 20 μM GGPP, and 5% glycerol at 0.5 pH units between 5.0 and 9.5 at 22°C. These enzymes were also assayed at pH 7.0 at 4°C, 16°C, 22°C, and 37°C for 135, 60, 45, and 30 min, respectively, to determine whether product profiles varied with temperature.
Kinetic Analysis of pmPaIso, pmPaLAS, pmPaIso L687W/H694Y/S721A/L725V, and pmPaLAS W679L/Y686H/A713S/V717L.
Single-vial assays were used as described above but with the addition of 0.1 mg/ml BSA to the buffer to stabilize the recombinant enzymes at the lower concentrations (≈10 nM). Assays were completed in triplicate with 0.5–42 μM GGPP for 10 min at 22°C. Assays were analyzed on the GC mass spectrometer by using selected ion monitoring of m/z 83 (for internal standard) and both 257 and 272 for diterpene products (abietadiene and isopimaradiene were used to quantify products). The response of the GC mass spectrometer at each substrate concentration was determined relative to the internal standard by using excess enzyme in the single-vial assay and allowing the reaction to proceed to completion for 2 h. Kinetic parameters were determined by nonlinear regression using the substrate inhibition model in ANEMONA (33).
Supplementary Material
ACKNOWLEDGMENTS.
We thank Dr. Gary Lesnicki and Adriana Cajiao of the University of British Columbia–Michael Smith Laboratories Process Pilot Plant Facility for assistance with protein expression and Lina Madilao for GCMS support. This research was supported by grants from the Natural Sciences and Engineering Research Council of Canada, Genome British Columbia, Genome Canada, the Province of British Columbia, and the Canada Foundation for Innovation (to J.B.). J.B. is supported in part by the University of British Columbia Distinguished University Scholars program and a Natural Sciences and Engineering Research Council of Canada Steacie Memorial Fellowship.
Note Added in Proof:
Wilderman and Peters (34) recently described results from site-directed mutagenesis of AgAS A723S confirming our observations with PaLAS A713S.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709466105/DC1.
References
- 1.Keeling CI, Bohlmann J. New Phytol. 2006;170:657–675. doi: 10.1111/j.1469-8137.2006.01716.x. [DOI] [PubMed] [Google Scholar]
- 2.Keeling CI, Bohlmann J. Phytochemistry. 2006;67:2415–2423. doi: 10.1016/j.phytochem.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 3.Trapp S, Croteau R. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:689–724. doi: 10.1146/annurev.arplant.52.1.689. [DOI] [PubMed] [Google Scholar]
- 4.Bohlmann J, Meyer-Gauen G, Croteau R. Proc Natl Acad Sci USA. 1998;95:4126–4133. doi: 10.1073/pnas.95.8.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin DM, Fäldt J, Bohlmann J. Plant Physiol. 2004;135:1908–1927. doi: 10.1104/pp.104.042028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trapp SC, Croteau RB. Genetics. 2001;158:811–832. doi: 10.1093/genetics/158.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christianson DW. Chem Rev. 2006;106:3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- 8.Greenhagen BT, O'Maille PE, Noel JP, Chappell J. Proc Natl Acad Sci USA. 2006;103:9826–9831. doi: 10.1073/pnas.0601605103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kampranis SC, et al. Plant Cell. 2007;19:1994–2005. doi: 10.1105/tpc.106.047779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Köllner TG, Schnee C, Gershenzon J, Degenhardt J. Plant Cell. 2004;16:1115–1131. doi: 10.1105/tpc.019877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshikuni Y, Ferrin TE, Keasling JD. Nature. 2006;440:1078–1082. doi: 10.1038/nature04607. [DOI] [PubMed] [Google Scholar]
- 12.Köllner TG, et al. Arch Biochem Biophys. 2006;448:83–92. doi: 10.1016/j.abb.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 13.Hyatt DC, Croteau R. Arch Biochem Biophys. 2005;439:222–233. doi: 10.1016/j.abb.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 14.Peters RJ, Croteau RB. Biochemistry. 2002;41:1836–1842. doi: 10.1021/bi011879d. [DOI] [PubMed] [Google Scholar]
- 15.Peters RJ, Croteau RB. Proc Natl Acad Sci USA. 2002;99:580–584. doi: 10.1073/pnas.022627099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stofer Vogel B, Wildung MR, Vogel G, Croteau R. J Biol Chem. 1996;271:23262–23268. doi: 10.1074/jbc.271.38.23262. [DOI] [PubMed] [Google Scholar]
- 17.Funk C, Croteau R. Arch Biochem Biophys. 1994;308:258–266. doi: 10.1006/abbi.1994.1036. [DOI] [PubMed] [Google Scholar]
- 18.Ro D-K, Arimura G, Lau SY, Piers E, Bohlmann J. Proc Natl Acad Sci USA. 2005;102:8060–8065. doi: 10.1073/pnas.0500825102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peters RJ, Carter OA, Zhang Y, Matthews BW, Croteau RB. Biochemistry. 2003;42:2700–2707. doi: 10.1021/bi020492n. [DOI] [PubMed] [Google Scholar]
- 20.Peters RJ, et al. Biochemistry. 2000;39:15592–15602. doi: 10.1021/bi001997l. [DOI] [PubMed] [Google Scholar]
- 21.Ravn MM, Coates RM, Flory JE, Peters RJ, Croteau R. Org Lett. 2000;2:573–576. doi: 10.1021/ol991230p. [DOI] [PubMed] [Google Scholar]
- 22.Peters RJ, Ravn MM, Coates RM, Croteau RB. J Am Chem Soc. 2001;123:8974–8978. doi: 10.1021/ja010670k. [DOI] [PubMed] [Google Scholar]
- 23.Ravn MM, Peters RJ, Coates RM, Croteau R. J Am Chem Soc. 2002;124:6998–7006. doi: 10.1021/ja017734b. [DOI] [PubMed] [Google Scholar]
- 24.Ro D-K, Bohlmann J. Phytochemistry. 2006;67:1572–1578. doi: 10.1016/j.phytochem.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 25.Schepmann HG, Pang J, Matsuda SP. Arch Biochem Biophys. 2001;392:263–269. doi: 10.1006/abbi.2001.2438. [DOI] [PubMed] [Google Scholar]
- 26.Guex N, Peitsch MC. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 27.Starks CM, Back KW, Chappell J, Noel JP. Science. 1997;277:1815–1820. doi: 10.1126/science.277.5333.1815. [DOI] [PubMed] [Google Scholar]
- 28.Xu M, Wilderman PR, Peters RJ. Proc Natl Acad Sci USA. 2007;104:7397–7401. doi: 10.1073/pnas.0611454104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirsch RD, Joly E. Nucleic Acids Res. 1998;26:1848–1850. doi: 10.1093/nar/26.7.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pham K, LaForge KS, Kreek MJ. BioTechniques. 1998;25:206–208. doi: 10.2144/98252bm04. [DOI] [PubMed] [Google Scholar]
- 31.O'Maille PE, Chappell J, Noel JP. Anal Biochem. 2004;335:210–217. doi: 10.1016/j.ab.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 32.Adams RP. Identification of Essential Oil Components by Gas Chromatography/Mass Spectrometry. Carol Stream, IL: Allured; 2007. [Google Scholar]
- 33.Hernandez A, Ruiz MT. Bioinformatics. 1998;14:227–228. doi: 10.1093/bioinformatics/14.2.227. [DOI] [PubMed] [Google Scholar]
- 34.Wilderman PR, Peters RJ. J Am Chem Soc. 2007;129:15736–15737. doi: 10.1021/ja074977g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.